Abstract
The genetic structure and antibiotic nonsusceptibility of all serotype 19A Streptococcus pneumoniae pediatric pneumococcal isolates received at the Spanish Pneumococcal Reference Laboratory (1990 to 2008) were analyzed. Of them, 410 (79.8%) isolates belonged to 14 sequence types (STs) with >10 isolates each, and 104 to 73 STs (with 21 new STs, ST5141 to ST5161, with one isolate each). Time trends in 2000 to 2008 (n = 471) were explored by lineal regression. Serotype 19A increased from 5.7% in 2000 to 16.8% in 2008 (R2 = 0.872; P = 0.001). Decreasing trends (P < 0.03) were found for ST202 (R2 = 0.774) and ST81 (R2 = 0.559), and increasing trends (P < 0.03) for ST878 (R2 = 0.544) and ST320 (R2 = 0.530), both belonging to the clonal complex (CC) Denmark14-32 and first detected in 2003 and 2007, respectively, and ST2013 (R2 = 0.704) and ST4461 (R2 = 0.707), both appearing in 2004. Penicillin nonsusceptibility was clustered in ST81, ST276, ST320, ST878, ST2013, and ST4461 (>90% nonsusceptibility), and amoxicillin and cefotaxime nonsusceptibility in ST320: 87% amoxicillin (MIC50/MIC90 = 8/8 μg/ml) and 43.5% cefotaxime (MIC50/MIC90 = 1/2 μg/ml) nonsusceptibility. No trends were found for erythromycin nonsusceptibility (ranging from 38.5% to 66.7%) and cefotaxime nonsusceptibility (ranging from 0.0% to 7.8%), but increasing trends (P < 0.02) were found for oral penicillin (from 16.7% in 2000 to 56.3% in 2008; R2 = 0.628) and amoxicillin (from 0.0% before 2007 to 13.8% in 2008; R2 = 0.628) nonsusceptibility. This study warns about the emergence of serotype 19A STs associated with high-level antibiotic nonsusceptibility, with a role for ST320 and ST878 occupying the niche left by some pneumococcal 7-valent conjugate vaccine (PCV7)-related resistant STs. The rapid expansion of serotype 19A and STs related to antibiotic resistance indicates that vaccines covering serotype 19A present advantages in countering invasive disease.
INTRODUCTION
Serotype 19A Streptococcus pneumoniae has been classified as having low invasive potential (4, 19, 23) but appears equally capable of causing nasopharyngeal colonization, acute otitis media, and invasive disease (24, 26). Due to this capability, serotype 19A was one of the most prevalent serotypes, together with the group included in the pneumococcal 7-valent conjugate vaccine (PCV7) prior to the introduction of PCV7, among both colonizing strains (17) and invasive isolates (12, 22). There has been an increase over time in the prevalence of serotype 19A among invasive and noninvasive Streptococcus pneumoniae isolates, and although the beginning of this increase occurred in some countries prior to PCV7 introduction (6, 8, 12, 15), in others it occurred after (21, 28). In addition to the fact that serotype 19A could fill the ecological niche left by the reduction in the number of PCV7 types after vaccine introduction (12, 14, 25), other facts have been postulated for the multifactorial explanation of the increasing 19A prevalence: (i) the macrolide and penicillin nonsusceptibility prevalence within this serotype that makes it selectable by antibiotic use (12), (ii) the antibiotic pressure together with capsular switching from a resistant clone (5), (iii) the emergence (within serotype 19A) of a minor resistant clone existing prior to introduction of PCV7 (16), (iv) the appearance of new resistant clones (2), or (v) any of them alone or in combination among the streptococcal population in which secular changes in serotype frequencies occur (6, 12). For all these reasons it is worthy to explore the evolution over time of antibiotic nonsusceptibility and its relation to clonality, since it may have preventive and therapeutic consequences.
The aim of this study was to analyze the genetic structure (through multilocus sequence typing [MLST]) of all serotype 19A pediatric pneumococcal isolates received at the Spanish Reference Laboratory for Pneumococci (SRLP) from 1990 to 2008 and its relation to antibiotic nonsusceptibility, in order to shed light on the epidemiological changes of serotype 19A in Spain.
MATERIALS AND METHODS
All serotype 19A isolates among the 4,714 pediatric (<15 years of age) invasive isolates received at the SRLP from January 1990 to December 2008 were considered. Serotyping was performed by the Quellung reaction and/or dot blot assay (13). Additionally, all 19A isolates were tested for serotype 19A/19F using a previously described method of real-time PCR (27). MLST was carried out as previously described (10). Briefly, internal fragments of the aroE, gdh, gki, recP, spi, xpt, and ddl genes were amplified by PCR from chromosomal DNA using the primer pairs described by Enright and Spratt (10). The amplified fragments were directly sequenced in each direction using the same primers of the initial amplification. The sequences at each of the seven loci were compared with the sequences of all of the known alleles at those loci. Sequences identical to the sequence of a known allele were assigned the same allele number. An allelic profile varying in one of the seven housekeeping genes is referred to as a single-locus variant (SLV). The assignment of alleles at each locus was carried out using the software available at the pneumococcal MLST website (http://www.mlst.net). Phylogenetic analysis of sequence types (STs) was performed using the program eBURST, which uses a model of bacterial evolution to produce clusters of closely related genotypes that are all descended from the founding STs; these clusters are identified as clonal complexes (CCs). STs and CCs were displayed graphically based on relationships to each other via SLVs, with the distance between STs and CCs based on the number of SLVs between them. Allelic combinations not described in the MLST database were submitted and assigned new ST numbers.
MIC determination was performed by agar dilution using Mueller-Hinton agar (Difco Laboratories, Detroit, MI) supplemented with 5% sheep blood (Biomedics, Madrid, Spain) as the culture medium and incubation under a 5% CO2 atmosphere, as previously described (11). S. pneumoniae ATCC 6303 and S. pneumoniae ATCC 49619 plus five clinical strains were used as quality controls, as in all determinations in the SRLP (11). CLSI breakpoints (7) were used for interpretation (susceptibility/resistance [μg/ml]): penicillin orally, ≤0.06/≥2; penicillin parenterally, ≤2/≥8; amoxicillin, ≤ 2/≥8; cefotaxime (nonmeningitis), ≤1/≥4; and erythromycin, ≤0.25/≥1.
Trends along time were explored using the lineal regression command (SPSS V14; SPSS Inc., Chicago, IL), with prevalence percentages plotted as dependent variables and time (year) as an independent variable. A P value of ≤0.05 was considered significant.
RESULTS
A total of 542 (11.5%) serotype 19A isolates were identified among the 4,714 pediatric invasive isolates received in the SRLP from 1990 to 2008: 49 out of 1,054 (4.6%) in the period 1990 to 1999 and 493 out of 3,660 (13.5%) from 2000 to 2008. Of the 542 serotype 19A isolates, 514 (43 from 1990 to 1999 and 471 from 2000 to 2008) could be recovered and confirmed as serotype 19A by real-time PCR for subsequent MLST and antibiotic susceptibility determinations.
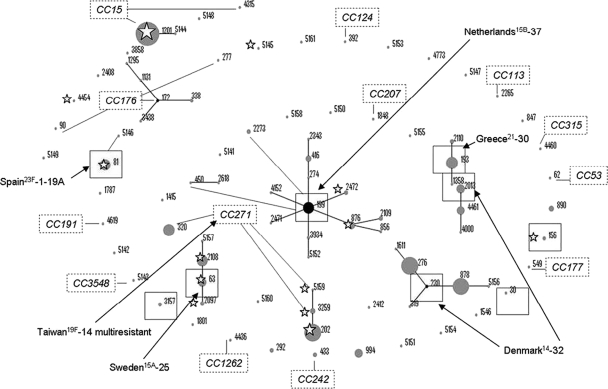
Of the 514 isolates, 30% had been isolated from children younger than 12 months, 35% from children aged 12 to 24 months, 24.7% from children aged 2 to 5 years, 3.5% from children older than 5 years, and 6.8% from children with ages not specified by the hospitals sending the isolates. A total of 80.0% of isolates were from blood samples, 10.5% from cerebrospinal fluid (CSF) samples, 8.0% from pleural fluid samples, and 1.6% from other types of samples (5 joint fluid, 2 peritoneal fluid, and 1 biopsy sample). Figure 1 shows the population snapshot of the 514 serotype 19A isolates from the period 1990 to 2008. A total of 410 (79.8%) isolates belonged to 14 STs with >10 isolates each, and 104 to 73 different STs (with 21 new assigned STs, numbered from ST5141 to ST5161, with one isolate each). The most frequent ST was ST1201 (18.9%) followed by ST202 and ST276 (9.9% each) and ST878 (8.9%). Table 1 shows the distribution by sample origin of STs with ≥20 isolates. While 72.3% of isolates from CSF and 68.1% of those from blood belonged to the nine STs shown in the table, 65.9% of isolates from pediatric parapneumonic empyema (PPE) belonged to two clonal complexes: CC271 (5 ST202, 5 ST320, and 1 ST1415) and CC230 (9 ST276 and 7 ST878) (Fig. 1). Interestingly, there were no isolates from PPE that belonged to ST81, ST193, and ST199 (all together accounting for 22.2% of isolates from CSF samples).
Fig. 1.
Population snapshot of the 514 serotype 19A isolates from the period 1990 to 2008. Clusters of related and unrelated STs are displayed as an eBURST diagram. STs are displayed graphically, with distances between STs based on number of SLVs between them. Dot sizes represent number of isolates. Founder STs are in boxes, primary founders (black dots) are positioned centrally in the cluster, and subgroup founders are shown as gray dots. Related outer primary founders of clonal complexes are labeled with open rectangles. STs found before the 2000s are marked with a star. Relationships with PMEN clones are indicated with arrows.
Table 1.
Distribution by sample origin of STs with ≥20 isolates in the study period (1990 to 2008)
| ST no. | No. of isolates (%)e |
|||
|---|---|---|---|---|
| CSF | PPE | Blood | Totala | |
| 81 | 6 (11.1) | 0 (0.0) | 14 (3.4) | 20 (3.9) |
| 193 | 5 (9.2) | 0 (0.0) | 19 (4.6) | 24 (4.7) |
| 199 | 1 (1.9) | 0 (0.0) | 19 (4.6) | 20 (3.9) |
| 202 | 2 (3.7) | 5 (12.2) | 44 (10.7) | 51 (9.9) |
| 276 | 7 (13.0) | 9 (22.0) | 35 (8.5) | 51 (9.9) |
| 320 | 1 (1.9) | 5 (12.2) | 15 (3.7) | 23b (4.5) |
| 878 | 7 (13.0) | 7 (17.1) | 31 (7.6) | 46c (8.9) |
| 1201 | 7 (13.0) | 4 (9.7) | 86 (20.9) | 97 (18.9) |
| 2108 | 3 (5.5) | 1 (2.4) | 17 (4.1) | 21 (4.1) |
| Other STs | 15 (27.7) | 10 (24.4) | 131 (31.9) | 161d (31.3) |
| Total | 54 (100) | 41 (100) | 411 (100) | 514a (100) |
Includes 8 isolates from other samples: 5 joint fluid, 2 peritoneal fluid, and 1 biopsy samples.
Includes 2 isolates from 2 joint fluid samples.
Includes 1 isolate from peritoneal fluid sample.
Includes 5 isolates from other samples: 3 joint fluid, 1 peritoneal fluid, and 1 biopsy sample.
CSF, cerebrospinal fluid; PPE, pediatric parapneumonic empyema.
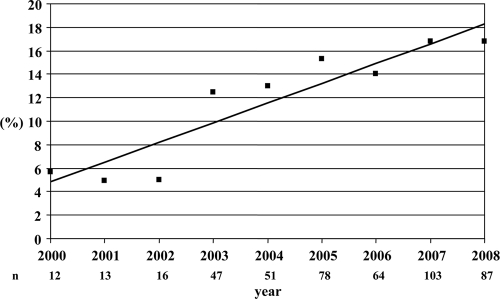
Since only 43 isolates were from the period 1990 to 1999 (18 ST202, 9 ST81, and 16 isolates belonging to other STs with one or two isolates each), with a scarce number of isolates per ST and year, trends of the different STs over time and susceptibility analysis were focused on isolates received from the year 2000 on. Figure 2 shows the yearly percentage of 19A isolates among all pediatric invasive isolates from the year 2000 on (n = 471). Serotype 19A increased from 5.7% in year 2000 to 16.8% in 2008 (R2 = 0.872; β = 1.682; P = 0.001).
Fig. 2.
Yearly percentages of 19A isolates among all pediatric invasive isolates from year 2000 on (n = 471).
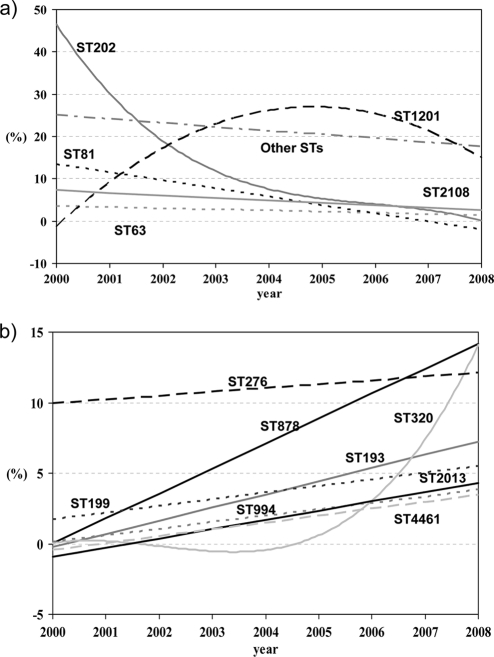
Considering only the 471 isolates from the period 2000 to 2008, Fig. 3a shows temporal trends for STs (individually those with >10 isolates) that were already present in the previous decade (1990 to 1999), and Fig. 3b trends for those STs without isolates in that period (1990 to 1999). As shown in Fig. 3a, a significant decreasing trend was found for ST202 (R2 = 0.774; β = −5.067; P = 0.002) and ST81 (R2 = 0.559; β = −1.950; P = 0.021). With respect to STs appearing in our study in the 2000s (not present in the period 1990 to 1999) (Fig. 3b), significant increasing trends were found for ST878 (R2 = 0.544; β = 1.767; P = 0.023), first detected in 2003, and ST320 (R2 = 0.530, β = 1.425, P = 0.026), first detected in 2007. Significant increasing trends were also found for ST2013 (R2 = 0.704; β = 0.657; P = 0.004) and ST4461 (R2 = 0.707; β = 0.492; P = 0.004), both STs appearing in 2004. Other STs included in Fig. 3b (ST276 appearing in 2000, ST199 appearing in 2002, and both ST193 and ST994 appearing in 2003) showed no significant increasing trends (near significance in the case of ST193; R2 = 0.428; β = 0.430; P = 0.056).
Fig. 3.
Temporal trends (2000 to 2008) for STs with >10 isolates. (a) STs already present in the period 1990 to 1999; (b) STs not present in the period 1990 to 1999.
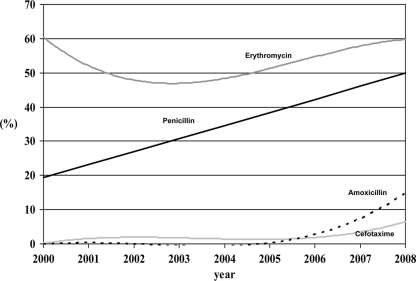
Figure 4 shows trends in the nonsusceptibility prevalence among all 19A isolates from the year 2000 on. Global nonsusceptibility rates of the 43 isolates received from 1990 to 1999 (with ≤8 isolates per year) were 32.6% for penicillin, 30.2% for erythromycin, and 0% for cefotaxime. No significant trends were found for erythromycin nonsusceptibility (ranging from 38.5% to 66.7%; R2 = 0.022; β = 0.548; P = 0.702) and cefotaxime nonsusceptibility (ranging from 0.0% to 7.8%; R2 = 0.196; β = 0.508; P = 0.232), but significant increasing trends were found for oral penicillin (from 16.7% in 2000 to 56.3% in 2008; R2 = 0.628; β = 3.797; P = 0.011) and amoxicillin nonsusceptibility (from 0.0% before 2007 to 13.8% in 2008; R2 = 0.628; β = 3.797; P = 0.011). Table 2 shows MIC50 and MIC90 values and percentages of nonsusceptibility to oral penicillin, amoxicillin, cefotaxime, and erythromycin for STs with ≥10 isolates from 2000 to 2008. Penicillin nonsusceptibility was clustered mainly in ST81, ST276, ST320, ST878, ST2013, and ST4461 (with >90% nonsusceptibility), while amoxicillin and cefotaxime nonsusceptibility was found almost exclusively in ST320: 87% nonsusceptibility to amoxicillin (MIC50/MIC90 values of 8/8 μg/ml) and 43.5% to cefotaxime (MIC50/MIC90 values of 1/2 μg/ml). By applying CLSI breakpoints for parenteral penicillin, only 27 out of the 471 (5.7%) isolates from 2000 on were nonsusceptible (MIC ≥ 2 μg/ml). Nonsusceptibility to parenteral penicillin was clustered in ST320 (17 out of the 27 isolates [63.0%]); most of the isolates from this ST320 were nonsusceptible (17 out of 23 [73.9%]). Only five STs showed ≥90% susceptibility to erythromycin: ST199, ST994, ST1201, ST2013, and ST4461.
Fig. 4.
Trends in nonsusceptibility prevalence among all 19A isolates from year 2000 on.
Table 2.
MIC50, MIC90, and percentage of nonsusceptibility to different compounds for STs with ≥10 isolates in the period 2000 to 2008a
| ST no. | Penicillin |
Amoxicillin |
Cefotaxime |
Erythromycin |
||||
|---|---|---|---|---|---|---|---|---|
| MIC50/MIC90 | % NS | MIC50/MIC90 | % NS | MIC50/MIC90 | % NS | MIC50/MIC90 | % NS | |
| 63 (n = 10) | 0.03/0.25 | 40.0 | ≤0.06/0.5 | 0.0 | 0.03/0.25 | 0.0 | ≥128/≥128 | 90.0 |
| 81 (n = 11) | 1/2 | 90.9 | 1/2 | 0.0 | 0.5/1 | 9.1 | ≥128/≥128 | 18.2 |
| 193 (n = 24) | ≤0.015/0.03 | 0.0 | ≤0.06/≤0.06 | 0.0 | ≤0.015/0.06 | 0.0 | ≥128/≥128 | 95.8 |
| 199 (n = 20) | ≤0.015/0.06 | 5.0 | ≤0.06/≤0.06 | 0.0 | ≤0.015/0.03 | 0.0 | ≤0.12/≤0.12 | 10.0 |
| 202 (n = 33) | ≤0.015/0.03 | 0.0 | ≤0.06/≤0.06 | 0.0 | ≤0.015/0.03 | 0.0 | ≥128/≥128 | 66.7 |
| 276 (n = 51) | 1/1 | 96.1 | 0.5/2 | 2.0 | 1/1 | 2.0 | ≥128/≥128 | 96.1 |
| 320 (n = 23) | 2/4 | 100 | 8/8 | 87.0 | 1/2 | 43.5 | ≥128/≥128 | 100 |
| 878 (n = 46) | 0.5/1 | 97.8 | 0.5/1 | 2.2 | 0.5/1 | 2.2 | ≥128/≥128 | 100 |
| 994 (n = 12) | ≤0.015/0.03 | 0.0 | ≤0.06/≤0.06 | 0.0 | ≤0.015/≤0.015 | 0.0 | ≤0.12/≤0.12 | 0.0 |
| 1201 (n = 95) | ≤0.015/≤0.015 | 0.0 | ≤0.06/≤0.06 | 0.0 | ≤0.015/≤0.015 | 0.0 | ≤0.12/≤0.12 | 5.3 |
| 2013 (n = 12) | 0.25/0.5 | 100 | 0.25/0.5 | 0.0 | 0.12/0.5 | 0.0 | ≤0.12/≤0.12 | 8.3 |
| 2108 (n = 20) | 0.03/0.25 | 40.0 | ≤0.06/0.25 | 0.0 | 0.03/0.25 | 0.0 | ≥128/≥128 | 95.0 |
| 4461 (n = 11) | 0.25/0.5 | 100 | 0.25/1 | 0.0 | 0.12/0.5 | 0.0 | ≤0.12/≤0.12 | 9.1 |
| Other (n = 103)b | 0.03/0.25 | 22.3 | ≤0.06/0.25 | 1.0 | 0.03/0.25 | 1.0 | 8/≥128 | 52.4 |
NS, nonsusceptibility.
Includes 73 different STs.
DISCUSSION
According to SRLP data, in Spain, serotype 19A represented 4.6% of all invasive isolates in the 1990s (1990 to 1999), with a significant increasing trend over the present decade, reaching 13.5% of all invasive isolates in the period 2000 to 2008. Furthermore, in parallel to the increase in the prevalence of serotype 19A among invasive isolates, penicillin nonsusceptibility (oral breakpoint of ≥0.12 μg/ml) among 19A invasive isolates also increased from 16.7% in year 2000 to 56.3% in 2008. This increase in penicillin nonsusceptibility was linked to significant increasing trends for ST320, ST878, ST2013, and ST4461 in the current decade, all of them belonging to clonal complexes related to antibiotic resistance. Therefore, the present study indicates an association between the increase in serotype 19A and penicillin nonsusceptibility in this serotype through the expansion of clonal complexes associated with this resistance trait. However, the expansion of serotype 19A may be multifactorial. Together with antibiotic resistance and antibiotic pressure (12), other factors have favored the expansion of serotype 19A, such as the lack of coverage of this serotype by PCV7 and the capability of this serotype of causing invasive disease, acute otitis media, and nasopharyngeal colonization (24, 26), consequently favoring capsular switching. All these factors have contributed to the genetic diversity of serotype 19A, providing significant survival advantages and making it one of the most prevalent serotypes among invasive isolates nowadays.
The most frequent ST found in our study was ST1201 (18.9%), a nonrelated Pneumococcal Molecular Epidemiology Network (PMEN) clone, with no significant trend from 2000 to 2008 and susceptibility to β-lactams and macrolides. The second most frequent ST was ST202 (9.9%), a double-locus variant of the Taiwan19F-14 PMEN clone, with a significant decreasing trend through all the study period and susceptibility to β-lactams but not to macrolides (66.7% nonsusceptibility to erythromycin).
All STs showing >90% penicillin nonsusceptibility (i.e., ST81, ST276, ST320, ST878, ST2013, and ST4461, exhibiting almost 100% nonsusceptibility) showed significant increasing trends in the period 2000 to 2008, except ST276 (96.1% nonsusceptibility; no significant trend) and ST81 (90.9% nonsusceptibility; significant decreasing trend). Among them, the most frequent ST was ST276 (9.9% of total 19A invasive isolates), closely followed by ST878 (8.9% of total), both single-locus variants of the clonal complex with ST230 as the founder and identifier of the Denmark14-32 clone. This clonal complex has also been reported as an increasing cause of pneumococcal infection in Portugal, which shares more than 1,000 km with Spain, and the spread of clones between countries may easily occur (1). ST276 had previously been reported in Spain (2, 18) and the United States (17), and in France the spread of isolates related to ST276 has been associated with the increase in pediatric invasive pneumococcal disease (IPD) by serotype 19A after PCV7 introduction (16). In Israel, ST276 has been associated with a 63.1% increase in the incidence of acute otitis media due to serotype 19A in Bedouin children, a population not vaccinated with PCV7 (8). Of note is closely related ST878, appearing in 2003 in our study. This ST was first detected in 2000 in Sri Lanka with a serotype 23F capsule, but isolates in Germany in 2004 already showed the serotype 19A capsule according to the MLST database (http://www.mlst.net).
Another ST associated with penicillin nonsusceptibility was ST81 (3.9% of total 19A invasive isolates but showing a significant decreasing trend over the current decade), the sadly famous Spain23F-1-19A clone of great concern in the 1980s. Considering the paradigm of capsular switching between serotypes 19A and 23F but also with the 19F, 14, and 9V serotypes, the significant decreasing trend of ST81 in the current study is in accordance with other studies (3, 9), showing a turndown in the prevalence of the Spain23F-1 (ST81) clone, associated with the dramatic decrease of serotype 23F in Spain (12).
Of great concern is the rapid spread of ST320, a double-locus variant of the Taiwan19F-14 PMEN clone showing fully nonsusceptibility to penicillin and erythromycin (with high MIC90 values) and high rates of nonsusceptibility to amoxicillin (87%) and cefotaxime (43.5%). Although ST320 had previously been reported in Spain (2, 18), it was first detected in 2007 in the present study, with a significant increase in the last few years. A South Korean study reported the presence of ST320 showing serotype 19A and 19F capsules in the early 1990s (6). The increase in serotype 19A in children was associated with the spread of ST320 before (1998 to 2003) and after (2004 to 2006) PCV7 introduction in South Korea (6). However, ST320 has been associated with the increase in multidrug-resistant serotype 19A invasive isolates isolated in the United States after PCV7 introduction (17, 21).
We also found in the present study the clonal cluster with ST199 as the ST founder belonging to the Netherlands15B-37 PMEN clone established worldwide, including ST199 (20 isolates), ST876 (8 isolates), ST416 (6 isolates), ST2109 (3 isolates), ST3934 (2 isolates), and ST2471, ST2472, ST4152, ST274, ST2343, and ST856 (one isolate each) in the current decade, most of them penicillin-susceptible isolates, in agreement with previous studies in Spain (18). In the United States, expansion of serotype 19A occurred through the clonal spread of ST199, which existed prior to universal vaccination (20, 25): ST199 (and its closely related variants) were predominant among 19A isolates from children aged <5 years prior to PCV7 and during 2003 and 2004, representing approximately 70% of invasive serotype isolates (20).
Globally, the analysis of the genetic structure of serotype 19A in the present study shows three related phenomena: (i) the low prevalence in the present decade of minor penicillin-nonsusceptible STs existing in the previous decade as ST81 (ST276 was detected in the year 2000, also prior to PCV7 introduction); (ii) the rapid spread of new penicillin-nonsusceptible STs as ST878 (first detected in 2003), ST4461 (in 2004), ST2013 (in 2006), and ST320 (in 2007); and (iii) the capsular switching in an ST81 clone from a vaccine serotype (23F) before PCV7 introduction to a nonvaccine serotype (19A) in the current decade.
A close look at the MLST database suggests that capsular switching is more frequent than expected, since many STs are shared by several serotypes. In this sense, eight STs (ST81, ST172, ST199, ST276, ST320, ST663, ST2012, and ST2013) express serotypes 19A or 19F, three STs (ST63, ST199, and ST2074) express serotype 19A or serogroup 15, ST93 expresses serotype 21 or 19A, and ST695 expresses serotype 19A or 4. Interestingly, capsular switching and acquisition of antibiotic resistance may occur through a single transformation event, as suggested by the proximity of the 19A-type capsular locus and the flanking pbp1a and pbp2x sequences (17, 29), linking the ability of pneumococci to exchange DNA to the success of some serotypes in colonizing and sharing the same ecologic niche.
In conclusion, our study warns about the emergence of serotype 19A STs associated with high levels of antibiotic nonsusceptibility and multidrug resistance. The outstanding role of ST320 and ST878 in occupying the niche left by some PCV7-related resistant STs should be highlighted. Because of the previously shown increasing trend of IPD due to serotype 19A (12) and the rapid expansion of STs related to antibiotic resistance shown in this study, the new conjugate vaccine (PCV13) covering serotype 19A represents an advantage in countering invasive disease, from both the serotype and resistance epidemiology perspectives. Since other serotypes with similar characteristics and disease potential may be the next in line to expand, surveillances and molecular epidemiology remain important after introduction of pneumococcal conjugate vaccines with broader coverage.
ACKNOWLEDGMENTS
This work was supported in part by an unrestricted grant from Pfizer S.A., Madrid, Spain, PRISM-AG S.L.L., and by Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación, (grant FIS PI 08539).
We are in debt to Cristina Mendez for her critical review of the manuscript.
Footnotes
Published ahead of print on 22 February 2011.
REFERENCES
- 1. Aguiar S. I., et al. 2010. Denmark14-230 clone as an increasing cause of pneumococcal infection in Portugal within a background of diverse serotype 19A lineages. J. Clin. Microbiol. 48:101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ardanuy C., et al. 2009. Emergence of a multidrug-resistant clone (ST320) among invasive serotype 19A pneumococci in Spain. J. Antimicrob. Chemother. 64:507–510 [DOI] [PubMed] [Google Scholar]
- 3. Ardanuy C., et al. 2009. Epidemiology of invasive pneumococcal disease among adult patients in Barcelona before and after pediatric 7-valent pneumococcal conjugate vaccine introduction, 1997–2007. Clin. Infect. Dis. 48:57–64 [DOI] [PubMed] [Google Scholar]
- 4. Brueggemann A. B., et al. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis. 187:1424–1432 [DOI] [PubMed] [Google Scholar]
- 5. Brueggemann A. B., Pai R., Crook D. W., Beall B. 2007. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog. 3:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choi E. H., et al. 2008. Streptococcus pneumoniae serotype 19A in children, South Korea. Emerg. Infect. Dis. 14:275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing; 19th informational supplement. M100-S19 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8. Dagan R., Givon-Lavi N., Leibovitz E., Greenberg D., Porat N. 2009. Introduction and proliferation of multidrug-resistant Streptococcus pneumoniae serotype 19A clones that cause acute otitis media in an unvaccinated population. J. Infect. Dis. 199:776–785 [DOI] [PubMed] [Google Scholar]
- 9. Davies T. A., Yee Y. C., Goldschmidt R., Sahm D. F., Evangelista A. T. 2008. Decline in the prevalence of pandemic clones Spain23F-1 and Spain9V-3 among US fluoroquinolone-resistant Streptococcus pneumoniae TRUST Surveillance isolates since 2001. Postgrad. Med. 120(3 Suppl. 1):39–45 [DOI] [PubMed] [Google Scholar]
- 10. Enright M. C., Spratt B. G. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049–3060 [DOI] [PubMed] [Google Scholar]
- 11. Fenoll A., et al. 2007. Influence of the beta-lactam resistance phenotype on the cefuroxime versus cefditoren susceptibility of Streptococcus pneumoniae and Haemophilus influenzae recovered from children with acute otitis media. J. Antimicrob. Chemother. 60:323–327 [DOI] [PubMed] [Google Scholar]
- 12. Fenoll A., et al. 2009. Temporal trends of invasive Streptococcus pneumoniae serotypes and antimicrobial resistance patterns in Spain from 1979 to 2007. J. Clin. Microbiol. 47:1012–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fenoll A., Jado I., Vicioso D., Casal J. 1997. Dot blot assay for the serotyping of pneumococci. J. Clin. Microbiol. 35:764–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hanage W. P., et al. 2007. Diversity and antibiotic resistance among nonvaccine serotypes of Streptococcus pneumoniae carriage isolates in the post-heptavalent conjugate vaccine era. J. Infect. Dis. 195:347–352 [DOI] [PubMed] [Google Scholar]
- 15. Harboe Z. B., et al. 2010. Temporal trends in invasive pneumococcal disease and pneumococcal serotypes over 7 decades. Clin. Infect. Dis. 50:329–337 [DOI] [PubMed] [Google Scholar]
- 16. Mahjoub-Messai F., et al. 2009. Population snapshot of Streptococcus pneumoniae serotype 19A isolates before and after introduction of seven-valent pneumococcal vaccination for French children. J. Clin. Microbiol. 47:837–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moore M. R., et al. 2008. Population snapshot of emergent Streptococcus pneumoniae serotype 19A in the United States, 2005. J. Infect. Dis. 197:1016–1027 [DOI] [PubMed] [Google Scholar]
- 18. Muñoz-Almagro C., et al. 2009. Emergence of invasive pneumococcal disease caused by multidrug-resistant serotype 19A among children in Barcelona. J. Infect. 59:75–82 [DOI] [PubMed] [Google Scholar]
- 19. Obando I., et al. 2008. Pediatric parapneumonic empyema, Spain. Emerg. Infect. Dis. 14:1390–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pai R., Moore M. R., Pilishvili T., Gertz R. E., Whitney C. G., Beall B. 2005. Postvaccine genetic structure of Streptococcus pneumoniae serotype 19A from children in the United States. J. Infect. Dis. 192:1988–1995 [DOI] [PubMed] [Google Scholar]
- 21. Pelton S. I., et al. 2007. Emergence of 19A as virulent and multidrug resistant pneumococcus in Massachusetts following universal immunization of infants with pneumococcal conjugate vaccine. Pediatr. Infect. Dis. J. 26:468–472 [DOI] [PubMed] [Google Scholar]
- 22. Robinson K. A., et al. 2001. Epidemiology of invasive Streptococcus pneumoniae infections in the United States, 1995–1998: opportunities for prevention in the conjugate vaccine era. JAMA 285:1729–1735 [DOI] [PubMed] [Google Scholar]
- 23. Sandgren A., et al. 2004. Effect of clonal and serotype-specific properties on the invasive capacity of Streptococcus pneumoniae. J. Infect. Dis. 189:785–796 [DOI] [PubMed] [Google Scholar]
- 24. Shouval D. S., Greenberg D., Givon-Lavi N., Porat N., Dagan R. 2006. Site-specific disease potential of individual Streptococcus pneumoniae serotypes in pediatric invasive disease, acute otitis media and acute conjunctivitis. Pediatr. Infect. Dis. J. 25:602–607 [DOI] [PubMed] [Google Scholar]
- 25. Singleton R. J., et al. 2007. Invasive pneumococcal disease caused by nonvaccine serotypes among Alaska native children with high levels of 7-valent pneumococcal conjugate vaccine coverage. JAMA 297:1784–1792 [DOI] [PubMed] [Google Scholar]
- 26. Sleeman K. L., et al. 2006. Capsular serotype-specific attack rates and duration of carriage of Streptococcus pneumoniae in a population of children. J. Infect. Dis. 194:682–688 [DOI] [PubMed] [Google Scholar]
- 27. Tarragó D., et al. 2008. Identification of pneumococcal serotypes from culture-negative clinical specimens by novel real-time PCR. Clin. Microbiol. Infect. 14:828–834 [DOI] [PubMed] [Google Scholar]
- 28. Techasaensiri C., et al. 2010. Epidemiology and evolution of invasive pneumococcal disease caused by multidrug resistant serotypes of 19A in the 8 years after implementation of pneumococcal conjugate vaccine immunization in Dallas, Texas. Pediatr. Infect. Dis. J. 29:294–300 [DOI] [PubMed] [Google Scholar]
- 29. Trzciński K., Thompson C. M., Lipsitch M. 2004. Single-step capsular transformation and acquisition of penicillin resistance in Streptococcus pneumoniae. J. Bacteriol. 186:3447–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]