Abstract
Human African trypanosomiasis continues to be an important public health threat in extensive regions of sub-Saharan Africa. Treatment options for infected patients are unsatisfactory due to toxicity, difficult administration regimes, and poor efficacy of available drugs. The aminoacyl-tRNA synthetases were selected as attractive drug targets due to their essential roles in protein synthesis and cell survival. Comparative sequence analysis disclosed differences between the trypanosome and mammalian methionyl-tRNA synthetases (MetRSs) that suggested opportunities for selective inhibition using drug-like molecules. Experiments using RNA interference on the single MetRS of Trypanosoma brucei demonstrated that this gene product was essential for normal cell growth. Small molecules (diaryl diamines) similar to those shown to have potent activity on prokaryotic MetRS enzymes were synthesized and observed to have inhibitory activity on the T. brucei MetRS (50% inhibitory concentration, <50 nM) and on bloodstream forms of T. brucei cultures (50% effective concentration, as low as 4 nM). Twenty-one compounds had a close correlation between enzyme binding/inhibition and T. brucei growth inhibition, indicating that they were likely to be acting on the intended target. The compounds had minimal effects on mammalian cell growth at 20 μM, demonstrating a wide therapeutic index. The most potent compound was tested in the murine model of trypanosomiasis and demonstrated profound parasite suppression and delayed mortality. A homology model of the T. brucei MetRS based on other MetRS structures was used to model binding of the lead diaryl diamine compounds. Future studies will focus on improving the pharmacological properties of the MetRS inhibitors.
INTRODUCTION
Drugs that meet modern standards for chemotherapeutics have, unfortunately, not yet been developed for human African trypanosomiasis (HAT). The currently used drugs for HAT include suramin (an injection drug introduced in 1922), melarsoprol (a highly toxic arsenical in use since 1949), pentamidine (introduced in the 1940s and given by painful intramuscular injections), eflornithine (introduced in the 1980s and given intravenously every 6 h in multigram quantities), and nifurtimox (a drug originally developed for Chagas' disease in the 1970s and now used in combination with eflornithine for HAT). With 60 million people in Africa at risk for HAT, the need for effective, safe, and affordable drugs remains as acute as ever. The completion of the Trypanosoma brucei genome sequencing project combined with advances in modern drug discovery techniques creates an unprecedented opportunity to develop overdue, modern drugs for this lethal disease.
Analysis of the sequenced T. brucei genome reveals hundreds of genes encoding enzymes, with some of the most interesting from a drug development standpoint being aminoacyl-tRNA synthetases (aaRSs). These enzymes are essential to the core biological process of translating gene nucleotide sequences into proteins. With a few notable exceptions (27), each aaRS must specifically recognize a single amino acid and attach it to a corresponding tRNA whose anticodon matches one or more of the codons for that amino acid. The sequence of reactions catalyzed consists of four essential steps: (i) recognition of the correct amino acid, (ii) reaction with ATP to form an aminoacyl-adenylate intermediate, (iii) recognition of a cognate tRNA, and (iv) transfer of the aminoacyl group to the terminal adenosine of the tRNA. Interference with any of these steps inhibits the formation of properly charged tRNA, leading to accumulation of uncharged tRNA on the ribosome. This results in disruption of normal protein chain elongation during translation. Not surprisingly, aaRS enzymes have been shown to be essential in genetic knockout or knockdown studies in a variety of organisms (3, 4, 10, 11).
T. brucei encodes 23 aaRS enzymes, one per amino acid, with the exception of 2 enzymes for aspartate (4), 2 for lysine (8), and 2 for tryptophan (3). The methionyl-RS (MetRS) of T. brucei has particularly attracted our interest because of significant differences with the mammalian orthologs (discussed herein) and because of the existence of compounds under development in Pharma targeting the homologous enzyme in bacteria. MetRS enzymes in general are divided into two major forms on the basis of sequence similarity and sensitivity to inhibitors (3, 12). MetRS1 is the form commonly found in Gram-positive bacteria, including Staphylococcus aureus and Streptococcus pyogenes. MetRS2 is found in archaea, eukaryotes, and many Gram-negative bacteria. Eukaryotic organisms contain both MetRS forms, with MetRS1 being the mitochondrial form and MetRS2 being the cytoplasmic form. In mammals, the mitochondrial aaRS enzymes are encoded in the nucleus and imported to the mitochondrion via import signals. The single MetRS of T. brucei is encoded in the nucleus and presumably functions in both the cytoplasm and the mitochondrion (26). On the basis of the sequence, the T. brucei MetRS enzyme groups with the MetRS1 form; thus, it is very distinct from the mammalian cytoplasmic MetRS enzyme. Differences with the human mitochondrial MetRS are discussed in this paper.
Synthetic compounds targeting bacterial MetRS enzymes are now in advanced preclinical development for treating bacterial infections caused by methicillin-resistant Staphylococcus aureus and Clostridium difficile (5, 6). These originated as diaryl diamine compounds under development by GlaxoSmithKline (10, 15, 16) and subsequently Replidyne (6, 13). We have synthesized compounds relating to the diaryl diamines and tested them for binding of the recombinant T. brucei MetRS, on T. brucei cell cultures, and in the murine model of T. brucei infection. This report also includes the results of molecular modeling studies of these compounds as well as RNA interference (RNAi) studies establishing the essentiality of the MetRS enzyme in T. brucei.
MATERIALS AND METHODS
Synthesis of diaryl diamines.
Synthesis of inhibitors followed the literature-reported route. Briefly, aniline was condensed with malonic acid in phosphorus oxychloride to give 2,4-dichloroquinoline (24). Subsequently, reaction with 4-methoxybenzyl alcohol activated with NaH was performed in dry tetrahydrofuran in the presence of 15-crown-5 to produce 2-chloro-4-(4-methoxybenzyloxy)-quinoline (16), which was reacted with excess 1,3-diaminopropane to give N1-[4-(4-methoxy-benzyloxy)-quinolin-2-yl]-propane-1,3-diamine (15). Deprotection was performed with 2:8 trifluoroacetic acid-dichloromethane, and condensation with a variety of substituted or unsubstituted benzaldehydes under reductive amination conditions (2.5% acetic acid in methanol and NaCNBH3) gave the final compounds, after purification (15, 16). The newly synthesized compounds gave satisfactory analytical data (details are provided with the supplemental material).
RNA interference.
The T. brucei MetRS homolog (Tb927.10.1500) was the target for the RNAi knockdown experiment. The region between bases 994 and 1615 of the open reading frame was amplified (primers 1 and 2 in Table S1 in the supplemental material) and cloned into 2T7TABlue (a gift of D. Horn, London School of Hygiene and Tropical Medicine) (1). Transfections and RNAi experiments were performed as previously described (23).
Northern analysis.
RNA was isolated from induced and noninduced cultures after 72 h of growth using a Qiagen RNEasy kit (no. 74104). Total RNA (11 μg per lane) was separated on a formaldehyde gel and blotted using standard procedures. The RNA membrane was then analyzed with a 32P-labeled DNA probe made to a region of the MetRS not overlapping with the portion used for RNA interference. The probe fragment was amplified with primers 3 and 4 (see Table S1 in the supplemental material), which correspond to bases 30 to 657 of the TbMetRS open reading frame. The blot was stripped and reprobed with DNA of the T. brucei β-tubulin gene for standardization (see primers 5 and 6 in Table S1 in the supplemental material).
PCR and ligase-independent cloning.
The gene coding for T. brucei methionyl-tRNA synthetase (Tb927.10.1500) was amplified from T. brucei brucei strain 427 genomic DNA using the primers 7 and 8 (see Table S1 in the supplemental material). The gene coding for amino acids 219 to 765 of the T. cruzi methionyl-tRNA synthetase (Tc00.1047053509247.50) was amplified from T. cruzi TCCL Brener genomic DNA with primers 9 and 10 (see Table S1 in the supplemental material). The PCR products were gel purified with a Qiagen kit and cloned into the AVA0421 vector by ligation-independent cloning as described previously (21). The AVA0421 vector is a derivative of the pET14b vector containing an N-terminal 6-histidine tag followed by a 3C protease cleavage site.
Production of recombinant T. brucei and T. cruzi MetRSs.
The expression of recombinant proteins was performed as previously described (21). The N-terminal 6-His fusion proteins were purified by nickel affinity chromatography followed by size exclusion gel chromatography (Superdex 75 26/60; GE Biosciences, Piscataway, NJ).
Thermal shift assay.
The thermal shift assay was performed as previously described (7), with the modifications that samples contained the MetRS enzyme (0.42 mg/ml for the T. brucei enzyme, 0.32 mg/ml for the T. cruzi enzyme), 50 μM inhibitor, 10 mM ATP magnesium salt, 10 mM l-methionine, and 5% dimethyl sulfoxide (DMSO). The inclusion of 10 mM ATP and methionine in the assay buffer was to suppress the overall thermal shift signal so that only tight-binding inhibitors could cause a significant shift in the melting temperature (Tm) of MetRS. The assays were performed twice independently in triplicate in each experiment.
Aminoacylation assays.
Enzyme activity was quantified by the attachment of [3H]methionine to tRNA in the presence of MetRS enzymes. Reactions were performed in 96-well filter plates with Durapore membranes (MSHVN4B10; Millipore) in volumes of 75 μl. The reaction was performed with 25 mM HEPES, pH 7.9, 10 mM MgCl2, 50 mM KCl, 0.2 mM spermine, 0.1 mg/ml bovine serum albumin, 2.5 mM dithiothreitol, and 10 U/ml pyrophosphatase (R4251; Sigma). Recombinant enzyme (4 nM) and compound inhibitors (50 nM) were mixed with the buffer and preincubated for 15 min. To start the reaction, 400 μg/ml bulk Escherichia coli tRNA (R4251; Sigma), 0.2 mM ATP, and 250 nM [3H]methionine (80 Ci/mmol) were added. The plate was incubated without shaking at room temperature for 120 min. The reactions were stopped by the addition of 100 μl cold 10% trichloroacetic acid. The reaction components were separated from tRNA by filtration through a vacuum manifold and washed three times with cold 10% trichloroacetic acid. The filter plates were dried overnight, scintillation fluid was added, and the counts on the plates were determined in a scintillation plate counter. Samples were run in quadruplicate, and the average activity of inhibitors was compared to that in control wells without inhibitors.
Growth inhibition assays of T. brucei, T. cruzi, and mammalian cell cultures.
T. brucei (bloodstream form strain 427 from K. Stuart, Seattle Biomedical Research Institute, Seattle, WA) was cultured in HMI-9 medium containing 10% fetal bovine serum, penicillin, and streptomycin at 37°C with 5% CO2 (14). Drug sensitivity of the T. brucei strain was determined in 96-well microtiter plates in triplicate with an initial inoculum of 1 × 104 trypomastigotes per well. Compound stock solutions were prepared in DMSO at 20 mM and added in serial dilutions for a final volume of 200 μl/well. Parasite growth was quantified at 48 h by the addition of AlamarBlue (Alamar Biosciences, Sacramento, CA) (25). Pentamidine isethionate (Aventis, Dagenham, United Kingdom) was included in each assay as a positive control. Standard errors within assays were consistently less than 15%.
Compounds were screened against the Tulahuen strain of T. cruzi expressing β-galactosidase in 96-well tissue culture plates as described previously (2). In this assay, T. cruzi proliferates as intracellular amastigotes within murine 3T3 fibroblasts. Compounds were screened in triplicate to determine 50% effective concentrations (EC50s). Standard errors within assays were consistently less than 15%.
The human lymphocytic cell line CRL-1855 (American Type Culture Collection) was used for the cytotoxicity assessment. The cells were grown in RPMI medium with 10% fetal bovine serum, penicillin, and streptomycin at 37°C with 5% CO2. Cells (5 × 103/well) were added to 96-well plates and incubated with serial dilutions of compounds for 48 h. At that time, cell viability was quantified by addition of AlamarBlue, and plates were incubated for an additional 4 h at 5% CO2, 37°C. Absorbance readings obtained at an optical density at 570 to 600 nm were used to calculate viability referenced against cells grown with no inhibitors.
Washout experiments on T. brucei cultures.
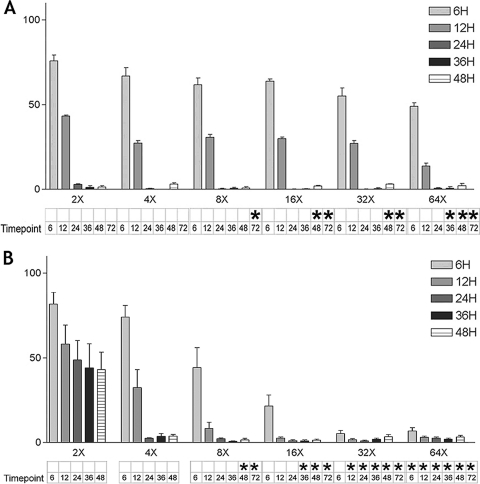
T. brucei cells (strain and culture conditions as described above) were grown in 1.5-ml volumes in 12-well plates in the presence of compound 2 or pentamidine. The concentrations of the compounds are represented as factors of their EC50s determined in a 48-h incubation. At defined times (6, 12, 24, 36, 48 h), the cells were removed, centrifuged, washed, and resuspended in medium without the compounds. The cultures were allowed to grow until the 48-h time point from the first exposure to the compounds, at which time the viability of the cultures was assessed using AlamarBlue as described above. The viability of cells was expressed as a percentage of the viability of the cells grown without the compounds for 48 h. In order to determine the time needed to kill all the parasites, aliquots of the same cultures (plus an additional aliquot taken from cells grown with the compounds for 72 h) were incubated in medium without the compounds for 10 days. The cultures were inspected by light microscopy daily for outgrowth of living parasites. This gave sufficient time for outgrowth of even a single surviving parasite, based on the findings of previous experiments. The conditions in which no parasite outgrowth was observed are marked (see Fig. 3).
Fig. 3.
Washout experiments of T. brucei with compound 2 and pentamidine. The bar graphs indicate the percent growth of the cultures compared to the level of growth of cultures grown without compound. The cultures were incubated with compound 2 (A) or with pentamidine (B) at concentrations 2 to 64 times the EC50 (established in 48-h experiments) and for various periods of exposure from 6 to 48 h. Below the bar graph, the cultures are marked with an asterisk if no cell growth was observed at 10 days following the washout from the compound.
Pharmacokinetic and efficacy studies in mice.
For pharmacokinetic studies, compound 1 was dissolved in 10% DMSO and injected by the intraperitoneal (i.p.) route at 50 mg/kg of body weight into BALB/c mice (7- to 8-week-old females, n = 3). At timed intervals, 20 μl of tail blood was collected in heparinized capillary tubes. Plasma was separated and frozen for later analysis by liquid chromatography-mass spectrometry (28). For efficacy studies, compound 1 was loaded into Alzet minipumps (model 1003D) and implanted subcutaneously into BALB/c mice (7- to 8-week-old females, n = 5 per group) for delivery at 25 mg/kg/day for 3 days. The control group was given the minipumps loaded with vehicle. Mice were infected with T. brucei strain 427 (1 × 104 i.p.) 1 day prior to implantation of the minipumps. Parasitemia was quantified on whole blood from tail bleeding using a hemacytometer. Blood levels of compound 1 were obtained from mice after the minipumps had been in place for 24 h. Concentrations of compound 1 were analyzed in plasma as described above.
Homology modeling and docking studies.
Homology modeling was done using the MODELLER software program (19). Docking studies utilized the FLO/QXP docking program and a Metropolis Monte Carlo scheme (20). Additional details of building the homology model are provided in the supplemental material.
RESULTS
Target validation by RNA interference.
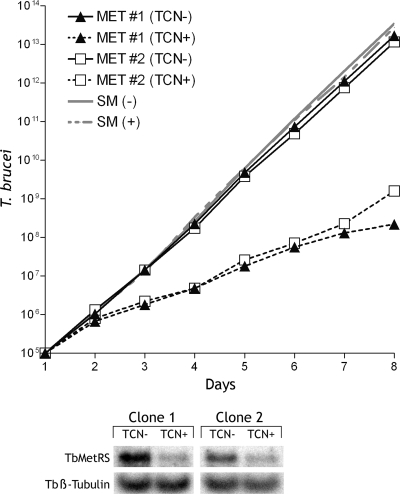
T. brucei is amenable to RNA interference analysis to establish the essentiality of gene targets for cell viability. Gene knockdown of the MetRS (Tb927.10.1500 gene) resulted in a dramatic growth inhibition phenotype; by day 8, growth was inhibited by a factor of ∼106 (Fig. 1). Total RNA was collected at 72 h, and Northern blots demonstrated that the MetRS mRNA was knocked down by between 57 and 77% compared to the level for control cells. Thus, even without complete knockdown of the MetRS transcript, cell growth was profoundly inhibited.
Fig. 1.
Growth of T. brucei undergoing RNAi against MetRS. (Top) A region of ∼500 nucleotides in the 3′ end of the gene was targeted using the p2T7 expression vector (1). RNAi was induced (+) by addition of tetracycline to the medium or left uninduced (−). The growth rate of two separate clones (MET 1 and MET 2) was tested and compared to that of the single-marker (SM) control, which does not contain the RNAi construct. Note that the y axis is log scale. (Bottom) Northern blots show the reduction in mRNA levels for the T. brucei MetRS (TbMetRS) in the two clones either induced with tetracycline (TCN+) or not induced with tetracycline (TCN−) for 72 h. Approximately equivalent loading of the samples is demonstrated by Northern blots for the constitutively expressed β-tubulin (Tbβ-Tubulin) mRNA. Densitometry of the T. brucei MetRS mRNA (normalized for β-tubulin) showed 77% and 57% signal knockdowns for clone 1 and clone 2, respectively.
Binding of compounds to T. brucei MetRS, inhibition of MetRS enzyme, and inhibition of trypanosome cell cultures.
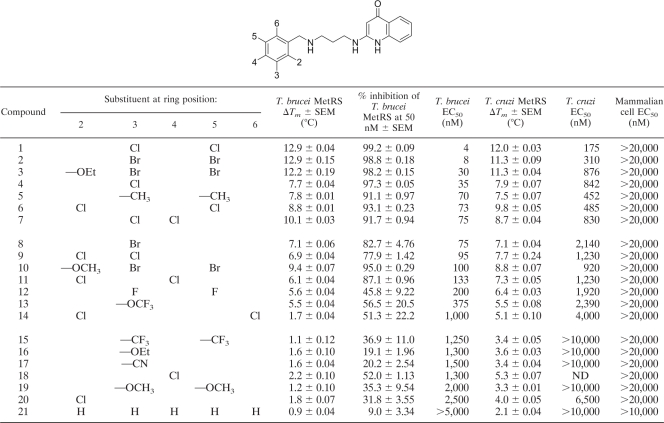
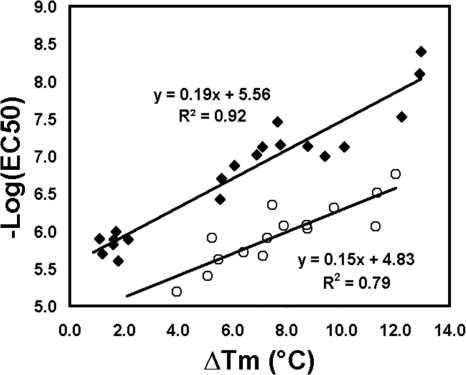
A set of diaryl diamines (compounds 2, 3, and 7) were synthesized and tested for binding to recombinant T. brucei MetRS using a thermal shift assay (Table 1). The initial set of compounds demonstrated large thermal shifts (ΔTm) of the T. brucei MetRS, exceeding 7°C. A ΔTm exceeding 2°C is usually considered significant. These compounds were also tested for activity on T. brucei cultures and found to have remarkable potency, particularly compound 2, with an EC50 of 8 nM. As a result, an additional 18 compounds were synthesized to investigate structure-activity relationships, particularly focusing on the phenyl ring on the left side of the scaffold, as shown in Table 1. By substituting the 3,5-bromines with 3,5-chlorines (compound 1), the activity against T. brucei cells was further improved to an EC50 of 4 nM. The activities all 21 compounds are displayed in Table 1. Binding of the compounds to the recombinant enzyme, as measured by thermal melting, and EC50s on T. brucei cultures were highly correlated (Fig. 2). An enzymatic assay was also developed to directly measure the aminoacylation activity of the T. brucei MetRS. The compounds were tested on the enzyme at 50 nM, with inhibition observed to be as high as 99% (compound 1; Table 1). Enzyme inhibition data were highly correlated with the thermal melt shifts (R2 = 0.821, P < 0.0001) and with the EC50s on T. brucei cultures (R2 = 0.646, P < 0.0001). The compounds did not inhibit growth of the mammalian cell line CRL-8155 at concentrations as high as 20 μM, indicating a very large selectivity index of as much as 5,000 (Table 1).
Table 1.
Binding and inhibitory activity of diaryl diamine compounds on MetRS enzymes and cell culturesa
Binding (ΔTm) was measured by the thermal melt method, enzyme inhibition (%) was measured in aminoacylation assays, and cell growth inhibition (EC50) was measured as described in Materials and Methods.
Fig. 2.
Correlation of cell growth inhibition (EC50) and binding to recombinant parasite MetRS. Binding was determined by the shift in the melting temperature (ΔTm) of the protein in the presence of compounds. R2 values are 0.92 (P < 0.0001) for T. brucei (solid diamonds) and 0.79 (P < 0.0001) for T. cruzi (open circles). Numeric data for this series of compounds are presented in Table 1.
In addition, binding of the compounds to the recombinant T. cruzi MetRS and inhibition of T. cruzi amastigote growth were evaluated (Table 1). Again, substantial ΔTm shifts of up to 12.0°C were observed when compounds interacted with the enzyme. There was also potent activity on T. cruzi amastigotes, although the EC50s were higher than those seen on T. brucei cells by a factor of ∼10 to 40.
Washout experiments.
In order to determine the exposure time required to kill T. brucei with a MetRS inhibitor, T. brucei cultures were incubated in the presence of compound 2 or pentamidine for defined time periods and then transferred to drug-free medium for growth quantification at 48 h (Fig. 3A). Incubation with compound 2 for 24 h at concentrations 2 times the EC50 resulted in >95% growth suppression, indicating that relative short exposure times lead to profound growth suppression. However, if the cultures were left to incubate for up to 10 days after the compound was washed out, parasites eventually grew out under most of the conditions, unless the cells were exposed at 8 times the EC50 for 72 h, 16 times the EC50 for 48 h, or 64 times the EC50 for 36 h (see values marked with asterisks in Fig. 3A). For comparison, the HAT drug pentamidine was completely trypanocidal when the cultures were exposed at 8 times the EC50 for 48 h or 16 times the EC50 for 36 h (Fig. 3B). Thus, the MetRS inhibitor is only slightly less rapid than pentamidine in exerting its trypanocidal effects.
Pharmacokinetic and efficacy experiments in mice.
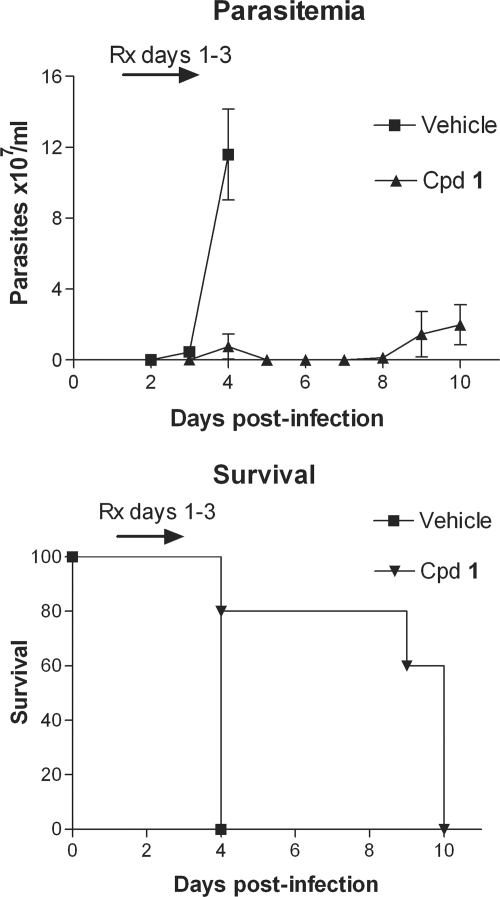
The most potent compound, compound 1, was subjected to a single-dose pharmacokinetic study in mice. Following an i.p. injection, blood was sampled at time intervals and analyzed by liquid chromatography-mass spectrometry. The terminal half-life was determined to be ∼1.0 h (data not shown). In order to provide a sustained blood level in mice well above the in vitro EC50, compound 1 was delivered by subcutaneous osmotic minipumps at 25 mg/kg/day for the efficacy experiment. The pumps are designed to deliver compound for 3 days. Blood levels on the day after implantation were measured to be 0.56 ± 0.12 μM (average ± standard error of the mean [SEM]; n = 5 mice). In T. brucei-infected mice, nearly complete suppression of parasitemia was observed by microscopic examination of blood smears for 8 days (Fig. 4). After 8 days, parasitemia became evident and the mice succumbed to infection. Average survival time was doubled in mice treated with compound 1 compared to that in mice treated with vehicle (Fig. 4). No apparent side effects were evident among the mice treated with compound 1 either in the single-dose pharmacokinetic studies or during the efficacy studies with the implanted minipumps.
Fig. 4.
Efficacy of compound 1 in the mouse model of acute T. brucei infection. Mice (n = 5 per group) were infected with bloodstream forms of T. brucei on day 0. On day 1, subcutaneous Alzet minipumps that delivered compound 1 (25 mg/kg/day) or vehicle for 3 days were implanted. Parasitemia (upper panel) rapidly rose in the vehicle group, and all of these mice were euthanized on day 4. In contrast, 4 of the 5 mice treated with compound 1 had dramatic suppression of parasitemia which gradually rebounded at about 7 or 8 days postinfection. They survived more than twice as long as the vehicle-treated mice (lower panel). Similar results were observed in a second experiment (data not shown).
Homology modeling and ligand docking studies of trypanosomatid MetRS enzymes.
Due to the absence of a crystal structure of T. brucei or T. cruzi MetRS, homology models were created for docking studies to gain insight into the binding mode of the diaryl diamine inhibitors. Three-dimensional structures of MetRS from 6 species were available as templates for homology modeling. The structures in the Protein Data Bank (PDB) originate from Aquifex aeolicus, Thermus thermophilus, Pyrococcus abyssi, Escherichia coli, and Mycobacterium smegmatis. In addition, the Medical Structural Genomics of Pathogenic Protozoa (MSGPP) consortium recently solved the structure of the Leishmania major enzyme, the first eukaryotic MetRS structure to be solved (18). Sequence comparisons of the T. cruzi and T. brucei MetRS proteins against the Leishmania MetRS and the homologs in the PDB showed overall amino acid sequence identities ranging from 24 to 60% (see Table S2 in the supplemental material). A more detailed analysis was performed to compare the residues constituting the methionine/ATP binding pockets of the respective enzymes (see Table S3 in the supplemental material). The crystal structure of the complex between E. coli MetRS and l-methionyladenylate served as the reference structure, with the active-site pocket defined by all residues within 5 Å of the l-methionyladenylate ligand. The pocket contained 24 residues, and the identity scores between the eight analyzed MetRS enzymes ranged from 58 to 100% (see Table S4 in the supplemental material). The three trypanosomatid organisms had identical methionine/ATP binding pockets.
An homology model for T. brucei and T. cruzi enzymes was built using the L. major structure, which has the highest sequence identity, and we modified the conformation of the binding site to mimic the arrangement seen in the A. aeolicus 2CSX structure. That conformational arrangement is required for inhibition of the bacterial MetRS by diaryl inhibitors (see Section S1 in the supplemental material). Upon docking into the homology model, compound 1 was found to fill two binding pockets (Fig. 5). The benzyl fragment occupies the methionine pocket and has one of the meta-chlorine atoms in the same position as the sulfur atom of methionine. The methionine pocket is formed from 11 residues, most of which are hydrophobic. The 2-amino-quinolinone fragment resides in a second, adjacent pocket and engages in two hydrogen bonds through its nitrogens with the carboxylate of Asp 287 and another hydrogen bond with the phenol hydroxyl of Tyr 472. Twelve other residues make up this second pocket. The aliphatic linker of the inhibitor mainly abuts Tyr 250 (Fig. 5). Comparison of the sequence of the human cytosolic enzyme to the sequences of the trypanosomatid enzymes reveals 13 differences of the 25 residues predicted to contact compound 1 (see Table S5 in the supplemental material). The human mitochondrial MetRS has five predicted differences in contact residues with compound 1 compared to the trypanosomatid enzymes (see Table S5 in the supplemental material). The significance of the similarity between the human mitochondrial MetRS and the T. brucei MetRS is discussed below.
Fig. 5.
Structural model of T. brucei MetRS with compound 1 docked in the active site. The dichlorinated benzyl fragment on the left resides in the methionine substrate pocket, while the 2-amino-quinolinone fragment occupies an adjacent pocket on the right and is predicted to interact with Asp 287. The protein molecular surface is shown in transparent gray. The atomic color code is cyan for protein C, yellow for ligand C, blue for N, red for O, and purple for Cl.
DISCUSSION
tRNA synthetases are established targets for antimicrobial chemotherapy, the foremost example being the bacterial isoleucyl-tRNA synthetase, which is the molecular target of mupirocin, an antibiotic used topically for the treatment of bacterial infections, including Staphylococcus aureus infections. This precedent indicated to us the potential for developing selective inhibitors of aaRS enzymes for other infectious pathogens such as trypanosomes. The MetRS enzyme was a particularly attractive target for piggyback drug development for neglected tropical diseases (9) because of the existence of potent MetRS inhibitors under development in Pharma for other indications (22). The similarities between the bacterial MetRS enzymes and those of T. brucei and T. cruzi led to our hypothesis that the existing compounds might bind trypanosomal MetRS enzymes and give rise to antitrypanosomal activity.
Before the activity of compounds was investigated, genetic experiments were conducted to establish that the MetRS enzyme was essential for trypanosome growth. The RNA interference method was used to knock down mRNA levels of the single MetRS gene identified in the T. brucei genome. The observed RNAi results (Fig. 1) confirmed our expectation that MetRS of T. brucei is critical for normal growth since protein synthesis is obviously dependent on minimum levels of charged tRNAMet. Cell growth was inhibited by a factor of 106; however, complete killing was not detected, possibly because the RNAi method resulted in incomplete knockdown of the MetRS transcript to only 57 to 77% below normal levels in two clones.
Three compounds (compounds 2, 3, and 7) were synthesized on the basis of published reports of MetRS inhibitors that had antibacterial activity (15, 16). These compounds were first tested on T. brucei cultures and observed to have remarkably potent activity, with EC50s of 8, 30, and 75 nM, respectively (Table 1). In order to determine if the compounds bound to the T. brucei MetRS enzyme, the thermal melt assay using recombinant enzyme was employed. Pronounced shifts in melting temperatures were observed (ΔTms, 12.9, 12.2, and 10.1°C, respectively). Generally, temperature shifts exceeding 2°C are considered significant; thus, the data indicated that the compounds were tightly bound by the recombinant MetRS enzyme. With these encouraging results, an additional 18 compounds were synthesized to further explore structure-activity relationships, primarily focusing on the phenyl group on the left side of the molecule (Table 1). The compound series resulted in a broad range of activities against T. brucei cultures from the most active compound, compound 1, with an EC50 of 4 nM, to the least active compound, compound 21, with an EC50 of >5,000 nM. All the compounds were tested in the thermal melt assay, and the correlation between ΔTm and EC50 was plotted (Fig. 2, upper line). A very high correlation was observed (R2 = 0.92), meaning that compounds that bind the enzyme with the highest affinity were the most potent at inhibiting T. brucei cell growth. This was evidence that the compounds act on the parasites through inhibition of the target enzyme and not by another off-target mechanism.
The compounds were also tested against the other major trypanosome pathogen, T. cruzi, and were again observed to have potent growth inhibition activity (Table 1). There was a high correlation between the compounds most active against T. brucei and T. cruzi, which is expected due to the 100% sequence identity in the predicted inhibitor binding sites (see Table S4 in the supplemental material). The EC50s against T. cruzi were generally higher (by a factor of 10 to 40) than those observed against T. brucei. Since T. cruzi amastigotes are grown intracellularly in mammalian 3T3 fibroblasts, the higher observed EC50s suggest that the compounds may be partially excluded from the intracellular environment of the host cells. Experiments of binding of the compounds to recombinant T. cruzi MetRS yielded results very similar to those observed with the T. brucei MetRS enzyme. As before, the binding affinity (ΔTm) and cell growth inhibition (EC50) were highly correlated (Fig. 2, bottom line).
In order to establish that binding to the recombinant T. brucei MetRS was related to inhibition of its enzymatic function, an aminoacylation assay was adopted for the T. brucei enzyme. The method assesses the complete enzyme reaction by measuring the incorporation of [3H]methionine into the tRNA substrate. The most potent compound, compound 1, inhibited the enzyme by 99.2% at 50 nM, suggesting that the 50% inhibitory concentration (IC50) is well below 50 nM, which is consistent with the low EC50 of 4 nM observed on T. brucei cells. An exact IC50 (possibly subnanomolar) for the highly potent compounds could not be determined because an enzyme concentration of 4 nM was necessary to give a robust signal in the assay. The correlation between enzyme inhibitory activity and the thermal melt results was very tight (R2 = 0.821, P < 0.0001) and supports the use of the thermal method before an enzyme assay has been developed and optimized (17).
The effect of the MetRS inhibitors on growth of the mammalian lymphocytic cell line CRL-8155 was also assessed. Since no inhibition of growth was observed at 20 μM for any of the compounds, this indicated a remarkably high selectivity index for the parasites over mammalian cells. The molecular modeling provides the explanation for why the compounds are unlikely to bind the human cytoplasmic MetRS enzyme (see Table S4 in the supplemental material). However, the differences between the trypanosomatid MetRS and the human mitochondrial MetRS are less pronounced (see Table S4 in the supplemental material) (discussed below). These data establish the potential to selectively inhibit the trypanosomatid MetRS and inhibit trypanosome growth while avoiding toxicity to mammalian cells.
In vitro washout experiments were conducted to determine the time required for compound 2 to lead to irreversible death of the parasites. When bloodstream forms of T. brucei were exposed to compound 2 at 8 times its EC50 for 72 h, the parasites did not recover. Similarly, when they were exposed to compound 2 at 16 times its EC50 for 48 h, they failed to recover. The conditions leading to irreversible growth arrest were similar to those of pentamidine, a drug in clinical use for early-stage HAT. Thus, sustained inhibition of MetRS completely kills T. brucei grown in vitro. This trypanocidal activity will be a necessary feature if the drug is to be used in late-stage HAT, where drugs that completely kill parasites are needed to clear infection from the central nervous system and cerebrospinal fluid.
Compound 1 had a short plasma half-life in mice (∼1 h). For the purposes of these studies, we were able to attain adequate plasma concentrations of the compound in mice by administering it via continuous infusion using osmotic minipumps. Plasma concentrations of ∼0.5 μM were measured in the mice receiving the minipumps. Using pumps designed to deliver compound for 3 days, profound suppression of parasitemia was observed. However, the parasites were not completely eradicated, as there was delayed recrudescence of parasitemia at about day 7 or 8 and the mice succumbed to the infection. There was also longer survival in the treated group (∼9 to 10 days) than the controls (4 days). These data indicate that compound 1 had a profound effect to suppress parasitemia and prolong survival in mice. It also suggests that either higher blood levels or longer exposure to compound 1 are required for cures. Since plasma levels were ∼100 times higher than the EC50 (4 nM), one might have expected complete clearance of the parasites on the basis of the results of the washout experiments described above. However, other factors, such as protein binding and access to parasites sequestered in tissues outside the blood compartment, may be reasons that complete cures were not observed.
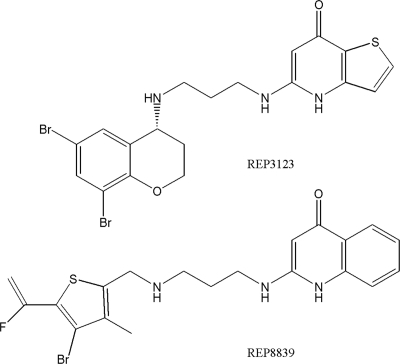
The analysis of the amino acids that form the predicted binding site of diaryl diamine compounds allowed comparisons between the trypanosomatid and the human MetRS enzymes (see Table S5 in the supplemental material). Substantial differences between the trypanosomatid and human cytosolic MetRS enzymes were observed; specifically, 13 of 25 residues are different, suggesting that the same inhibitor would be unlikely to bind tightly to this pocket in both enzymes. This fits with published data that the diaryl diamine REP8839 (Fig. 6) is essentially inactive on the human cytosolic enzyme, with Ki being >20,000 nM (12). Fewer differences in the inhibitor binding site were observed between trypanosomatid and human mitochondrial MetRS enzymes, with only 5 of 25 residues being different. This also fits with published data that REP8839 inhibits the human mitochondrial enzyme with a Ki of 10 nM (12). Of note, REP8839 inhibits the S. aureus MetRS with a Ki of 10 pM (12), suggesting that extremely tight binding is possible. Unfortunately, we do not know the Kis of diaryl diamines against the trypanosomatid enzymes because the methods for accurately measuring the Kis of highly active compounds against MetRS (involving an adaptation of an ATP-PPi exchange assay [12]) have not been developed for the trypanosomatid enzymes. If the Ki on trypanosomatid MetRS is very low (≪1 nM), then it is possible that the greater activity of the diaryl diamines on trypanosomatid cells than mammalian cells (Table 1) is due to selective activity on the trypanosomatid enzyme. However, it seems likely that there are other factors contributing to the lack of toxicity of the diaryl diamines against mammalian cells and live animals. One possibility is that the compounds may not penetrate into mammalian cells as well as they do into trypanosomatid cells. The observation that T. cruzi (grown inside mammalian fibroblasts) has EC50s as much as 44-fold higher than those for T. brucei (grown in axenic culture), despite having identical binding pockets in the MetRS enzyme (see Table S4 in the supplemental material), suggests that the compounds do not efficiently penetrate the mammalian cell membrane. It could also be that so little compound passes through the second barrier (the mitochondrial membrane) that the mitochondrial MetRS is not significantly inhibited. As new compounds are made to alter the pharmacological properties, the changes could affect the permeability properties. As a result, it will be important to carefully test for toxicity on mammalian cells that may be mediated through inhibition of the mitochondrial MetRS.
Fig. 6.
Structures of MetRS inhibitors in clinical trials for bacterial infections (5, 6, 25).
In summary, the MetRS enzyme has been genetically and chemically validated to be a drug target for T. brucei and T. cruzi. Importantly, we provide a proof of concept that the T. brucei MetRS can be targeted for antitrypanosomal chemotherapy in the mouse model of T. brucei infection. More work is required to improve the pharmacological properties of the compounds obtained so far, including improving oral bioavailability and penetration through the blood-brain barrier. The pharmacological issues are critical, as the highest priority with respect to African sleeping sickness is to discover new therapeutics (preferably orally administered) that will effectively treat stage 2 disease, i.e., when the parasites have entered the central nervous system (http://www.dndi.org/diseases/hat/target-product-profile.html). The reported research is an exciting first step toward our goal of developing safe and effective new treatments for HAT and perhaps Chagas' disease.
Supplementary Material
ACKNOWLEDGMENTS
Support for this research was provided by National Institute of Allergy and Infectious Diseases grants AI067921 and AI084004.
We are grateful to Colin McMartin and Regine Bohaceck for providing the FLO/QXP software.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 31 January 2011.
REFERENCES
- 1. Alibu V. P., Storm L., Haile S., Clayton C., Horn D. 2005. A doubly inducible system for RNA interference and rapid RNAi plasmid construction in Trypanosoma brucei. Mol. Biochem. Parasitol. 139:75–82 [DOI] [PubMed] [Google Scholar]
- 2. Buckner F. S., Verlinde C. L. M. J., La Flamme A. C., Van Voorhis W. C. 1996. Efficient technique for screening drugs for activity against Trypanosoma cruzi using parasites expressing β-galactosidase. Antimicrob. Agents Chemother. 40:2592–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Charriere F., Helgadottir S., Horn E. K., Soll D., Schneider A. 2006. Dual targeting of a single tRNA(Trp) requires two different tryptophanyl-tRNA synthetases in Trypanosoma brucei. Proc. Natl. Acad. Sci. U. S. A. 103:6847–6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Charriere F., et al. 2009. Dual targeting of a tRNA ASP requires two different aspartyl-tRNA synthetases in Trypanosoma brucei. J. Biol. Chem. 284:16210–16217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Critchley I. A., et al. 2009. Spectrum of activity and mode of action of REP3123, a new antibiotic to treat Clostridium difficile infections. J. Antimicrob. Chemother. 63:954–963 [DOI] [PubMed] [Google Scholar]
- 6. Critchley I. A., et al. 2005. Antibacterial activity of REP8839, a new antibiotic for topical use. Antimicrob. Agents Chemother. 49:4247–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crowther G. J., et al. 2009. Buffer optimization of thermal melt assays of Plasmodium proteins for detection of small-molecule ligands. J. Biomol. Screen. 14:700–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Espanol Y., Thut D., Schneider A., de Pouplana L. R. 2009. A mechanism for functional segregation of mitochondrial and cytosolic genetic codes. Proc. Natl. Acad. Sci. U. S. A. 106:19420–19425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gelb M. H., et al. 2003. Protein farnesyl and N-myristoyl transferases: piggy-back medicinal chemistry targets for the development of antitrypanosomatid and antimalarial therapeutics. Mol. Biochem. Parasitol. 126:155–163 [DOI] [PubMed] [Google Scholar]
- 10. Gentry D. R., et al. 2003. Variable sensitivity to bacterial methionyl-tRNA synthetase inhibitors reveals subpopulations of Streptococcus pneumoniae with two distinct methionyl-tRNA synthetase genes. Antimicrob. Agents Chemother. 47:1784–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Giaever G., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391 [DOI] [PubMed] [Google Scholar]
- 12. Green L. S., et al. 2009. Inhibition of methionyl-tRNA synthetase by REP8839 and effects of resistance mutations on enzyme activity. Antimicrob. Agents Chemother. 53:86–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guiles J., Critchley I., Sun X. 2008. New agents for Clostridium difficile-associated disease. Expert Opin. Invest. Drugs 17:1671–1683 [DOI] [PubMed] [Google Scholar]
- 14. Hirumi H., Hirumi K. 1989. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 75:985–989 [PubMed] [Google Scholar]
- 15. Jarvest R. L., et al. 2002. Nanomolar inhibitors of Staphylococcus aureus methionyl tRNA synthetase with potent antibacterial activity against gram-positive pathogens. J. Med. Chem. 45:1959–1962 [DOI] [PubMed] [Google Scholar]
- 16. Jarvest R. L., et al. 2003. Optimisation of aryl substitution leading to potent methionyl tRNA synthetase inhibitors with excellent gram-positive antibacterial activity. Bioorg. Med. Chem. Lett. 13:665–668 [DOI] [PubMed] [Google Scholar]
- 17. Kroe R. R., et al. 2003. Thermal denaturation: a method to rank slow binding, high-affinity P38alpha MAP kinase inhibitors. J. Med. Chem. 46:4669–4675 [DOI] [PubMed] [Google Scholar]
- 18. Larson E. T., et al. 2011. Structure of Leishmania major methionyl-tRNA synthetase in complex with intermediate products methionyladenylate and pyrophosphate. Biochimie 93:570–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marti-Renom M. A., et al. 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29:291–325 [DOI] [PubMed] [Google Scholar]
- 20. McMartin C., Bohacek R. S. 1997. QXP: powerful, rapid computer algorithms for structure-based drug design. J. Comput. Aided Mol. Des. 11:333–344 [DOI] [PubMed] [Google Scholar]
- 21. Mehlin C., et al. 2006. Heterologous expression of proteins from Plasmodium falciparum: results from 1000 genes. Mol. Biochem. Parasitol. 148:144–160 [DOI] [PubMed] [Google Scholar]
- 22. Ochsner U. A., Sun X., Jarvis T., Critchley I., Janjic N. 2007. Aminoacyl-tRNA synthetases: essential and still promising targets for new anti-infective agents. Expert Opin. Invest. Drugs 16:573–593 [DOI] [PubMed] [Google Scholar]
- 23. Ojo K. K., et al. 2008. Glycogen synthase kinase 3 is a potential drug target for African trypanosomiasis therapy. Antimicrob. Agents Chemother. 52:3710–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Osborne A. G., Buley J. M., Clarke H., Dakin R. C. H., Price P. I. 1993. 2,4-Dihaloquinolines. Synthesis, orientation effects and proton and carbon-13 NMR spectral studies. Organic Bio-Organic Chem. 22:2747–2755 [Google Scholar]
- 25. Raz B., Iten M., Grether-Buhler Y., Kaminsky R., Brun R. 1997. The Alamar Blue assay to determine drug sensitivity of African trypanosomes (T.b. rhodesiense and T.b. gambiense) in vitro. Acta Trop. 68:139–147 [DOI] [PubMed] [Google Scholar]
- 26. Rinehart J., Horn E. K., Wei D., Soll D., Schneider A. 2004. Non-canonical eukaryotic glutaminyl- and glutamyl-tRNA synthetases form mitochondrial aminoacyl-tRNA in Trypanosoma brucei. J. Biol. Chem. 279:1161–1166 [DOI] [PubMed] [Google Scholar]
- 27. Sheppard K., et al. 2008. From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res. 36:1813–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Van Voorhis W. C., et al. 2007. Efficacy, pharmacokinetics, and metabolism of tetrahydroquinoline inhibitors of Plasmodium falciparum protein farnesyltransferase. Antimicrob. Agents Chemother. 51:3659–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.