Abstract
Among 1,349 Streptococcus pneumoniae invasive isolates, 45 (3.3%) were levofloxacin resistant. Serotype distribution was as follows: 8 (n = 32 isolates), 19A (n = 4 isolates), 7F (n = 3 isolates), 9V (n = 2 isolates), 10A (n = 1 isolate), 19F (n = 1 isolate), 6B (n = 1 isolate), and nontypeable (n = 1 isolate). Levofloxacin-resistant isolates had dual mutations in the gyrA and parC genes. Serotype 8 strains corresponded to a capsular switching of the Sweden15A-25 clone. Levofloxacin resistance was also detected among multiresistant (ST27619A, Spain9V-ST156, ST8819F, and ST15426B) and among usually antibiotic-susceptible (Netherlands7F-ST191, ST120119A, and ST263910A) clones.
INTRODUCTION
Dissemination of non-antimicrobial-susceptible clones has been a major factor in the emergence of resistance in Streptococcus pneumoniae (10, 15). Moreover, the introduction of the pneumococcal 7-valent conjugate vaccine (PCV7) has brought about a shift (9, 13, 16), with an increase in the incidence of non-PCV7 nonsusceptible serotypes, such as 19A, which can complicate the selection of empirical treatments. Recently, the 13-valent pneumococcal conjugate vaccine (PCV13) has been granted marketing authorization. Currently, most of the circulating serotypes in Spain are covered by this vaccine (9).
Resistance to fluoroquinolones among pneumococci is primarily caused by mutations in the quinolone resistance-determining regions (QRDRs) of the parC and gyrA genes (7, 18, 20). Although at present the prevalence of fluoroquinolone resistance in S. pneumoniae remains low (5, 17), ongoing surveillance is necessary.
In this study, we describe the serotypes, antimicrobial susceptibilities, fluoroquinolone resistance mutations, and molecular characterizations of levofloxacin-resistant invasive pneumococci isolated in our area over a 2-year period.
From February 2007 to January 2009, a total of 1,349 invasive strains (one isolate for every invasive pneumococcal disease [IPD] episode) from 1,324 patients (age range, <1 to 101 years; 785 males) were studied. Nineteen patients had two recurrent infections, and three had three recurrences. The strains were recovered from 34 hospitals in the area of Madrid (Spain).
Identification of the capsular serotypes was determined by the Pneumotest-Latex kit (Statens Serum Institut, Copenhagen, Denmark) and by Quellung reaction using commercial factor antisera (Statens Serum Institut, Copenhagen, Denmark).
Susceptibilities to penicillin, erythromycin, clindamycin, cefotaxime, vancomycin, and levofloxacin were determined by the Etest method (AB bioMérieux, Solna, Sweden). Inducible resistance to macrolides was detected by the double-disk diffusion method using erythromycin (15 μg) and clindamycin (2 μg) disks.
Levofloxacin-resistant (MIC ≥ 8 μg/ml) S. pneumoniae isolates were characterized by pulsed-field gel electrophoresis (PFGE) after restriction with SmaI (14). Multilocus sequence typing (MLST) was performed as previously described (8). The QRDR regions of the gyrA and parC genes were amplified and sequenced as described elsewhere (18).
Among 1,349 S. pneumoniae invasive isolates, 45 levofloxacin-resistant strains (3.3%; all showing a levofloxacin MIC of ≥32 μg/ml) were detected. Only two strains (0.15%) showed intermediate susceptibility (levofloxacin MIC of 4 μg/ml). Overall, the MIC50 and MIC90 of levofloxacin were 0.75 and 1.75 μg/ml, respectively. All levofloxacin-resistant strains were isolated from adult patients (age range, 35 to 98 years). Serotype distribution was as follows: 8 (n = 32 isolates), 19A (n = 4 isolates), 7F (n = 3 isolates), 9V (n = 2 isolates), 10A (n = 1 isolate), 19F (n = 1 isolate), 6B (n = 1 isolate), and nontypeable (n = 1 isolate). Most serotype 8 levofloxacin-resistant strains (n = 19) were isolated from a single hospital (the remaining 13 strains were isolated from eight different hospitals).
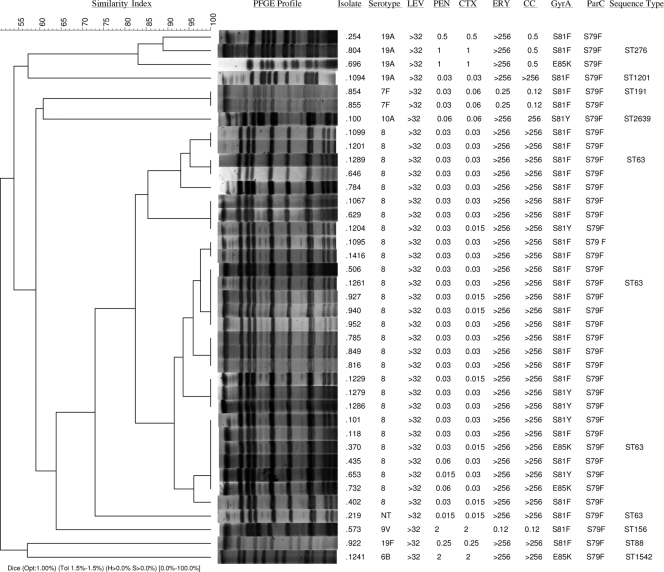
Thirty-nine of the 45 levofloxacin-resistant strains, serotypes 8 (n = 28), 19A (n = 4), 7F (n = 2), 10A (n = 1), 19F (n = 1), 6B (n = 1), 9V (n = 1), and nontypeable (n = 1), were available for molecular characterization. Results of serotyping, antimicrobial susceptibility, QRDR characterization, PFGE, and sequence type (ST) are shown in Fig. 1.
Fig. 1.
PFGE dendrogram and information of 39 levofloxacin-resistant S. pneumoniae isolates, including serotype, MICs (μg/ml) of the antimicrobial agents tested, sequence type (ST), and QRDR substitutions in GyrA and ParC. LEV, levofloxacin; PEN, penicillin; CTX, cefotaxime; ERY, erythromycin; CC, clindamycin.
The 28 serotype 8 strains showed one identical PFGE type, were susceptible to penicillin and cefotaxime, and showed erythromycin and clindamycin resistance with the constitutive macrolide-lincosamide-streptogramin B (cMLSB) phenotype. Characterization of gyrA and parC QRDRs identified the ParC S79F mutation in all isolates. In addition, 21 isolates presented the S81F mutation, five the S81Y mutation, and two the E85K mutation in GyrA. Three randomly selected isolates from this PFGE type were shown to be ST63, which corresponds to a capsular switching event of the Sweden15A-25-ST63 clone. The nontypeable strain was grouped in the same PFGE type as serotype 8 isolates, and MLST analysis identified this strain as belonging to ST63.
Among the four serotype 19A isolates, two PFGE types were identified, and one included three erythromycin-resistant and clindamycin-intermediate isolates, expressing the inducible MLSB (iMLSB) phenotype. All three isolates presented the ParC S79F mutation, and two of them harbored the S81F mutation and one the E85K mutation in GyrA. MLST of one of the three isolates identified the strain as ST276, a single-locus variant (SLV) of ST230, representative of the Denmark14-32 clone. The remaining 19A isolate, identified as ST1201, was susceptible to penicillin, cefotaxime, erythromycin, and clindamycin and showed S79F ParC and S81F GyrA substitutions. The two serotype 7F isolates were shown to be ST191. The only 9V strain available was identified as ST156 (Spain9V-3 clone). The 6B isolate was identified as ST1542, an SLV of Spain6B-2-ST90. Isolates expressing 19F and 10A serotypes were identified as ST88 (minor Spanish 19F clone) and ST2639, respectively.
In this study, the prevalence of fluoroquinolone resistance among pneumococcal invasive isolates was 3.3%, which is higher than previously reported in Spain (4). It has been proposed that most fluoroquinolone-resistant strains of S. pneumoniae arise from heterogeneous mutations (11, 12). However, levofloxacin resistance in this study was limited to a few serotypes and was due mainly to a clonal spread of a serotype 8 ST63 (Sweden15A-25). These isolates were fully susceptible to penicillin and resistant to erythromycin and clindamycin. This clone, not included in the conjugate vaccine, has been previously detected among fluoroquinolone-resistant S. pneumoniae in Spain (4), and capsular switching events have also been described in this clone (4). The serotype 19A ST276 clone, an SLV of Denmark14-32-ST230, was also responsible for resistance to levofloxacin in our series. This multiresistant clone included in the PCV13 is disseminated worldwide (9, 13, 16). However, to our knowledge, this is the first report of resistance to levofloxacin in strains belonging to this clone. In addition, it is noteworthy that we have detected resistance to fluoroquinolones among usually antibiotic-susceptible clones circulating in Spain: Netherlands7F-ST191, ST1201 (19A), and ST2639 (10A) (1, 6, 19).
DNA sequence analysis of the QRDRs of the gyrA and parC genes showed that all isolates contained the S79F mutation in ParC. All isolates also presented amino acid changes in GyrA, with most changes occurring at S81, whereas changes at E85 were rare. We found heterogeneity in the GyrA QRDR amino acid substitutions among isolates belonging to the same PFGE type. This suggests that dissemination of these organisms is not only due to clonal spread but probably also due to independent selection.
Strains with first-step mutation to quinolones (mutation in only one of the target genes) are at a higher risk for developing resistance (2) and may determine the treatment outcome (3). However, the very low proportion of strains with intermediate susceptibility to levofloxacin detected in this study (0.15%) suggests that isolates with first-step mutations to quinolones are not very frequent in our area.
Acknowledgments
Contributing members of the Madrid Streptococcus pneumoniae Microbiological Group (MSPSG) are Mercedes Marín (Hospital General Universitario Gregorio Marañón); Francisca Sanz, Fernando Chaves, Maria Angeles Orellana (Hospital Universitario 12 de Octubre); Elia Gómez, Elena Loza, María Antonia Meseguer (Hospital Universitario Ramón y Cajal); Fernando González-Romo, Carmen Betriu (Hospital Clínico San Carlos); Margarita Sánchez-Concheiro, Juana Cacho (Hospital Universitario de Getafe); Beatriz Orden, Pilar Mendaza, Isabel Sanchez (Hospital Universitario Puerta de Hierro Majadahonda); María del Carmen de las Cuevas (Hospital Universitario de La Princesa); Isabel Wilhelmi (Hospital Universitario Severo Ochoa); Maria Pilar Romero (Hospital Universitario La Paz); Alberto Delgado, José Valverde (Hospital Universitario Fundación Alcorcón); Peña Gómez-Herrúz (Hospital Universitario Príncipe de Asturias); Ricardo Fernández Roblas, María Carolina Isea Peña (Fundación Jiménez Díaz); Belén Hernández-Milán (Hospital Infantil Universitario Niño Jesús); Jose Luis Gómez Garcés (Hospital Universitario de Móstoles); Santiago Salso (Hospital Universitario Madrid Montepríncipe); Elena García-Peñuela (Hospital Sanitas La Zarzuela); Dolores Martín (Hospital Infanta Cristina); Diana Monclús (Hospital San Rafael); Maria Jose Uría (Hospital del Sureste); Andrea González Prieto (Hospital del Tajo); Almudena Alambra (Hospital Universitario Madrid, Madrid Sanchinarro y Madrid Torrelodones); Aída Sanchez-García, Elena Sáez (Hospital Infanta Sofía); Luis Ruiz Velasco (Hospital Moncloa); Luisa García-Picaz o (Hospital El Escorial); Carolina Camplelo (Hospital Infanta Leonor); Laura Molina, Isabel García-Arata (Hospital de Fuenlabrada); Mercedes Martínez-Arroyo (Hospital Sanitas La Moraleja); Esteban Aznar (Hospital del Henares); Ana Enríquez (Hospital Carlos III); Amparo Catón (Sanatorio Nuestra Señora del Rosario); Isabel Jiménez (Clínica Fuensanta); Laura Navas, Pilar Sanchez de la Blanca (Hospital Los Madroños); Angeles Gutiérrez, Araceli Arce (Servicio de Epidemiología de la Comunidad de Madrid).
Footnotes
Published ahead of print on 7 March 2011.
REFERENCES
- 1. Ardanuy C., et al. 2009. Epidemiology of invasive pneumococcal disease among adult patients in Barcelona before and after pediatric 7-valent pneumococcal conjugate vaccine introduction, 1997-2007. Clin. Infect. Dis. 48:57–64 [DOI] [PubMed] [Google Scholar]
- 2. Davidson R., et al. 2002. Resistance to levofloxacin and failure of treatment of pneumococcal pneumonia. N. Engl. J. Med. 346:747–750 [DOI] [PubMed] [Google Scholar]
- 3. de Cueto M., et al. 2008. Fatal levofloxacin failure in treatment of a bacteremic patient infected with Streptococcus pneumoniae with a preexisting parC mutation. J. Clin. Microbiol. 46:1558–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de la Campa A. G., et al. 2009. Changes in fluoroquinolone-resistant Streptococcus pneumoniae after 7-valent conjugate vaccination, Spain. Emerg. Infect. Dis. 15:905–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de la Campa A. G., et al. 2003. Genetic characterization of fluoroquinolone-resistant Streptococcus pneumoniae strains isolated during ciprofloxacin therapy from a patient with bronchiectasis. Antimicrob. Agents Chemother. 47:1419–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de la Pedrosa E. G., et al. 2008. Polyclonal population structure of Streptococcus pneumoniae isolates in Spain carrying mef and mef plus erm(B). Antimicrob. Agents Chemother. 52:1964–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eliopoulos G. M. 2004. Quinolone resistance mechanisms in pneumococci. Clin. Infect. Dis. 38:S350–S356 [DOI] [PubMed] [Google Scholar]
- 8. Enright M. C., Spratt B. G. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049–3060 [DOI] [PubMed] [Google Scholar]
- 9. Fenoll A., et al. 2009. Temporal trends of invasive Streptococcus pneumoniae serotypes and antimicrobial resistance patterns in Spain from 1979 to 2007. J. Clin. Microbiol. 47:1012–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klugman K. P. 2002. The successful clone: the vector of dissemination of resistance in Streptococcus pneumoniae. J. Antimicrob. Chemother. 50(Suppl. S2):1–5 [DOI] [PubMed] [Google Scholar]
- 11. Lynch J. P., III, Zhanel G. G. 2010. Streptococcus pneumoniae: epidemiology and risk factors, evolution of antimicrobial resistance, and impact of vaccines. Curr. Opin. Pulm. Med. 16:217–225 [DOI] [PubMed] [Google Scholar]
- 12. Lynch J. P., III, Zhanel G. G. 2009. Streptococcus pneumoniae: does antimicrobial resistance matter? Semin. Respir. Crit. Care Med. 30:210–238 [DOI] [PubMed] [Google Scholar]
- 13. Mahjoub-Messai F., et al. 2009. Population snapshot of Streptococcus pneumoniae serotype 19A isolates before and after introduction of seven-valent pneumococcal vaccination for French children. J. Clin. Microbiol. 47:837–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McEllistrem M. C., Stout J. E., Harrison L. H. 2000. Simplified protocol for pulsed-field gel electrophoresis analysis of Streptococcus pneumoniae. J. Clin. Microbiol. 38:351–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McGee L., et al. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39:2565–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moore M. R., et al. 2008. Population snapshot of emergent Streptococcus pneumoniae serotype 19A in the United States, 2005. J. Infect. Dis. 197:1016–1027 [DOI] [PubMed] [Google Scholar]
- 17. Morrissey I., Colclough A., Northwood J. 2007. TARGETed surveillance: susceptibility of Streptococcus pneumoniae isolated from community-acquired respiratory tract infections in 2003 to fluoroquinolones and other agents. Int. J. Antimicrob. Agents 30:345–351 [DOI] [PubMed] [Google Scholar]
- 18. Muñoz R., de la Campa A. G. 1996. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob. Agents Chemother. 40:2252–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Muñoz-Almagro C., et al. 2009. Emergence of invasive pneumococcal disease caused by multidrug-resistant serotype 19A among children in Barcelona. J. Infect. 59:75–82 [DOI] [PubMed] [Google Scholar]
- 20. Pan X. S., Ambler J., Mehtar S., Fisher L. M. 1996. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 40:2321–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]