Abstract
Overexpression of the multidrug efflux pump Mdr1 causes increased fluconazole resistance in the pathogenic yeast Candida albicans. The transcription factors Mrr1 and Cap1 mediate MDR1 upregulation in response to inducing stimuli, and gain-of-function mutations in Mrr1 or Cap1, which render the transcription factors hyperactive, result in constitutive MDR1 overexpression. The essential MADS box transcription factor Mcm1 also binds to the MDR1 promoter, but its role in inducible or constitutive MDR1 upregulation is unknown. Using a conditional mutant in which Mcm1 can be depleted from the cells, we investigated the importance of Mcm1 for MDR1 expression. We found that Mcm1 was dispensable for MDR1 upregulation by H2O2 but was required for full MDR1 induction by benomyl. A C-terminally truncated, hyperactive Cap1 could upregulate MDR1 expression both in the presence and in the absence of Mcm1. In contrast, a hyperactive Mrr1 containing a gain-of-function mutation depended on Mcm1 to cause MDR1 overexpression. These results demonstrate a differential requirement for the coregulator Mcm1 for Cap1- and Mrr1-mediated MDR1 upregulation. When activated by oxidative stress or a gain-of-function mutation, Cap1 can induce MDR1 expression independently of Mcm1, whereas Mrr1 requires either Mcm1 or an active Cap1 to cause overexpression of the MDR1 efflux pump. Our findings provide more detailed insight into the molecular mechanisms of drug resistance in this important human fungal pathogen.
INTRODUCTION
Overexpression of the multidrug efflux pump Mdr1 is one mechanism by which the pathogenic yeast Candida albicans can develop increased resistance to the antifungal drug fluconazole, which is widely used to treat Candida infections (reviewed in reference 13). The MDR1 gene is not significantly expressed in drug-susceptible C. albicans strains under standard growth conditions, but its transcription can be induced by certain toxic compounds, like benomyl or H2O2 (6, 7, 9, 17). Many fluconazole-resistant clinical C. albicans isolates constitutively upregulate MDR1 under noninducing conditions, and all such isolates studied to date contain gain-of-function mutations in the zinc cluster transcription factor Mrr1 (3, 14). MRR1 plays a central role in MDR1 expression, as its deletion abolishes both the inducible activation of the MDR1 promoter in drug-susceptible strains and constitutive MDR1 overexpression in fluconazole-resistant clinical isolates (14).
In addition to Mrr1, other transcription factors have also been implicated in the regulation of MDR1 expression (1, 4, 16, 17, 20, 26, 27). The bZip transcription factor Cap1, which mediates oxidative-stress responses in C. albicans, is required for the induction of MDR1 transcription by H2O2 and also contributes to benomyl-induced MDR1 expression (17, 18a, 26). Therefore, Mrr1 and Cap1 upregulate MDR1 in response to inducing chemicals in a cooperative fashion. However, Cap1 is dispensable for constitutive MDR1 overexpression in strains containing gain-of-function mutations in Mrr1 (1, 17, 18a). Conversely, a C-terminally truncated, hyperactive Cap1 can also activate the MDR1 promoter in the absence of MRR1, albeit less efficiently than in a wild-type background (18a). Hyperactive forms of either Mrr1 or Cap1 can therefore promote MDR1 overexpression independently of each other.
Deletion analyses of the MDR1 promoter identified a region that is important for benomyl-induced MDR1 upregulation, as well as for constitutive MDR1 overexpression (16, 17). This region, which was termed BRE (for benomyl response element) or MDRE (for MDR1 drug resistance element), contains a binding site for the MADS box transcription factor Mcm1. When inserted into a heterologous promoter, the BRE/MDRE rendered the promoter responsive to benomyl in a drug-susceptible strain and constitutively active in an MDR1-overexpressing strain background (16, 17). Mcm1 is involved in a variety of cellular processes and mediates both repressing and activating functions, presumably by recruiting coregulatory proteins to the respective promoters (18, 23). Mcm1 has been shown to bind to the MDR1 promoter in vivo (11, 23), where it may act together with Mrr1 and/or Cap1 to control expression of the efflux pump in response to inducing stimuli and enable constitutive MDR1 overexpression in strains containing hyperactive MRR1 or CAP1 alleles.
As MCM1 is essential in C. albicans, the role of this transcription factor in MDR1 regulation cannot be addressed by deleting the gene. A conditional mcm1 mutant that contains a single copy of MCM1 under the control of a tetracycline-repressible promoter has therefore been used to analyze the function of Mcm1 in C. albicans. The addition of doxycycline to the conditional mutant results in depletion of Mcm1 within 3 h, but the cells remain viable for a prolonged period, allowing the study of their behavior in the absence of Mcm1 (18). It was shown that depletion of Mcm1 from the cells resulted in the loss of binding activity of protein extracts to an MDR1 promoter fragment containing the MDRE (16). However, whether Mcm1 is indeed important for the regulation of MDR1 expression and whether it acts as an activator or a repressor of the MDR1 promoter were not directly tested in that study. To understand in more detail how the expression of this multidrug efflux pump is controlled in C. albicans, we investigated if Mcm1 is necessary for the upregulation of MDR1 in response to inducing chemicals and if hyperactive forms of Mrr1 and Cap1 require Mcm1 as a coregulator to mediate MDR1 overexpression.
MATERIALS AND METHODS
Strains and growth conditions.
The C. albicans strains used in this study are listed in Table 1. All strains were stored as frozen stocks with 15% glycerol at −80°C and subcultured on YPD agar plates (10 g yeast extract, 20 g peptone, 20 g glucose, 15 g agar per liter) at 30°C. Strains were routinely grown in YPD liquid medium at 30°C in a shaking incubator. For selection of nourseothricin-resistant transformants, 200 μg/ml nourseothricin (Werner Bioagents, Jena, Germany) was added to YPD agar plates. To obtain nourseothricin-sensitive derivatives in which the SAT1 flipper cassette was excised by FLP-mediated recombination, transformants were grown overnight in YPM medium (10 g yeast extract, 20 g peptone, 20 g maltose per liter) without selective pressure to induce the MAL2 promoter that controls expression of the Candida-adapted FLP (caFLP) gene in the SAT1 flipper cassette. One hundred to 200 cells were then spread on YPD plates containing 10 μg/ml nourseothricin and grown for 2 days at 30°C. Nourseothricin-sensitive clones were identified by their small colony size and confirmed by restreaking them on YPD plates containing 200 μg/ml nourseothricin.
Table 1.
C. albicans strains used in this study
| Strain | Parent strain | Relevant characteristics or genotypea | Reference |
|---|---|---|---|
| SC5314 | Wild-type reference strain | 5 | |
| SCMPG2A and -B | SC5314 | ACT1/act1::PMDR1-GFP-caSAT1 | 14 |
| MRcan42 | SC5314 | ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 | 18 |
| ENO1/eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2 | |||
| mcm1::URA3::97t::MCM1::myc/mcm1::FRT | |||
| MRcan43 | SC5314 | ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 | 18 |
| ENO1/eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2 | |||
| MCM1::myc-URA3::97t::MCM1/mcm1::FRT | |||
| can42MPG2A and -B | MRcan42 | ACT1/act1::PMDR1-GFP-caSAT1 | This study |
| can43MPG2A and -B | MRcan43 | ACT1/act1::PMDR1-GFP-caSAT1 | This study |
| can42MRR1R31A and -B | MRcan42 | MRR1/MRR1P683S-SAT1-FLIP | This study |
| can42MRR1R32A | can42MRR1R31A | MRR1/MRR1P683S-FRT | This study |
| can42MRR1R32B | can42MRR1R31B | MRR1/MRR1P683S-FRT | This study |
| can42MRR1R33A | can42MRR1R32A | MRR1P683S-SAT1-FLIP/MRR1P683S-FRT | This study |
| can42MRR1R33B | can42MRR1R32B | MRR1P683S-SAT1-FLIP/MRR1P683S-FRT | This study |
| can42MRR1R34A | can42MRR1R33A | MRR1P683S-FRT/MRR1P683S-FRT | This study |
| can42MRR1R34B | can42MRR1R33B | MRR1P683S-FRT/MRR1P683S-FRT | This study |
| can43MRR1R31A and -B | MRcan43 | MRR1/MRR1P683S-SAT1-FLIP | This study |
| can43MRR1R32A | can43MRR1R31A | MRR1/MRR1P683S-FRT | This study |
| can43MRR1R32B | can43MRR1R31B | MRR1/MRR1P683S-FRT | This study |
| can43MRR1R33A | can43MRR1R32A | MRR1P683S-SAT1-FLIP/MRR1P683S-FRT | This study |
| can43MRR1R33B | can43MRR1R32B | MRR1P683S-SAT1-FLIP/MRR1P683S-FRT | This study |
| can43MRR1R34A | can43MRR1R33A | MRR1P683S-FRT/MRR1P683S-FRT | This study |
| can43MRR1R34B | can43MRR1R33B | MRR1P683S-FRT/MRR1P683S-FRT | This study |
| can42CAP1R11A | MRcan42 | CAP1ΔC333-SAT1-FLIP/CAP1-2 | This study |
| can42CAP1R11B | MRcan42 | CAP1-1/CAP1ΔC333-SAT1-FLIP | This study |
| can42CAP1R12A | can42CAP1R11A | CAP1ΔC333-FRT/CAP1-2 | This study |
| can42CAP1R12B | can42CAP1R11B | CAP1-1/CAP1ΔC333-FRT | This study |
| can42CAP1R13A | can42CAP1R12A | CAP1ΔC333-FRT/CAP1ΔC333-SAT1-FLIP | This study |
| can42CAP1R13B | can42CAP1R12B | CAP1ΔC333-SAT1-FLIP/CAP1ΔC333-FRT | This study |
| can42CAP1R14A | can42CAP1R13A | CAP1ΔC333-FRT/CAP1ΔC333-FRT | This study |
| can42CAP1R14B | can42CAP1R13B | CAP1ΔC333-FRT/CAP1ΔC333-FRT | This study |
| can43CAP1R11A and -B | MRcan43 | CAP1-1/CAP1ΔC333-SAT1-FLIP | This study |
| can43CAP1R12A | can43CAP1R11A | CAP1-1/CAP1ΔC333-FRT | This study |
| can43CAP1R12B | can43CAP1R11B | CAP1-1/CAP1ΔC333-FRT | This study |
| can43CAP1R13A | can43CAP1R12A | CAP1ΔC333-SAT1-FLIP/CAP1ΔC333-FRT | This study |
| can43CAP1R13B | can43CAP1R12B | CAP1ΔC333-SAT1-FLIP/CAP1ΔC333-FRT | This study |
| can43CAP1R14A | can43CAP1R13A | CAP1ΔC333-FRT/CAP1ΔC333-FRT | This study |
| can43CAP1R14B | can43CAP1R13B | CAP1ΔC333-FRT/CAP1ΔC333-FRT | This study |
Apart from the indicated features, all strains are identical to their parents. SAT1-FLIP denotes the SAT1 flipper cassette; FRT is the FLP recombination target sequence, one copy of which remains in the genome after recycling of the SAT1 flipper cassette. The CAP1 alleles in strain SC5314 were distinguished by a BglII restriction site polymorphism. The CAP1 allele containing the polymorphic BglII site in the upstream region was designated CAP1-2. caSAT1, Candida-adapted SAT1.
Strain construction.
C. albicans strains were transformed by electroporation (10) with the following gel-purified linear DNA fragments. A KpnI-SacII fragment from pMPG2S (14) was used to integrate a PMDR1-GFP reporter fusion at the ACT1 locus of strains MRcan42 and MRcan43, resulting in strains can42MPG2A and -B and can43MPG2A and -B, respectively. A SacI-ApaI fragment from pMRR1R3 (18a) was used to insert the hyperactive MRR1P683S allele in place of one of the MRR1 wild-type alleles of strains MRcan42 and MRcan43 with the help of the SAT1 flipper cassette, generating strains can42MRR1R31A and -B and can43MRR1R31A and -B, respectively. The SAT1 flipper cassette was then recycled to obtain strains can42MRR1R32A and -B and can43MRR1R32A and -B. A second round of transformation and marker recycling resulted in strains can42MRR1R34A and -B and can43MRR1R34A and -B, in which both endogenous MRR1 alleles had been replaced by the hyperactive MRR1P683S allele. In an analogous fashion, a SacI-ApaI fragment from pCAP1R1 (18a) was used to substitute the wild-type CAP1 alleles of strains MRcan42 and MRcan43 for the hyperactive CAP1ΔC333 allele in two rounds of transformation and recycling of the SAT1 flipper cassette, generating strains can42CAP1R14A and -B and can43CAP1R14A and -B, respectively. Nourseothricin-resistant transformants were selected as described previously (15), and correct integration of all constructs was confirmed by Southern hybridization with the upstream and downstream flanking sequences. The introduction of the P683S mutation into the first and second MRR1 allele of the transformants was confirmed by reamplification and direct sequencing of the PCR products.
Isolation of genomic DNA and Southern hybridization.
Genomic DNA from C. albicans strains was isolated as described previously (15). The DNA was digested with appropriate restriction enzymes, separated on a 1% agarose gel, and, after ethidium bromide staining, transferred by vacuum blotting onto a nylon membrane and fixed by UV cross-linking. Southern hybridization with enhanced-chemiluminescence-labeled probes was performed with the Amersham ECL Direct Nucleic Acid Labeling and Detection System (GE Healthcare, Braunschweig, Germany) according to the instructions of the manufacturer.
Analysis of MDR1 promoter activity by FACS.
YPD overnight cultures of green fluorescent protein (GFP) reporter and parental control strains were each diluted 10−2 in six Erlenmeyer flasks containing 50 ml YPD medium (three without and three with 20 μg/ml doxycycline). After 3 h of growth at 30°C, 50 μg/ml benomyl or 0.005% H2O2 was added to two of the cultures (in both cases with and without doxycycline) to induce MDR1 expression; the third culture in each case was left untreated. After 60 min of further incubation, the mean fluorescence of the cells was determined by flow cytometry. Fluorescence-activated cell sorter (FACS) analysis was performed with a FACSCalibur cytometry system equipped with an argon laser emitting at 488 nm (Becton Dickinson, Heidelberg, Germany). Fluorescence was measured on the FL1 fluorescence channel equipped with a 530-nm band-pass filter. Twenty thousand cells were analyzed per sample. Fluorescence data were collected by using logarithmic amplifiers. The mean fluorescence (arbitrary values) was determined with CellQuest Pro (Becton Dickinson) software.
Analysis of doxycycline-induced MCM1 repression by Western immunoblotting.
Strains can42MPG2A and -B were grown as described above for determination of GFP expression. Whole-cell protein extracts were prepared from two cultures grown for 3 h in the presence or absence of doxycycline and from the remaining cultures after further incubation for 60 min in the presence of benomyl or H2O2 (with and without doxycycline). Cells were collected by centrifugation, washed twice in water, and broken by vortexing them for 10 min at 4°C with 300 μl 0.5-mm glass beads in 300 μl breaking buffer (100 mM Tris-Cl [pH 7.5], 200 mM NaCl, 20% glycerol, 5 mM EDTA, 4% Complete EDTA-free Protease Inhibitor Cocktail stock solution [Roche Diagnostics GmbH, Mannheim, Germany], 0.1% β-mercaptoethanol). Samples were centrifuged at 13,000 rpm for 5 min at 4°C, the supernatant was collected, and the protein concentration was quantified with a NanoDrop 1000 (Thermo Fisher Scientific, Wilmington, MA). Extracts were heated at 65°C for 10 min, and 400 μg total protein from each sample was separated on an SDS-12% polyacrylamide gel. Proteins were transferred onto a nitrocellulose membrane with a Trans-Blot SD Semi-Dry transfer apparatus (Bio-Rad, Munich, Germany). For detection of Mcm1-Myc, a monoclonal anti-c-Myc antibody (purified mouse immunoglobulin, clone 9E10, product number M 4439; Sigma-Aldrich Chemie GmbH) was used as the primary antibody at a dilution of 1:6,000, and goat anti-mouse IgG (Fab-specific) peroxidase conjugate (Sigma A 9917) was used as the second antibody at a dilution of 1:6,000. Blots were developed using a chemiluminescence detection system (GE Healthcare UK Limited, Chalfont, United Kingdom) under conditions recommended by the manufacturer.
Analysis of MDR1 expression by quantitative real-time reverse transcription (RT)-PCR.
YPD overnight cultures of strains SC5314, MRcan42, MRcan43, can42MRR1R34A and -B, can43MRR1R34A and -B, can42CAP1R14A and -B, and can43CAP1R14A and -B were diluted 10−2 in 50 ml fresh YPD medium with or without 20 μg/ml doxycycline and incubated for 4 h at 30°C. RNA was extracted by the hot-phenol method (2) combined with a purification step with an RNeasy kit (Qiagen, Hilden, Germany). Contaminating DNA was removed by treatment with the Ambion Turbo DNA-free kit (Applied Biosystems, Darmstadt, Germany), and cDNA was prepared from the RNA with the SuperScript III Reverse Transcriptase kit (Invitrogen, Karlsruhe, Germany). Generation of cDNA and absence of genomic DNA was controlled by PCR with the primers EFB1A (5′-ATTGAACGAATTCTTGGCTGAC-3′) and EFB1B (5′-CATCTTCTTCAACAGCAGCTTG-3′), which bind outside the EFB1 intron and yield a PCR product of 0.55 kb from cDNA and a 0.8-kb PCR product from genomic DNA. Quantitative PCR (qPCR) was performed with the iQ SYBR green Supermix (Bio-Rad) and the primer pairs MDR5RT (5′-ATTTGTTCAGATCAGTCATTGCTTCAGTGT-3′) and MDR6RT (5′-GGTCCGTTCAAGTAAAACAAAACTGGAATA-3′) for MDR1 and ACT1RT (5′-AGTGTGACATGGATGTTAGAAAAGAATTATACGG-3′) and ACT2RT (5′-ACAGAGTATTTTCTTTCTGGTGGAGCA-3′) for ACT1, which served as the reference, under the following conditions: initial denaturation for 3 min at 95°C; 40 cycles of 30 s at 95°C, 40 s at 50°C, and 10 s at 72°C; and 1 cycle of 30 s at 95°C and 30 s at 57°C. Melt curves were generated and threshold cycle (Ct) values were calculated by the Real Time program, Bio-Rad iQ5 V2.0 optical systems software. The Ct values obtained with the software were then used to calculate the relative MDR1 mRNA levels, adjusted to the ACT1 mRNA levels, using MDR1 expression in the wild-type strain SC5314 (set to 1) as a reference. Two independent RNA extractions, each with two technical replicates, were used to calculate means and standard deviations of the final relative expression values.
RESULTS
Mcm1 is required for benomyl-induced, but not H2O2-induced, MDR1 expression.
To monitor activation of the MDR1 promoter in response to inducing chemicals in the presence and absence of Mcm1, we introduced a PMDR1-GFP reporter fusion into the conditional mcm1 mutant MRcan42, which contains a single Myc-tagged MCM1 allele under the control of a tetracycline-repressible promoter. In addition, the reporter fusion was introduced into the control strain MRcan43, which contains both a tetracycline-repressible MCM1 copy and a Myc-tagged MCM1 allele with its own promoter. Two independent transformants of each parental strain were used for further analysis.
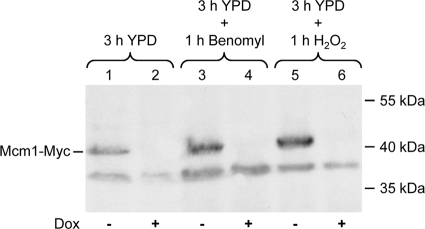
We first confirmed that Mcm1 could be efficiently depleted by addition of doxycycline to the conditional mutants carrying the reporter fusion and that the presence of the MDR1 inducers benomyl and H2O2 did not affect doxycycline-mediated repression. As can be seen in Fig. 1, Mcm1 was not detectably expressed after 3 h of growth in the presence of doxycycline, and the addition of benomyl or H2O2 did not alleviate this repression.
Fig. 1.
Depletion of Mcm1 from reporter strains by treatment with doxycycline. Strains can42MPG2A and -B were grown in the absence (−) or presence (+) of doxycycline (Dox) and treated with benomyl or H2O2 as described in Materials and Methods. Whole-cell protein extracts were prepared and analyzed by Western immunoblotting with an anti-Myc antibody. The position of Myc-tagged Mcm1 is indicated; the lower band is a nonspecific cross-reacting protein. The positions of molecular mass markers are given on the right of the blot. Shown are the results for strain can42MPG2A; the same results were obtained with can42MPG2B.
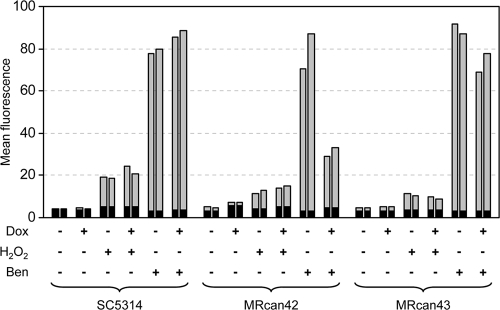
We then determined MDR1 promoter activity by quantifying the fluorescence of the reporter strains. As previously reported (14), MDR1 was not detectably expressed in the wild-type strain SC5314 in YPD medium but could be induced by H2O2 and, even more efficiently, by benomyl (Fig. 2). The presence of doxycycline did not affect the activity of the MDR1 promoter under noninducing or inducing conditions in the wild-type background. H2O2 also induced the MDR1 promoter in the conditional mutant MRcan42, albeit somewhat less efficiently than in strain SC5314, and this induction was also observed after depletion of Mcm1. In contrast, Mcm1 depletion resulted in reduced induction of the MDR1 promoter by benomyl, as the fluorescence of the conditional mutants was decreased in the presence of doxycycline. Doxycycline had little effect on benomyl-induced MDR1 expression in the control strain MRcan43. These results demonstrate that Mcm1 is dispensable for the induction of MDR1 expression by H2O2 but is required for full MDR1 induction by benomyl.
Fig. 2.
Activation of the MDR1 promoter by H2O2 and benomyl in the wild-type strain, SC5314; the conditional mcm1 mutant, MRcan42; and the control strain, MRcan43. Parental strains and transformants carrying a PMDR1-GFP reporter fusion were grown in the absence (−) or presence (+) of doxycycline (Dox) and treated with H2O2 or benomyl (Ben) as described in Materials and Methods. The mean fluorescence of the cells was determined by flow cytometry. The results obtained with two independently generated reporter strains (SCMPG2A and -B, can42MPG2A and -B, or can43MPG2A and -B) are shown in each case. The background fluorescence of the parental strains, which do not contain the GFP gene, is indicated by the black part of each bar.
Mcm1 is required for MDR1 overexpression by hyperactive Mrr1, but not by hyperactive Cap1.
The induction of the MDR1 promoter by H2O2 requires the bZip transcription factor Cap1, which is activated by oxidative stress (17, 18a, 26). Cap1 also contributes to, but is not essential for, benomyl-induced MDR1 expression (17, 18a, 26). In contrast, the zinc cluster transcription factor Mrr1 is indispensable for MDR1 expression in the presence of either of these inducers (14). As Mcm1 was necessary for full benomyl-induced, but not for H2O2-induced, MDR1 expression, we investigated whether Mcm1 was required for the constitutive MDR1 overexpression caused by hyperactive forms of Mrr1 and Cap1. To this end, we introduced the P683S gain-of-function mutation (14) into both resident MRR1 alleles of the conditional mcm1 mutant MRcan42 and the control strain MRcan43. In addition, both wild-type CAP1 alleles of these strains were replaced by the C-terminally truncated, hyperactive CAP1ΔC333 allele. Homozygous strains were generated, because the activating mutations in MRR1 and CAP1 have a stronger effect on MDR1 expression when they are present in both alleles (18a). Two independent series of mutants (A and B) were constructed in each case and used for further analysis.
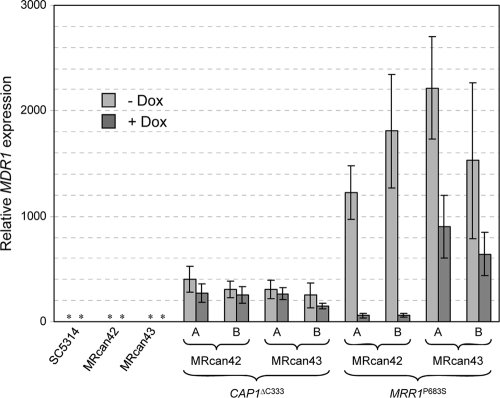
As the MDR1 promoter is constitutively activated in strains expressing hyperactive Mrr1 or Cap1, GFP was not useful as a reporter gene to measure the dependence of MDR1 expression on Mcm1, because considerable amounts of the relatively stable GFP would remain in the cells after MCM1 expression was shut off by the addition of doxycycline. We therefore measured MDR1 mRNA levels in the presence and absence of doxycycline by quantitative RT-PCR. As can be seen in Fig. 3, MDR1 was constitutively expressed at high levels in strains carrying the CAP1ΔC333 allele (between 250- and 400-fold higher than in the control strain SC5314), and Mcm1 depletion in the conditional mcm1 mutants by doxycycline did not significantly reduce MDR1 mRNA levels. MDR1 expression was even higher in strains containing the hyperactive MRR1P683S allele (between 1,200- and 2,200-fold higher than in the control strain, SC5314). Mcm1 depletion by doxycycline resulted in a drastic reduction of MDR1 mRNA levels in the conditional mutants (between 20- and 30-fold). A moderate reduction of MDR1 mRNA levels (ca. 2.5-fold) was also observed in the control strains, presumably because strain MRcan43 also strongly overexpresses MCM1 from the Tet promoter and exhibits only low MCM1 expression levels from the wild-type allele after the addition of doxycycline. These results indicate that a hyperactive Cap1 can mediate MDR1 overexpression independently of Mcm1, whereas a hyperactive Mrr1 requires Mcm1 to promote MDR1 expression.
Fig. 3.
Effect of Mcm1 depletion on MDR1 overexpression mediated by hyperactive forms of Cap1 and Mrr1. The wild-type strain, SC5314; the conditional mcm1 mutant, MRcan42; the control strain, MRcan43; and independent derivatives (A and B) of MRcan42 and MRcan43 that were rendered homozygous for the CAP1ΔC333 or the MRR1P683S allele were grown for 4 h in the absence (light-gray bars) or presence (dark-gray bars) of doxycycline as described in Materials and Methods. MDR1 mRNA levels were determined by real-time RT-PCR and are presented as relative expression levels compared to those of the reference strain, SC5314, in the absence of doxycycline, which were set to 1. The graph shows the means and standard deviations of two independent experiments, with duplicate measurements performed with each strain. *, MDR1 expression levels in SC5314 and the parental strains MRcan42 and MRcan43 were too low to be visible in the graph.
DISCUSSION
The transcription factors Mrr1 and Cap1 are both involved in the induction of MDR1 expression in response to inducing chemicals, and activating mutations in either of the two transcription factors result in constitutive overexpression of the efflux pump (1, 3, 14, 17, 26). However, little is known about how Mrr1 and Cap1 activate MDR1 transcription and which additional regulatory factors may be required. In this study, we have addressed the role of the MADS box transcription factor Mcm1, which has previously been implicated in the regulation of MDR1 expression and was recently shown to bind to the MDR1 promoter in vivo (11, 16, 17, 23).
Mcm1 can act as both a positive and a negative regulator of transcription (18, 23). We found that depletion of Mcm1 did not result in constitutive activity of the MDR1 promoter, indicating that the role of Mcm1 is not that of a repressor of MDR1 expression in the absence of inducing conditions (Fig. 2 and 3). Depletion of Mcm1 also did not affect the inducibility of the MDR1 promoter by H2O2 or the constitutive MDR1 overexpression caused by a hyperactive Cap1. On the other hand, benomyl-induced MDR1 expression was reduced, but not abolished, after depletion of Mcm1, and a hyperactive Mrr1 could not upregulate MDR1 in the absence of Mcm1. The MDR1 transcripts that were still present after Mcm1 depletion in strains expressing the hyperactive Mrr1 may represent mRNAs that were produced before Mcm1 depletion and not yet degraded. Alternatively, the cells might have contained residual amounts of Mcm1 that were not detectable by Western blot analysis (Fig. 1). These results suggest that the induction of MDR1 expression by H2O2 occurs mainly via activation of Cap1, which acts independently of Mcm1, and that induction by benomyl occurs partially via Mrr1, which depends on Mcm1 to activate the MDR1 promoter, and partially via Cap1. Like H2O2, benomyl causes oxidative stress and activates Yap1, the homolog of Cap1, in Saccharomyces cerevisiae (12). In response to oxidative stress, conserved cysteine residues in the C terminus of Yap1 form disulfide bonds, resulting in conformational changes that interrupt the interaction of Yap1 with the nuclear export protein Crm1, so that Yap1 can accumulate in the nucleus and activate its target genes (24). In C. albicans, Cap1 activity is also regulated by nuclear-cytoplasmic shuttling, and mutation of one of the conserved cysteines in Cap1 or removal of the C terminus results in constitutive Cap1 activity (1, 25).
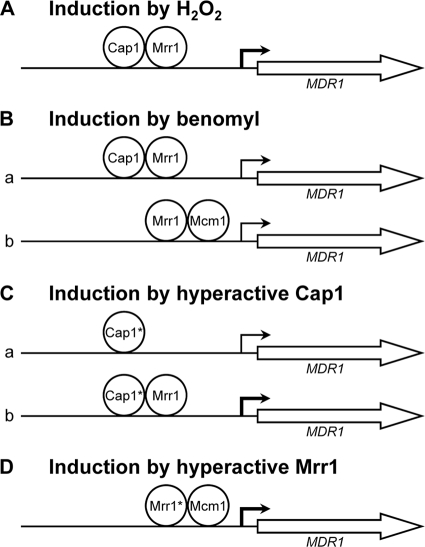
Based on the results of this and previous studies (1, 8, 11, 14, 16, 17, 18a, 23, 26), the model shown in Fig. 4 depicts the involvement of Mrr1, Cap1, and Mcm1 in four scenarios in which MDR1 expression is upregulated. In the presence of H2O2 (Fig. 4A), Cap1 is activated, accumulates in the nucleus, and induces MDR1 expression. Under these conditions, Cap1 requires Mrr1 to activate the MDR1 promoter, but Mcm1 is dispensable. In the presence of benomyl (Fig. 4B), Cap1 is also activated by oxidative stress and can induce MDR1 expression, together with Mrr1, independently of Mcm1 (a). However, MDR1 induction by benomyl can also occur at a reduced level in the absence of Cap1 in an Mrr1-dependent fashion, and this requires the presence of Mcm1 (b). How benomyl activates Mrr1 is not known. It is possible that Mrr1 is activated by direct drug binding, similar to the zinc cluster transcription factors Pdr1 and Pdr3, which regulate efflux pump expression in S. cerevisiae and Candida glabrata (22), but benomyl may also activate Mrr1 in an indirect fashion. The C-terminally truncated, hyperactive Cap1 can upregulate MDR1 expression, even in the absence of Mrr1, but not as efficiently as in a wild-type background (18a) (Fig. 4C). In this respect, the hyperactive Cap1 differs from activated wild-type Cap1, which requires Mrr1 to induce the MDR1 promoter in response to H2O2 or benomyl. Similar to oxidative stress-activated Cap1, however, the hyperactive Cap1 does not depend on Mcm1 to induce MDR1 expression. Finally, hyperactive Mrr1 can promote MDR1 overexpression independently of Cap1, but in this case, it requires Mcm1 (Fig. 4D). Altogether, these results indicate that Mrr1 can upregulate MDR1 expression in cooperation with either Cap1 or Mcm1. In response to oxidative stress, Mrr1 cooperates mainly with activated Cap1 to induce the MDR1 promoter, and Mcm1 is largely dispensable. In the absence of inducers, Cap1 remains in the cytoplasm, and hyperactive Mrr1 depends on the presence of Mcm1 to cause MDR1 overexpression. These observations are in accordance with previous findings that deletion of the putative Mcm1 binding sequence (the BRE/MDRE [see the introduction]) from a truncated MDR1 promoter abolished benomyl-induced or constitutive MDR1 overexpression (16, 17). However, deletion of the BRE/MDRE from the full-length MDR1 promoter did not affect its constitutive activity in strains that contained gain-of-function mutations in Mrr1 (3, 8, 16). The result of our present study showing that hyperactive Mrr1 requires Mcm1 to mediate MDR1 overexpression indicates that Mcm1 can contribute to MDR1 expression in a manner that is independent of its binding site in the BRE/MDRE. This may occur either by binding of Mcm1 to an additional region in the MDR1 promoter or by interacting with bound Mrr1 without itself binding to the DNA. Mcm1 has also been found to bind to the MRR1 promoter, indicating that Mcm1 can also affect MDR1 expression indirectly, by modulating the expression of MRR1 (23). In contrast, transcriptional-profiling studies showed that the hyperactive Mrr1 did not affect the expression of MCM1 or CAP1, and the activated Cap1 also did not affect MCM1 or MRR1 expression levels (14, 18a, 26).
Fig. 4.
Model of the roles of the transcription factors Cap1, Mrr1, and Mcm1 in MDR1 upregulation by inducing chemicals or gain-of-function mutations in Cap1 and Mrr1. The thinner bent arrows indicate reduced MDR1 promoter activity in the absence of the missing transcription factor. (A) H2O2 activates Cap1, resulting in accumulation of the transcription factor in the nucleus, where it can induce MDR1 expression, together with Mrr1, in an Mcm1-independent fashion. (B) Benomyl activates Mrr1 in an unknown way and also, at least partially, Cap1. When Cap1 is available at the MDR1 promoter, Mrr1 and Cap1 can induce MDR1 expression independently of Mcm1 (a); in the absence of Cap1, Mrr1 requires Mcm1 to induce MDR1 expression (b). (C) A hyperactive form of Cap1 (labeled Cap1*) can induce the MDR1 promoter in the absence of inducing stimuli and independently of Mcm1 and Mrr1 (a), but full induction requires the presence of Mrr1 (b). (D) A hyperactive Mrr1 containing a gain-of-function mutation (labeled Mrr1*) can induce the MDR1 promoter independently of Cap1 (which is localized in the cytoplasm in the absence of inducing stimuli) but requires the coregulator Mcm1.
An important goal for future research will be to unravel how Mrr1, Cap1, and Mcm1 activate MDR1 transcription. Depending on the conditions, Pdr1 and Pdr3 of S. cerevisiae interact with different subunits of the mediator complex to recruit RNA polymerase II to the promoters of their target genes (21, 22). Mrr1 may act in a similar fashion, but it has also recently been shown that both Mrr1 and Cap1 recruit Ada2, a subunit of the SAGA-ADA coactivator complex, to induce transcription (19). It is likely that, depending on the inducing conditions and the combination of transcription factors involved, C. albicans uses different mechanisms to upregulate expression of the MDR1 efflux pump.
ACKNOWLEDGMENTS
We thank Steffen Rupp for providing strains MRcan42 and MRcan43 and for helpful advice on the use of the strains.
This study was supported by the Deutsche Forschungsgemeinschaft (DFG grants MO 846/3 and SFB 630) and the National Institutes of Health (NIH grant AI058145). S.M. was supported by the University of Pisa.
Footnotes
Published ahead of print on 22 February 2011.
REFERENCES
- 1. Alarco A. M., Raymond M. 1999. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 181:700–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ausubel F., et al. 1989. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 3. Dunkel N., Blaß J., Rogers P. D., Morschhäuser J. 2008. Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol. Microbiol. 69:827–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dunkel N., et al. 2008. A gain-of-function mutation in the transcription factor Upc2p causes upregulation of ergosterol biosynthesis genes and increased fluconazole resistance in a clinical Candida albicans isolate. Eukaryot. Cell 7:1180–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gillum A. M., Tsay E. Y., Kirsch D. R. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179–182 [DOI] [PubMed] [Google Scholar]
- 6. Gupta V., et al. 1998. Identification of polymorphic mutant alleles of CaMDR1, a major facilitator of Candida albicans which confers multidrug resistance, and its in vitro transcriptional activation. Curr. Genet. 34:192–199 [DOI] [PubMed] [Google Scholar]
- 7. Harry J. B., et al. 2005. Drug-induced regulation of the MDR1 promoter in Candida albicans. Antimicrob. Agents Chemother. 49:2785–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hiller D., Stahl S., Morschhäuser J. 2006. Multiple cis-acting sequences mediate upregulation of the MDR1 efflux pump in a fluconazole-resistant clinical Candida albicans isolate. Antimicrob. Agents Chemother. 50:2300–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Karababa M., Coste A. T., Rognon B., Bille J., Sanglard D. 2004. Comparison of gene expression profiles of Candida albicans azole-resistant clinical isolates and laboratory strains exposed to drugs inducing multidrug transporters. Antimicrob. Agents Chemother. 48:3064–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Köhler G. A., White T. C., Agabian N. 1997. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J. Bacteriol. 179:2331–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lavoie H., Sellam A., Askew C., Nantel A., Whiteway M. 2008. A toolbox for epitope-tagging and genome-wide location analysis in Candida albicans. BMC Genomics 9:578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lucau-Danila A., et al. 2005. Early expression of yeast genes affected by chemical stress. Mol. Cell. Biol. 25:1860–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morschhäuser J. 2010. Regulation of multidrug resistance in pathogenic fungi. Fungal Genet. Biol. 47:94–106 [DOI] [PubMed] [Google Scholar]
- 14. Morschhäuser J., et al. 2007. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog. 3:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reuß O., Vik Å., Kolter R., Morschhäuser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127 [DOI] [PubMed] [Google Scholar]
- 16. Riggle P. J., Kumamoto C. A. 2006. Transcriptional regulation of MDR1, encoding a drug efflux determinant, in fluconazole-resistant Candida albicans strains through an Mcm1p binding site. Eukaryot. Cell 5:1957–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rognon B., Kozovska Z., Coste A. T., Pardini G., Sanglard D. 2006. Identification of promoter elements responsible for the regulation of MDR1 from Candida albicans, a major facilitator transporter involved in azole resistance. Microbiology 152:3701–3722 [DOI] [PubMed] [Google Scholar]
- 18. Rottmann M., Dieter S., Brunner H., Rupp S. 2003. A screen in Saccharomyces cerevisiae identified CaMCM1, an essential gene in Candida albicans crucial for morphogenesis. Mol. Microbiol. 47:943–959 [DOI] [PubMed] [Google Scholar]
- 18a. Schubert S., et al. 2011. Regulation of efflux pump expression and drug resistance by the transcription factors Mrr1, Upc2, and Cap1 in Candida albicans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sellam A., et al. 2009. Genome-wide mapping of the coactivator Ada2p yields insight into the functional roles of SAGA/ADA complex in Candida albicans. Mol. Biol. Cell 20:2389–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sellam A., Tebbji F., Nantel A. 2009. Role of Ndt80p in sterol metabolism regulation and azole resistance in Candida albicans. Eukaryot. Cell 8:1174–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shahi P., Gulshan K., Naar A. M., Moye-Rowley W. S. 2010. Differential roles of transcriptional mediator subunits in regulation of multidrug resistance gene expression in Saccharomyces cerevisiae. Mol. Biol. Cell 21:2469–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thakur J. K., et al. 2008. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452:604–609 [DOI] [PubMed] [Google Scholar]
- 23. Tuch B. B., Galgoczy D. J., Hernday A. D., Li H., Johnson A. D. 2008. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 6:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yan C., Lee L. H., Davis L. I. 1998. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 17:7416–7429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang X. T., de Micheli M., Coleman S. T., Sanglard D., Moye-Rowley W. S. 2000. Analysis of the oxidative stress regulation of the Candida albicans transcription factor, Cap1p. Mol. Microbiol. 36:618–629 [DOI] [PubMed] [Google Scholar]
- 26. Znaidi S., et al. 2009. Identification of the Candida albicans Cap1p regulon. Eukaryot. Cell 8:806–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Znaidi S., et al. 2008. Genomewide location analysis of Candida albicans Upc2p, a regulator of sterol metabolism and azole drug resistance. Eukaryot. Cell 7:836–847 [DOI] [PMC free article] [PubMed] [Google Scholar]