Abstract
The association of an IS440-sul3 platform with Tn21 class 1 integrons carried by IncI1 plasmids encoding extended-spectrum β-lactamases (ESBLs; mainly SHV-12 and CTX-M-14) among worldwide Escherichia coli clones of phylogroups A (ST10, ST23, and ST46), B1 (ST155, ST351, and ST359), and D/B2 (ST131) is reported. An in silico comparative analysis of sul3 elements available in the GenBank database shows the evolution of sul3 platforms by hosting different transposable elements facilitating the potential genesis of IS26 composite transposons and further insertion element-mediated promoted arrangements.
INTRODUCTION
Acquired resistance to sulfonamides is due to the presence of dihydropteroate synthase (DHPS) genes located on class 1 integrons (sul1 and sul3) or genetic islands bearing Tn5393 and ISCR2 (sul2) (14, 23, 25). Since its first description in the early 1960s, there is evidence of the spread of plasmids containing sul1 and sul2 among different hosts (10, 12, 13, 19, 26). The sul3 integrons have been increasingly reported among animals and, to a lesser extent, among humans since they were identified in the mid-1990s (2, 4, 11, 23, 24, 32). However, the genetic elements linked to their spread remain scarcely explored (2, 23, 32).
We analyzed 344 clonally unrelated Enterobacteriaceae isolates (249 Escherichia coli, 56 Klebsiella pneumoniae, 20 Enterobacter cloacae, 3 Enterobacter aerogenes, 7 Klebsiella oxytoca, 6 Salmonella enterica serovar Paratyphi, and 3 Citrobacter isolates). They included 244 extended-spectrum-β-lactamase (ESBL) or metallo-β-lactamase (MBL) producers obtained from hospitalized and healthy humans (1988 to 2006) and 100 non-ESBL producers (66 obtained from blood samples from inpatients and 34 obtained from feces samples from healthy volunteers without recent exposure to antibiotics or hospital environments; 1988 to 2006) (see Table 2). Species identification and susceptibility testing were performed by using the automated WIDER system (Fco. Soria Melguizo, Madrid, Spain) and standard methods (7). Clonal relatedness among Escherichia coli isolates was established by pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST) (30; http://www.mlst.net), and determination of phylogenetic groups by a multiplex PCR assay (6).
Table 2.
Epidemiological data of isolates producing sul3 integrons in Hospital Ramón y Cajala
| Integron type (no. of isolates) | sul3 plasmid (kb)b | Inc group(s) (no. of isolates) | RFLP type (no. of isolates)c | pMLST IncI1d |
ESBL(s) (no. of isolates) | E. coli MLST result(s) | Yr | Origin (no. of isolates) | Antibiotic resistancee | sul gene(s) (no. of isolates)g | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ardA | trbA-pndA | sogS | pilV | ||||||||||
| I (2) | 100 | I1, B/O | A (2) | 4 | 3 | DLV 4 | 1 | CTX-M-14 | ST57, ST350 | 2000–2001 | Urine/P | Sm, Na, Su, (W), (Te), Ch, Km | sul1 (1) |
| II (3) | 125 | Y | G | SHV-12 | ST48/ST10 | 2000 | Blood/P | Sm, (Sp), (Cip), (Na), Su, W, Ch, (Ap), (Te) | sul2 | ||||
| 125, 55 | B/O and NTf | ND | SHV-12 | 2000 | Feces/HV | ||||||||
| 100 | I1, B/O | F | 4 | 16 | 9 | 2 | CTX-M-1 | ST131 | 2002 | Urine/P | |||
| III (15) | 100 | I1 (6) | B (6) | 4 | 3 | DLV 4 | 1 | SHV-12, CTX-M-14 (2), CTX-M-15 (1), VIM-1 (1) | ST359, ST155, DLV ST155, ST156, ST46; ND | 1997–2006 | Urine (2)/P, blood/P, feces/HV, gangrene/P | (Sm), (Sp), (Na), (Su), (W), (Te), (Ch), (Nt), (Km), (Gm), (Tb), (Ak) | sul1, sul2 |
| 150 | FIB+B/Of | D | 4 | 3 | DLV 4 | 1 | SHV-12 | ST695 | 2002 | Urine/P | sul1, sul2 | ||
| 100 | I1+B/Of | ||||||||||||
| 100 | I1, B/O | C1 | 4 | 3 | DLV 4 | 1 | SHV-12 | ST10 | 2002 | Urine/P | sul1, sul2 | ||
| 100 | I1, B/O | C2 | 4 | 13 | 2 | 1 | CTX-M-14 | ST359 | 2002 | Urine/P | sul1 | ||
| 50 | I1, K, FIB, F | ND | CTX-M-14, SHV-12 | ST167 | 2001–2002 | Urine (3)/P, cutaneous/P, genital/P, epidemic surveillance/P | sul1, sul2 | ||||||
| 70 | Y, FIA, F | CTX-M-9 | ST351, ST648 | ||||||||||
| 90 | K, B/O, F | CTX-M-14 | ST624 | ||||||||||
| 105 | A/C2, F | TEM-24 | ST131, ST648; ST46 (2) | ||||||||||
| 150 | FIA, (no F) | SHV-12 | |||||||||||
| 220 | I1, N, B/O | CTX-M-14 | |||||||||||
| IV (1) | 100 | I1 | E | 1 | 4 | 1 | 2 | SHV-12 | ST23 | 2002 | Urine/P | Sm, Sp, Na, Su, W, Ch, Te | sul1, sul2 |
sul3 was detected among a collection of strains which included (i) 244 ESBL or MBL producers [TEM (−4, −12, −24, −27, and −52), SHV (−2, −2a, −5, −12, and −13), CTX-M (−1, −3, −9, −10, −14, −15, and −32), VIM-1, and OXA-30], representatives of ESBL clones recovered in Hospital Ramón y Cajal from 1988 to 2006, a collection partially explored in other studies with other purposes (18, 19, 21, 30) (1988 to 2006) and (ii) 66 non-ESBL isolates from blood samples obtained from different inpatients and 34 fecal isolates obtained from healthy volunteers living in the Madrid area (18). Abbreviations: Sm, streptomycin; Sp, spectinomycin; Na, nalidixic acid; Cip, ciprofloxacin; Te, tetracycline; Sul, sulfonamide; W, trimethoprim; Ch, chloramphenicol; Ak, amikacin; Km, kanamycin; Gm, gentamicin; Nt, netilmicin; Tb, tobramycin; Ap, apramycin; DLV, double-locus variant; NT, not typeable; ND, not done; P, hospitalized patient; HV, healthy volunteer. Names in boldface indicate either an ESBL encoded by bla genes or a sulz gene located on plasmids carrying sul3.
Plasmids transferred by conjugation to E. coli K-12 BM21 (rifampin and nalidixic acid resistant, Lac+, and plasmid free) are underlined (18).
Detailed analysis of 13 IncI1-like transferable plasmids revealed 8 RFLP patterns, designated with capital letters (A to G). Plasmids from six isolates were not studied further since they were cotransferred with other plasmids carrying different ESBL genes.
Diversity of five genes corresponding to the backbone of IncI plasmids (repY = replicase, see the text; ardA = gene encoding a type I restriction-modification enzyme; trbA = gene involved in maintenance and plasmid transfer; pndA = gene involved in plasmid transfer; sogS = gene coding for a DNA primase; pill = gene associated with type IV pilus biogenesis) following the allele designation given and used in the MLST scheme proposed by Alessandra Carattoli (http://pubmlst.org/perl/bigsdb/bigsdb.pl?db=pubmlst_plasmid_seqdef). Numbers represent different alleles for each gene.
Susceptibility to non-beta-lactam antibiotics was determined by using the standard disk diffusion method (7). Antibiotics in parentheses indicate that resistance was not present in all isolates. Cotransference with sul3 is indicated by underlining.
Strains containing two copies of a given sul3 integron carried by different plasmids.
Underlined genes were cotransferred with sul3.
The sul3 gene was detected in 6% of the strains (22 strains from 12 hospitalized patients, 8 outpatients, and 2 healthy humans; 1997 to 2006). All were ESBL/MBL-producing E. coli strains showing different PFGE patterns and sequence types (ST) linked to phylogroups A (n = 9; ST10, ST23, ST46, ST156, and ST695), B1 (n = 6; ST155, ST351, and ST359), and D/B2, (n = 7; ST131, ST350, ST624, and ST648). The most common ESBLs/MBLs were SHV-12 (n = 13) and CTX-M-14 (n = 8), followed by CTX-M-15 (n = 2), CTX-M-1, CTX-M-9, VIM-1, and TEM-24 (n = 1 each) (see Table 2). Four strains produced two ESBL/MBL enzymes (CTX-M-14, CTX-M-15, or VIM-1 plus SHV-12). The sul3 strains often carried other sul genes (sul1, sul2, and sul3, n = 16/22; sul1 and sul3, n = 2; sul2 and sul3, n = 1; sul3 only, n = 3) and expressed resistance to sulfonamides (86%), streptomycin (86%), trimethoprim (77%), tetracycline (77%), and chloramphenicol (64%). The low prevalence of the sul3 gene is similar to that reported by other studies (2, 4, 15), in contrast to that reported for the sul1 or sul2 gene (14, 19, 26). The association of sul3 with ESBL E. coli producers with zoonotic potential (phylogroup B2 E. coli O25:H4-ST131 and phylogroup D E. coli O25a-ST648, -ST69, and -ST393) (8, 31) highlights the role of these frequent clones in the evolution of antibiotic resistance to sulfonamides and beta-lactams in areas of common exposure to these antibiotics, such as farms or hospitals.
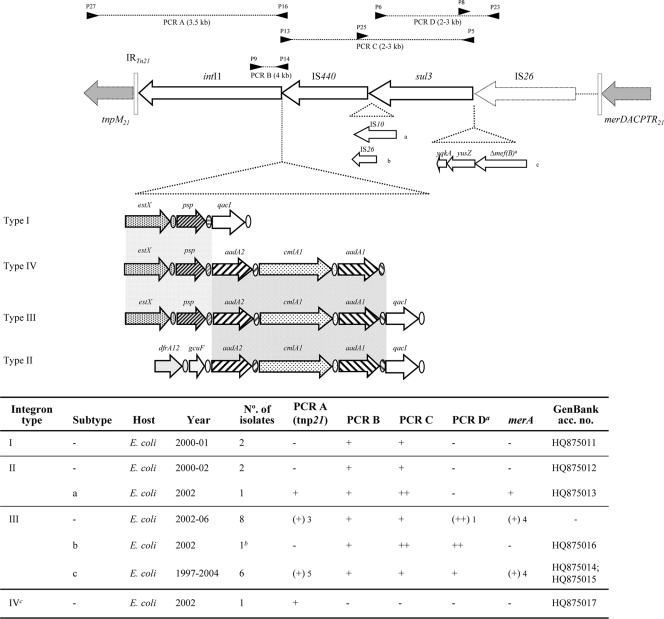
Characterization of sul3 class 1 integrons and linkage to Tn21 derivatives were accomplished by analyzing the presence of intI1, sul3, qacI, tnpM21, and mer21 by PCR/hybridization and further PCR mapping (Fig. 1 and Table 1). The diversity of sul3 platforms was established by comparison of restriction fragment length polymorphism (RFLP) patterns of HindIII-, EcoRI-, or PstI-digested amplicons, primer walking sequencing of representatives types, and further analysis of all sul3 elements available in the GenBank database. We detected four sul3 integron arrangements arbitrarily designated types IS3 to IVS3 (Table 2; Fig. 1). The predominant one was type IIIS3, intI1-estX-psp-aadA2-cmlA1-aadA1-qacI-IS440-sul3, present in 15 ESBL/MBL E. coli strains in our collection since 1997 and also globally distributed among Enterobacteriaceae of different origins (2, 3, 16, 17, 28). Two integrons similar to type IIIS3 were intI1-estX-psp-qacI-IS440-sul3 (type IS3) and intI1-estX-psp-aadA2/aadA1a-qacI-IS440-sul3 (from farm animals in Switzerland; GenBank accession no. FM242710) (Fig. 2). Differences in attC carried by the psp and aadA2 gene cassettes of different integrons suggest arrangements. Type IIS3, intI1-dfrA12-gcuF-aadA2-cmlA1-aadA1-qacI-IS440-sul3, has also been detected in humans, animals, and foods in Europe and Asia since 2003. Finally, a few sul3 platforms lacking qacI included intI1-estX-psp-aadA2-cmlA1-aadA1-IS440-sul3 (type IVS3; GenBank accession no. HQ875017), intI1-dfrA12-gcuF-aadA2-cmlA1-aadA1-IS440-sul3 (27), and partial sequences aadA1-IS440-sul3-orf1-mefB and aadA3-IS440-sul3-orf1-mefB (GenBank accession no. FJ196384 and FJ196388, respectively). The sul3 integrons obtained from E. coli and Salmonella enterica isolates from humans and farm animals were associated with Tn21 platforms often interrupted by IS26 at different positions (Fig. 1 and 2). Analysis of sequences revealed that a genetic platform, IS440-sul3-orf1-mefB, is specifically located beyond the attC sites of the qacI, aadA1, and aadA3 gene cassettes of integrons (Fig. 2). The lack of identifiable boundaries for IS440, a putative IS256 family member, reflects either a single insertion downstream of the last gene cassette (most probably qacI) of an ancestral integron, followed by further excisions and insertions of different gene cassettes or independent insertions at the end of different attC sites. The existence of identical integron arrangements lacking and containing IS26 and IS10 surrounded by direct repeats of the original target sequence indicates that the IS440-sul3 platform, once acquired, has been the target of further lateral transfer events. Moreover, complete or partial copies of IS26 at the boundaries of different integrons led to the genesis of putative IS26 composite transposons (Fig. 1 and 2). The lack of target site duplication at the sides of these IS26 copies might indicate the transference of either IS26 composite elements by homologous recombination among diverse plasmids or larger platforms in which they were located.
Fig. 1.
Schematic representation of sul3 genetic elements characterized by PCR mapping based on the Tn402 and Tn21 sequences. (Top) The locations of the primers used for the PCR mapping assay are represented with black arrowheads. P13 and P14 hybridize in the qacI gene. (Middle) The attC sites or 59-base elements were represented by circles patterned as corresponding gene cassettes. (Bottom) Preliminary screening of sul3 elements related to Tn402 and Tn21 was performed as shown in the table. ++, amplicon of larger size; (+), variable amplification results, with the number of positive results indicated; a, mefB was complete in only two isolates, and in most cases, the gene was truncated at different points by IS26 (GenBank accession no. HQ875016, HQ875014, and HQ875015); b, this isolate harbors an integron with insertions b and c; c, integron amplification was reached using primers P9 and P5. tnp21 was found upstream of the integrases of different sul3 integron types (n = 10), and a copy of IS26 was detected downstream of the sul3 gene (n = 8). Coexistence of both of the boundaries was detected for 7 isolates. The presence of the mer operon of Tn21 was inferred by merA.
Table 1.
Oligonucleotides used in this studyb
| Primer no. | Primer | Sequence (5′–3′) | GenBank accession no. | Nucleotide positions | Source/reference(s) |
|---|---|---|---|---|---|
| 1 | sul1F | CGGCGTGGGCTACCTGAACG | EU622038 | 3508–3528 | 15 |
| 2 | sul1R | GCCGATCGCGTGAAGTTCCG | EU622038 | 3921–3940 | 15 |
| 3 | sul2F | GCGCTCAAGGCAGATGGCATT | M36657 | 534–555 | 15 |
| 4 | sul2R | GCGTTTGATACCGGCACCCGT | M36657 | 819–798 | 15 |
| 5 | sul3F | GAGCAAGATTTTTGGAATCGT | AJ459418 | 2980–3001 | 23 |
| 6 | sul3R | CTAACCTAGGGCTTTGGATA | AJ459418 | 3770–3750 | This study |
| 7 | sul3F2 | TATCCAAAGCCCTAGGTTAG | AJ459418 | 3750–3770 | This study |
| 8 | sul3R2 | GAACTACGACTGGTTTC | AJ459418 | 2797–2780 | This study |
| 9 | IntI1F | GGGTCAAGGATCTGGATTTCG | AF071413 | 4775–4755 | 21 |
| 10 | IntI1R | ACATGCGTGTAAATCATCGTCG | AF071413 | 4333–4312 | 21 |
| 11 | 5′CS | GGCATCCAAGCAGCAAG | AF174129 | 1236–1252 | 21 |
| 12 | 3′CS | AAGCAGACTTGACCTGAT | AF174129 | 2813–2830 | 21 |
| 13 | qacIF | ACTGGCTCTTTCTGGCTATT | EF051039 | 5064–5084 | This study |
| 14 | qacIR | TAACGATAAGTCCCATGCCA | EF051039 | 5343–5323 | This study |
| 15 | cmlA1F | CACTTCCAAGAACGCAGACA | EF051039 | 2621–2641 | This study |
| 16 | cmlA1R | TTCCGATGCTTCCTAGCAGT | U12338 | 8020–8000 | This study |
| 17 | cmlA1FR | ACTGCTAGGAAGCATCGGAA | EF051039 | 3823–3803 | This study |
| 18 | aadA2R | TGACTTGATGATCTCGCC | AF174129 | 2692–2709 | 21 |
| 19 | estX | TTCCTTATGTGCATGGGTT | EF051039 | 794–813 | This study |
| 20 | dfrA12F | TTACGTCCAACGTTAGCAC | EF051037 | 1088–1107 | This study |
| 21 | aadA1R | ATTGCGCTGCCATTCTCCA | EF051037 | 4179–4160 | This study |
| 22 | psp R | ATCAGGGTGCCAGACAAGA | EF051039 | 1189–1170 | This study |
| 23 | IS26F | AGCGGTAAATCGTGGAGTGA | AF205943 | 324–344 | 21 |
| 24 | IS26R | AGGCCGGCATTTTCAGCGTG | AF205943 | 979–960 | 21 |
| 25 | IS440R | TGCGGGTACTTACTCCTTG | FJ587511 | 6415–6396 | This study |
| 26 | Orf1R | GCAATCCATTAGATTCATAC | FJ196385 | 10347–10327 | This study |
| 27 | TnpM Fw | CCGTGGTGGTGCATAGCAT | AF071413 | 4020–4002 | 21 |
| 28 | merA1 | ACCATCGGCGGCACCTGCGT | AF071413 | 17597–17578 | 21 |
| 29 | merA5 | ACCATCGTCAGGTAGGGGAACAA | AF071413 | 16360–16382 | 21 |
| 30 | copAF | ATGCGCCATAAGGCATTCA | NC_0050144 | 215–234 | 30 |
| 31 | repAR | AGTCGCTTCAGATGGTCAT | NC_005014 | 1427–1408 | 30 |
| 32 | mobP12F | GCAAAAGATGACACTGAYCCYGTTTT | NC_005014 | 67717–67743 | 1, 30 |
| 33 | mobP12R | AGCGATGTGGATGTGAAGGTTATCHGTRTC | NC_002122 | 31165–31120 | 1, 30 |
| 34 | ardAF | ATGTCTGTTGTTGCACCTGC | AP005147 | 61469–61811 | PubMLSTa |
| 35 | ardAR | TCACCGACGGAACACATGACC | AP005147 | 61926–61906 | PubMLST |
| 36 | trbAF | GCGGTTATCGGGCTACTA | AP005147 | 74976–74959 | This study |
| 37 | pndAF | GAATTCGTTGTCTGTAGCA | AP005147 | 73503–73521 | This study |
| 38 | sogSF | TTCCGGGGCGTAGACAATACT | AP005147 | 93088–93108 | PubMLST |
| 39 | sogSR | AACAGTGATATGCCGTCGC | AP005147 | 93378–93360 | PubMLST |
| 40 | pilVF | CCATATGACCATCCAGTGCG | AP005147 | 114765–114784 | PubMLST |
| 41 | pilVR | AACCACTATCTCGCCAGCAG | AP005147 | 115080–115061 | PubMLST |
The primer sequences used in primer walking assays are available upon request.
Fig. 2.
Diversity of the sul3 integrons based on published studies and sequences deposited in the GenBank database. In silico comparative analysis was made using BLAST and CLUSTAL softwares available at the BLAST (http://www.ncbi.nlm.nih.gov/blast//) and ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html) websites. The sul3 gene is associated with class 1 integrons that differ from class 1 sul1 integrons in the 3′ region, which comprise the gene cassette initially annotated qacH but called qacI afterward in order to distinguish it from the unrelated qacH gene of Staphylococcus aureus (GenBank accession no. AF205943) (20, 22). a, the first described sul3 platform recovered from an E. coli swine isolate in Switzerland in 2002 (GenBank accession no. AJ459418); b, orf1 is also annotated as yqkA-yusZ or orfA-orfB (GenBank accession no. AY509004, EU219534, FM242710, FM244708, FM244709, FM244710, FM244712, and FM244713); c, insertions of IS10 or IS26 within IS440-sul3 were found in some isolates (GenBank accession no. FJ587511, HQ875013, and HQ875016) (Fig. 1); d, the orf1-mefB sequence was absent or not determined; e, the presence of sequences upstream of intI1 or downstream of tniC was not determined or was absent; f, mefB is absent or truncated. aadA2 corresponding to FM242710 is a hybrid of aadA1a and aadA2 containing attCaadA1a. It is annotated aadA2 in the GenBank database.
Plasmids carrying sul3 belonged to a diversity of IncI1, IncY, and IncB/O groups (55 to 220 kb), mostly conjugative ones (86%). They were identified by PCR typing methods targeting replication and conjugation sequences of plasmids of Enterobacteriaceae (1, 5) and further hybridization of S1 nuclease-digested genomic DNA from E. coli transconjugants or wild-type strains with appropriate probes (30). Characterization of IncI1 plasmids also required RFLP profile comparison and sequencing of the whole replication region, relaxase (nikB) (1, 30), and genes included in the plasmid MLST scheme (http://pubmlst.org/perl/bigsdb/bigsdb.pl?db=pubmlst_plasmid_seqdef). Type IIIS3 was identified on blaSHV-12-IncI1 plasmids designated B, C1, C2, and D (100 to 150 kb). Genes linked to replication (cop, repY, and repA), transfer (nikB, trbA-pndA, sogS, and pilV), or restriction-modification systems (ardA) of plasmids B and D were identical to those carried by pCVM29188_101 from Salmonella enterica, an IncI1 plasmid carrying sul3 and blaCMY-2 (GenBank accession no. CP001121). Plasmid B was identified from 1997 to 2006 in ESBL strains from different continents (data not shown). Plasmids C1 and C2 were considered IncI1-mosaic plasmids, as repA carried by plasmid C1 was similar to that carried by IncK plasmid R387, and plasmid C2 harbors repA and sogS alleles that are different from those harbored by pCVM29188_101. The repY and nikB genes carried by plasmid E were identical to those carried by IncI1 pSL476_91 and pRYC106 (GenBank accession no. NC_011081 and GQ892053, respectively). The type IS3 integron was located on IncI1 plasmids similar to those of types B and D, while the type IIS3 integron was linked to nontransferable IncY, IncI1/Iγ, or IncB/O plasmids.
In summary, this work describes the recent spread of the IS440-sul3 platform, facilitated by its location on highly IncI1-transferable plasmids and by IS26-promoted rearrangements among multidrug resistance transposons and/or plasmids harbored by frequent E. coli clones. Coselection exerted by other antibiotics and/or biocides to which sulfonamide-resistant strains are also resistant, and the low fitness cost of plasmids in which sul genes are located, might be responsible for the spread and persistence of sul3, as suggested for other genes encoding trimethoprim-sulfonamide resistance (9, 29). The concurrent emergence and dissemination of the DHPS sul3 and ESBL genes among human Enterobacteriaceae since the early 1990s suggest the recent recruitment of adaptive mechanisms in different environments (animals and humans exposed to beta-lactams and sulfonamides) by the same predominant genetic platforms.
Acknowledgments
T.C. and M.P.G.-B. are supported by fellow research contracts from the Spanish Ministry of Science and Innovation (grant FI09/00901) and from Consejo Superior de Investigaciones Científicas (JAE-Doc), respectively. This study and the work done by the IRYCIS group were funded by research grants from the European Commission (LSHMCT-2008-223031 and KBBE-2008-2B-227258) and by the CIBERESP Network for Biomedical Research in Epidemiology and Public Health (Instituto Carlos III, Spanish Ministry of Science and Innovation; reference no. CB06/02/0053). Work completed in the FdlC laboratory was supported by grants BFU2008-00995/BMC (Spanish Ministry of Education), REIPI RD06/0008/1012 (RETICS research network, Instituto de Salud Carlos III, Spanish Ministry of Science and Innovation), and LSHM-CT-2005_019023 (European VI Framework Program). We also are grateful to the Spanish Network for the Study of Plasmids and Extrachromosomal Elements (REDEEX) for encouraging and funding cooperation among Spanish microbiologists working on the biology of mobile genetic elements (grant BFU 2008-0079-E/BMC; Spanish Ministry of Science and Innovation).
Footnotes
Published ahead of print on 22 February 2011.
REFERENCES
- 1. Alvarado A., Garcillán-Barcia M. P., de la Cruz F. 2008. A PCR-based method for classification of conjugative plasmids based on relaxase sequences, p. 134 In Proceedings of the International Plasmid Biology Conference International Plasmid Biology Society, Gdansk, Poland [Google Scholar]
- 2. Antunes P., Machado J., Peixe L. 2007. Dissemination of sul3-containing elements linked to class 1 integrons with an unusual 3′ conserved sequence region among Salmonella isolates. Antimicrob. Agents Chemother. 51:1545–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bischoff K. M., White D. G., Hume M. E., Poole T. L., Nisbet D. J. 2005. The chloramphenicol resistance gene cmlA is disseminated on transferable plasmids that confer multiple-drug resistance in swine Escherichia coli. FEMS Microbiol. Lett. 243:285–291 [DOI] [PubMed] [Google Scholar]
- 4. Byrne-Bailey K. G., et al. 2009. Prevalence of sulfonamide resistance genes in bacterial isolates from manured agricultural soils and pig slurry in the United Kingdom. Antimicrob. Agents Chemother. 53:696–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carattoli A., et al. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 6. Clermont O., Bonacorsi S., Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing; 19th informational supplement. CLSI document M100-S19. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8. Cortés P., et al. 2010. Isolation and characterization of potentially pathogenic antimicrobial-resistant Escherichia coli strains from chicken and pig farms in Spain. Appl. Environ. Microbiol. 76:2799–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Enne V. I., Livermore D. M., Stephens P., Hall L. M. 2001. Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357:1325–1328 [DOI] [PubMed] [Google Scholar]
- 10. Fricke W. F., et al. 2009. Comparative genomics of the IncA/C multidrug resistance plasmid family. J. Bacteriol. 191:4750–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guerra B., Junker E., Helmuth R. 2004. Incidence of the recently described sulfonamide resistance gene sul3 among German Salmonella enterica strains isolated from livestock and food. Antimicrob. Agents Chemother. 48:2712–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heuer H., Kopmann C., Binh C. T., Top E. M., Smalla K. 2009. Spreading antibiotic resistance through spread manure: characteristics of a novel plasmid type with low % G+C content. Environ. Microbiol. 11:937–949 [DOI] [PubMed] [Google Scholar]
- 13. Heuer H., Smalla K. 2007. Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ. Microbiol. 9:657–666 [DOI] [PubMed] [Google Scholar]
- 14. Huovinen P., Sundström L., Swedberg G., Sköld O. 1995. Trimethoprim and sulfonamide resistance. Antimicrob. Agents Chemother. 39:279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kerrn M. B., Klemmensen T., Frimodt-Moller N., Espersen F. 2002. Susceptibility of Danish Escherichia coli strains isolated from urinary tract infections and bacteraemia, and distribution of sul genes conferring sulphonamide resistance. J. Antimicrob. Chemother. 50:513–516 [DOI] [PubMed] [Google Scholar]
- 16. Lee M. F., Chen Y. H., Peng C. F. 2009. Molecular characterisation of class 1 integrons in Salmonella enterica serovar Choleraesuis isolates from southern Taiwan. Int. J. Antimicrob. Agents 33:216–222 [DOI] [PubMed] [Google Scholar]
- 17. Liu J., Keelan P., Bennett P. M., Enne V. I. 2009. Characterization of a novel macrolide efflux gene, mef(B), found linked to sul3 in porcine Escherichia coli. J. Antimicrob. Chemother. 63:423–426 [DOI] [PubMed] [Google Scholar]
- 18. Machado E., et al. 2005. Integron content of extended-spectrum-beta-lactamase-producing Escherichia coli strains over 12 years in a single hospital in Madrid, Spain. Antimicrob. Agents Chemother. 49:1823–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Machado E., et al. 2007. Preservation of integron types among Enterobacteriaceae producing extended-spectrum beta-lactamases in a Spanish hospital over a 15-year period (1988 to 2003). Antimicrob. Agents Chemother. 51:2201–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Naas T., Mikami Y., Imai T., Poirel L., Nordmann P. 2001. Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 183:235–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Novais A., et al. 2006. Dissemination and persistence of blaCTX-M-9 are linked to class 1 integrons containing CR1 associated with defective transposon derivatives from Tn402 located in early antibiotic resistance plasmids of IncHI2, IncP1-alpha, and IncFI groups. Antimicrob. Agents Chemother. 50:2741–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Partridge S. R., Tsafnat G., Coiera E., Iredell J. R. 2009. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 33:757–784 [DOI] [PubMed] [Google Scholar]
- 23. Perreten V., Boerlin P. 2003. A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob. Agents Chemother. 47:1169–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Phuong Hoa P. T., Nonaka L., Hung Viet P., Suzuki S. 2008. Detection of the sul1, sul2, and sul3 genes in sulfonamide-resistant bacteria from wastewater and shrimp ponds of north Vietnam. Sci. Total Environ. 405:377–384 [DOI] [PubMed] [Google Scholar]
- 25. Rådström P., Swedberg G. 1988. RSF1010 and a conjugative plasmid contain sulII, one of two known genes for plasmid-borne sulfonamide resistance dihydropteroate synthase. Antimicrob. Agents Chemother. 32:1684–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rådström P., Swedberg G., Sköld O. 1991. Genetic analyses of sulfonamide resistance and its dissemination in gram-negative bacteria illustrate new aspects of R plasmid evolution. Antimicrob. Agents Chemother. 35:1840–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sáenz Y., et al. 2010. Class 1 integrons lacking qacEDelta1 and sul1 genes in Escherichia coli isolates of food, animal and human origins. Vet. Microbiol. 144:493–497 [DOI] [PubMed] [Google Scholar]
- 28. Soufi L., et al. 2009. Prevalence and diversity of integrons and associated resistance genes in Escherichia coli isolates from poultry meat in Tunisia. Foodborne Pathog. Dis. 6:1067–1073 [DOI] [PubMed] [Google Scholar]
- 29. Sundqvist M., et al. 2010. Little evidence for reversibility of trimethoprim resistance after a drastic reduction in trimethoprim use. J. Antimicrob. Chemother. 65:350–360 [DOI] [PubMed] [Google Scholar]
- 30. Valverde A., et al. 2009. Spread of bla(CTX-M-14) is driven mainly by IncK plasmids disseminated among Escherichia coli phylogroups A, B1, and D in Spain. Antimicrob. Agents Chemother. 53:5204–5212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Woodford N., Turton J., Livermore D. M. 2011. Multi-resistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 344:3964–4100 [DOI] [PubMed] [Google Scholar]
- 32. Wu S., Dalsgaard A., Hammerum A. M., Porsbo L. J., Jensen L. B. 2010. Prevalence and characterization of plasmids carrying sulfonamide resistance genes among Escherichia coli from pigs, pig carcasses and human. Acta Vet. Scand. 52:47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]