Abstract
It is important to develop new anti-HIV drugs that are effective against the existing drug-resistant mutants. Because the excision mechanism is an important pathway for resistance to nucleoside analogs, we are preparing analogs that retain a 3′-OH and can be extended after they are incorporated by the viral reverse transcriptase. We show that 4′-C-alkyl-deoxyadenosine (4′-C-alkyl-dA) compounds can be phosphorylated in cultured cells and can inhibit the replication of HIV-1 vectors: 4′-C-methyl- and 4′-C-ethyl-dA show both efficacy and selectivity against HIV-1. The compounds are also effective against viruses that replicate using reverse transcriptases (RTs) that carry nucleoside reverse transcriptase inhibitor resistance mutations, with the exception of the M184V mutant. Analysis of viral DNA synthesis in infected cells showed that viral DNA synthesis is blocked by the incorporation of either 4′-C-methyl- or 4′-C-ethyl-2′-deoxyadenosine. In vitro experiments with purified HIV-1 RT showed that 4′-C-methyl-2′-dATP can compete with dATP and that incorporation of the analog causes pausing in DNA synthesis. The 4′-C-ethyl compound also competes with dATP and shows a differential ability to block DNA synthesis on RNA and DNA templates. Experiments that measure the ability of the compounds to block DNA synthesis in infected cells suggest that this differential block to DNA synthesis also occurs in infected cells.
INTRODUCTION
Nucleoside reverse transcriptase (RT) inhibitors (NRTIs) are a key component of highly active antiretroviral therapy (HAART) in HIV-1-infected individuals. All of the NRTIs approved for treatment lack the 3′-hydroxyl on the deoxyribose sugar (or sugar analog) and, once they are incorporated by the RT of HIV-1, inhibit viral replication by immediately blocking the extension of the viral DNA chain. Unfortunately, treatment with all of the available drugs, including the NRTIs, can select for mutations that make the virus drug resistant. NRTIs select for mutations that enhance the ability of RT to discriminate NRTIs from normal nucleosides. NRTI resistance mutations can cause this enhanced discrimination by two mechanisms: excision and exclusion (5, 31, 33, 34). In HIV-1, excision is the primary pathway for zidovudine (AZT) resistance. AZT-resistant HIV-1 isolates from AZT-treated patients carry a number of resistance mutations, which can include M41L, D67N, K70R, T215F/Y, and K219Q (17, 36). The mechanism of AZT excision is related to pyrophosphorolysis, the reverse of the reaction by which AZT triphosphate is incorporated by HIV-1 RT, except that in the AZT excision reaction, ATP takes the place of pyrophosphate. The AZT resistance mutations enhance the ability of HIV-1 RT to bind the excision substrate, ATP (5, 40). The product of the ATP-mediated excision reaction is AZTppppA. Because the AZT monophosphate is removed from the end of the primer strand in the excision reaction, RT can resume viral DNA synthesis (1, 5, 21, 29, 30, 33). Additional mutations (for example, insertions in the fingers subdomain) which extend the ability of HIV-1 RT to excise a wider range of NRTIs convert an AZT-resistant RT into a multidrug-resistant RT (4, 20, 39).
The other mechanism of NRTI resistance, exclusion, involves mutations that enhance discrimination at the point at which the triphosphate form of the analog is incorporated into DNA. The best-studied exclusion mutation is the M184V/I variant, which causes high-level resistance to lamivudine (3TC) (37) and emtricitabine (FTC). Position 184 is immediately adjacent to two of the three active-site aspartates, D185 and D186. Introduction of a β-branched-chain amino at position 184 creates a steric clash with the pseudosugar moieties of 3TC and FTC, which interferes with their incorporation (11, 31).
Other mutations that result in NRTI exclusion include L74V, K65R, and Q151M, the last of which is usually found with accompanying mutations. The resistance mutations L74V and K65R both participate in crucial hydrogen bond networks at the RT polymerase active site which stabilize the incoming deoxynucleoside triphosphate (dNTP) (10, 14, 41); however, these mutations rarely occur together in the clinical setting (13, 44). The Q151M mutation changes the hydrogen bond network that involves the 3′-OH group of dNTP, thus reducing the incorporation of NRTIs, which lack a 3′-OH group (14, 35). In patient isolates, the Q151M mutation is typically found in concert with A62V, V75I, F77L, and F116Y. Data derived from patient isolates indicate that the Q151M mutation is the primary mutation in the resistance pathway; its acquisition is followed by the acquisition of mutations at A62, V75, F77, and F116 (38). The Q151M mutation alone is sufficient to impart resistance to AZT, dideoxyinosine, stavudine (d4T), and dideoxycytosine (ddC), but the accompanying mutations, A62V, V75I, F77L, and F116Y (Q151M complex), enhance drug resistance (15). The mutations at positions A62, V75, and F77 help stabilize the hydrophobic core of the fingers domain of RT to better accommodate the Gln → Met change at position 151, while the F116Y mutation restores hydrogen bond interactions in the active site that are altered by the Q151M mutation (14).
The major challenges in designing new NRTIs involve issues associated with activation of the compound by host cell kinases, toxicity, and drug resistance. It is important to design compounds that have little or no susceptibility to any of the known resistance mutations. To be active, NRTIs must be reasonably well phosphorylated by host cell kinases. If the triphosphate form of an NRTI can be incorporated by host polymerases, blocking either nuclear or mitochondrial DNA synthesis, there can be undesirable cytotoxicity (7). Because excision is a major pathway for NRTI resistance, we wanted to develop NRTIs that are poorly excised. The existence of multidrug-resistant RTs that can excise a wide range of NRTIs makes this a difficult problem. One possible solution is to develop nucleoside analogs that are not immediate chain terminators (3, 34). The excision mechanism depends on the fact that NRTIs that lack a 3′-OH remain at the end of the primer, where they can be excised by an NRTI-resistant RT. To avoid this problem, we have focused on developing nucleoside analogs that retain a 3′-OH with the expectation that, if additional normal nucleotides are added after the analog has been incorporated, the additional normal nucleotides would protect the analog from excision. This approach raises a question about whether this type of analog is more likely than the current NRTIs, which lack a 3′-OH, to be incorporated into host DNA, to cause toxicity. We have tested several compounds which retain a 3′-OH for toxicity in cultured cells. Some of the compounds show significant toxicity at concentrations that are relatively close to the concentrations needed to inhibit HIV replication; however, others, including some of the compounds we report here, show little toxicity in cell culture. Ohrui and Mitsuya also found that some of the nucleoside analogs that they tested which retain a 3′-OH showed little or no toxicity in cell culture (26). The proposal that some compounds that retain a 3′-OH can be relatively nontoxic is also supported by the fact that the approved anti-hepatitis B virus drug entecavir has a 3′-OH and has been used successfully in the clinic. The entecavir results shows that nucleoside analogs that retain a 3′-OH are not necessarily too toxic to be used to treat human diseases.
We previously asked whether conformationally locked nucleoside analogs can be used to inhibit HIV-1 reverse transcription (3, 6, 8, 9, 18). The conformationally locked nucleoside analog north-methanocarbathymidine (N-MCT) is poorly phosphorylated by host cell thymidine kinases (TKs). In cultured cells that express the herpes simplex virus (HSV) TK, N-MCT is an effective inhibitor of HIV-1 vectors that replicate using excision-proficient RTs. In vitro assays indicated that the triphosphate form of N-MCT acts as a delayed chain terminator, blocking viral DNA extension 2 nucleotides after the N-MCT monophosphate is incorporated by HIV-1 RT.
Ohrui and Mitsuya suggested that 4′-C-deoxyribose-modified nucleoside analogs would be viable candidates for NRTI development (26). We previously tested the ability of 4′-C-methyl- and 4′-C-ethyl thymidine to inhibit the replication of wild-type (WT) and drug-resistant HIV-1 vectors, and we tested their effects on the polymerase activity of HIV-1 RT in vitro (2). The data showed that both compounds act as chain terminators; however, their effects on viral DNA synthesis are different. 4′-C-methyl-dTTP can be incorporated by HIV RT, but after the analog has been incorporated, the next nucleotide is added slowly, resulting in a pause during DNA synthesis referred to as kinetic chain termination. Despite the fact that 4′-C-ethyl-thymidine has a 3′-OH, it behaves like a conventional (immediate) chain terminator in vitro. Cell culture data indicated that both of these compounds are poorly phosphorylated by cellular kinases. However, experiments done using HSV TK-expressing cells showed that the phosphorylated versions of these compounds were able to block HIV-1 replication. Hayakawa et al. suggested that 4′-C-substituted purines have more favorable selective indices than the corresponding pyrimidines in cells infected with HIV-1 (12). We changed the nucleobase on the 4′-C analogs from thymidine to adenine to determine whether this change would increase the efficacy of the compound. Here, we show that 4′-C-methyl- and 4′-C-ethyl-2′-deoxyadenosine (4′-C-Me-dA and 4′-C-Et-dA, respectively) (Fig. 1) are reasonably well phosphorylated and that these compounds can inhibit the replication of HIV-1 vectors that replicate using WT and several NRTI-resistant RTs, although the M184V mutation does reduce the susceptibility of HIV-1 vectors to these compounds.
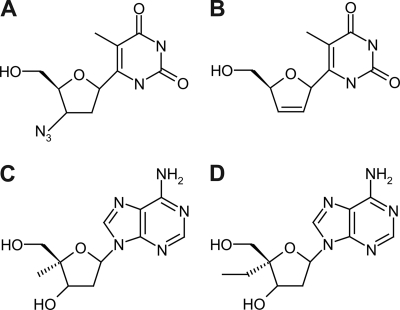
Fig. 1.
Structures of the nucleoside analogs AZT (A), d4T (B), 4′-C-methyl-2′-deoxyadenosine (compound 1) (C), and 4′-C-ethyl-2′-deoxyadenosine (compound 2) (D).
MATERIALS AND METHODS
Synthesis of compounds. (i) General techniques.
All reagents and solvents purchased were of the highest commercial quality and were used without further purification unless otherwise stated. Column chromatography was performed on a silica gel 60, 230- to 400-mesh column (E. Merck). 1H and 13C nuclear magnetic resonance (NMR) spectra were recorded on a Varian Unity Inova instrument at 400 and 100 MHz, respectively. Spectra are referenced to the solvent in which they were run, and NMR signals are identified with nucleoside numbering. Positive-ion fast-atom bombardment mass spectra (FAB-MS) were obtained on a VG 70-SE double-focusing mass spectrometer operated at an accelerating voltage of 8 kV under the control of a MASPEC-II32 data system for Windows (MasCom GmbH, Bremen, Germany). Either glycerol or 3-nitrobenzyl alcohol was used as the sample matrix, and ionization was effected by a beam of xenon atoms generated in a saddle-field ion gun at 8.0 ± 0.5 kV. Elemental analyses were performed by Atlantic Microlab, Inc., Atlanta, GA.
(ii) (2R,3R,4S)-4-(Benzyloxy)-5-[(tert-butyldiphenylsilyloxy)methyl]-2-(6-phenylamido-9H-purin-9-yl)-5-vinyltetrahydrofuran-3-yl ethanoate (compound 7).
A stirred suspension of compound 6 (3.01 g, 5.11 mmol) and N6-benzoyladenine (1.47 g, 6.14 mmol) in anhydrous acetonitrile (100 ml) was cooled over an ice bath. Trimethylsilyl trifluoromethanesulfonate (1.92 ml, 10.54 mmol) was added slowly, and the reaction mixture was stirred for 1 h. After quenching of the reaction with solid NaHCO3 (3 g), the mixture was filtered and concentrated in vacuo and the residue was partitioned between ethyl acetate (200 ml) and water (70 ml). The organic layer was dried (MgSO4), reduced to dryness, and purified by flash column chromatography (silica gel) using 25% ethyl acetate in hexanes to give 2.38 g (60.7%) of compound 7 as a colorless foam. 1H NMR (400 MHz, CDCl3) δ 8.63 (s, 1H, H-8), 8.19 (s, 1H, H-2), 7.96 (m, 2H, phenyl [Ph]), 7.23 to 7.56 (m, 18H, Ph), 6.22 (d, J = 4.1 Hz, 1H, H-1′), 5.94 (dd, J = 17.4, 11.0 Hz, 1H, CH CHH), 5.78 (dd, J = 5.9, 4.2 Hz, 1H, H-2′), 5.44 (dd, J = 17.4, 1.5 Hz, 1H, CH
CHH), 5.78 (dd, J = 5.9, 4.2 Hz, 1H, H-2′), 5.44 (dd, J = 17.4, 1.5 Hz, 1H, CH CHH), 5.22 (dd, J = 11.0, 1.4 Hz, CH
CHH), 5.22 (dd, J = 11.0, 1.4 Hz, CH CH2), 4.86 (d, J = 5.9 Hz, 1H, H-3′), 4.54 (ABq, J = 11.3 Hz, 2H, OCH2Ph), 3.64 (ABq, J = 11.5 Hz, 2H, CH2OSi), 2.00 (s, 3H, acetate [OAc]), 0.98 [s, 9H, SiC(CH3)3]; 13C NMR (100 MHz, CDCl3) δ 169.3, 164.7, 152.84, 151.51, 149.63, 141.75, 137.41, 135.60, 135.47, 134.24, 133.68, 132.68, 132.56, 132.53, 129.93, 129.85, 128.77, 128.43, 128.01, 127.89, 127.85, 127.81, 123.40, 116.17, 88.63, 86.37, 86.31, 74.44, 74.42, 74.06, 74.02, 66.03, 26.88, 20.67, 20.65, 19.21; FAB-MS m/z 768.3 (7.1%) [M + H]+. Analysis calculated for C44H45N5O6Si: C, 68.82; H, 5.91; N, 9.12. Found: C, 68.49; H, 6.08; N, 8.88.
CH2), 4.86 (d, J = 5.9 Hz, 1H, H-3′), 4.54 (ABq, J = 11.3 Hz, 2H, OCH2Ph), 3.64 (ABq, J = 11.5 Hz, 2H, CH2OSi), 2.00 (s, 3H, acetate [OAc]), 0.98 [s, 9H, SiC(CH3)3]; 13C NMR (100 MHz, CDCl3) δ 169.3, 164.7, 152.84, 151.51, 149.63, 141.75, 137.41, 135.60, 135.47, 134.24, 133.68, 132.68, 132.56, 132.53, 129.93, 129.85, 128.77, 128.43, 128.01, 127.89, 127.85, 127.81, 123.40, 116.17, 88.63, 86.37, 86.31, 74.44, 74.42, 74.06, 74.02, 66.03, 26.88, 20.67, 20.65, 19.21; FAB-MS m/z 768.3 (7.1%) [M + H]+. Analysis calculated for C44H45N5O6Si: C, 68.82; H, 5.91; N, 9.12. Found: C, 68.49; H, 6.08; N, 8.88.
(iii) (2R,3R,4S)-2-(6-Amino-9H-purin-9-yl)-4-(benzyloxy)-5-[(tert-butyldiphenyl-silyloxy)methyl]-5-vinyltetrahydrofuran-3-ol (compound 8).
Compound 7 was suspended in methanolic saturated ammonia (30 ml) and stirred in a sealed vessel at room temperature for 20 h. After removal of all volatiles in vacuo, the residue was purified by flash column chromatography (silica gel) using 50% ethyl acetate in hexanes, followed by 100% ethyl acetate, to give 0.27 g (95.6%) of compound 8 as a colorless foam. 1H NMR (400 MHz, dimethyl sulfoxide [DMSO]-d6 + D2O) δ 8.22 (s, 1H, H-8), 7.90 (s, 1H, H-2), 7.24 to 7.52 (m, 15H, Ph), 6.02 (dd, J = 17.4, 11.0 Hz, 1H, CH CHH), 5.93 (d, J = 5.6 Hz, 1H, H-1′), 5.32 (dd, J = 17.4, 1.8 Hz, 1H, CH
CHH), 5.93 (d, J = 5.6 Hz, 1H, H-1′), 5.32 (dd, J = 17.4, 1.8 Hz, 1H, CH CHH), 5.13 (dd, J = 11.0, 1.8 Hz, 1H, CH
CHH), 5.13 (dd, J = 11.0, 1.8 Hz, 1H, CH CH2), 4.99 (t, J ≈ 5.5 Hz, 1H, H-2′), 4.67 (ABq, J = 12.1 Hz, 2H, OCH2Ph), 4.43 (d, J = 5.4 Hz, 1H, H-3′), 3.57 (ABq, J = 10.7 Hz, 2H, CH2OSi), 0.88 [s, 9H, SiC(CH3)3]; 13C NMR (100 MHz, CDCl3) δ 161.25, 157.77, 154.63, 144.89, 143.58, 141.46, 140.27, 140.20, 137.71, 137.51, 135.09, 133.36, 133.02, 132.73, 132.63, 124.37, 119.48, 118.00, 110.00, 106.051, 101.87, 92.44, 92.24, 83.59, 77.54, 77.40, 72.21, 31.77, 23.98; FAB-MS m/z 622.3 (82.8%) [M + H]+. Analysis calculated for C35H39N5O4Si: C, 67.61; H, 6.32; N, 11.26. Found: C, 67.27; H, 6.29; N, 11.19.
CH2), 4.99 (t, J ≈ 5.5 Hz, 1H, H-2′), 4.67 (ABq, J = 12.1 Hz, 2H, OCH2Ph), 4.43 (d, J = 5.4 Hz, 1H, H-3′), 3.57 (ABq, J = 10.7 Hz, 2H, CH2OSi), 0.88 [s, 9H, SiC(CH3)3]; 13C NMR (100 MHz, CDCl3) δ 161.25, 157.77, 154.63, 144.89, 143.58, 141.46, 140.27, 140.20, 137.71, 137.51, 135.09, 133.36, 133.02, 132.73, 132.63, 124.37, 119.48, 118.00, 110.00, 106.051, 101.87, 92.44, 92.24, 83.59, 77.54, 77.40, 72.21, 31.77, 23.98; FAB-MS m/z 622.3 (82.8%) [M + H]+. Analysis calculated for C35H39N5O4Si: C, 67.61; H, 6.32; N, 11.26. Found: C, 67.27; H, 6.29; N, 11.19.
(iv) O-(2R,3R,4S)-2-(6-Amino-9H-purin-9-yl)-4-(benzyloxy)-5-[(tert-butyldiphenyl-silyloxy)methyl]-5-vinyltetrahydrofuran-3-yl O-phenyl carbonothioate (compound 9).
Phenyl chlorothionoformate (0.20 ml, 1.44 mmol) was added to a suspension of compound 8 (0.350 g, 0.56 mmol) and 4-(dimethylamino)pyridine (DMAP; 0.30 g, 2.45 mmol) in anhydrous acetonitrile (50 ml). The resulting mixture was stirred at room temperature for 20 h, after which all volatiles were removed in vacuo. The residue was taken up ethyl acetate (100 ml), washed with water (twice with 50 ml each time), and dried (MgSO4). Purification was by flash column chromatography (silica gel) using a gradient of 0% to 70% ethyl acetate in hexanes to give 0.348 g (81.9%) of compound 9 as a colorless foam. 1H NMR (400 MHz, CDCl3) δ 8.16 (s, 1H, H-8), 7.94 (s, 1H, H-2), 7.52 (m, 4H, Ph), 7.16 to 7.22 (m, 19H, Ph), 6.88 (br d, 2H, Ph), 6.30 (dd, J ≈ 5.7, 4.0 Hz, 1H, H-2′), 6.26 (d, J = 3.9 Hz, 1H, H-1′), 6.0 (dd, J = 17.4, 11.1 Hz, 1H, CH CHH), 5.63 (br s, 2H, NH2), 5.45 (dd, J = 17.4, 1.4 Hz, 1H, CH
CHH), 5.63 (br s, 2H, NH2), 5.45 (dd, J = 17.4, 1.4 Hz, 1H, CH CHH), 5.21 (dd, J = 11.0, 1.4 Hz, 1H, CH
CHH), 5.21 (dd, J = 11.0, 1.4 Hz, 1H, CH CH2), 5.13 (d, J = 5.8 Hz, 1H, H-3′), 4.66 (ABq, J = 11.3 Hz, 2H, OCH2Ph), 3.66 (ABq, J = 11.3 Hz, 2H, CH2OSi), 0.96 [s, 9H, SiC(CH3)3]; 13C NMR (100 MHz, CDCl3) δ 139.59, 137.39, 135.60, 135.54, 134.07, 132.66, 132.64, 129.85, 129.78, 129.47, 128.46, 128.05, 127.99, 127.75, 127.71, 126.65, 121.68, 120.06, 116.46, 88.44, 85.90, 82.21, 74.28, 66.02, 26.84, 19.23; FAB-MS m/z 758.2 (18.1%) [M + H]+. Analysis calculated for C42H43N5O5SSi: C, 66.55; H, 5.72; N, 9.24. Found: C, 66.38; H, 5.77; N, 9.17.
CH2), 5.13 (d, J = 5.8 Hz, 1H, H-3′), 4.66 (ABq, J = 11.3 Hz, 2H, OCH2Ph), 3.66 (ABq, J = 11.3 Hz, 2H, CH2OSi), 0.96 [s, 9H, SiC(CH3)3]; 13C NMR (100 MHz, CDCl3) δ 139.59, 137.39, 135.60, 135.54, 134.07, 132.66, 132.64, 129.85, 129.78, 129.47, 128.46, 128.05, 127.99, 127.75, 127.71, 126.65, 121.68, 120.06, 116.46, 88.44, 85.90, 82.21, 74.28, 66.02, 26.84, 19.23; FAB-MS m/z 758.2 (18.1%) [M + H]+. Analysis calculated for C42H43N5O5SSi: C, 66.55; H, 5.72; N, 9.24. Found: C, 66.38; H, 5.77; N, 9.17.
(v) 9-[(2R,4S)-4-(Benzyloxy)-5-[(tert-butyldiphenyl-silyloxy)methyl]-5-vinyltetrahydrofuran-2-yl]-9H-purin-6-amine (compound 10).
n-Tributyltin hydride (1.2 ml, 4.49 mmol) was added to a solution of 2,2′-azobisisobutyronitrile (AIBN; 0.22 g, 1.33 mmol) in anhydrous toluene (40 ml). The solution was purged with argon for 15 min, and then a solution of compound 9 (1.0 g, 1.32 mmol) in toluene (40 ml) was added slowly. The resulting mixture was heated at reflux for 45 min and cooled. The volatiles were removed in vacuo, and the crude product was purified by flash column chromatography (silica gel) using 25% ethyl acetate in hexanes and then 100% ethyl acetate to give 0.722 g (90.3%) of compound 10 as a colorless foam. 1H NMR (400 MHz, CDCl3) δ 8.2 (br 2, 2H, H-2, H-8), 7.52 (m, 4H, Ph), 7.22 to 7.37 (m, 16H, Ph), 6.35 (dd, J = 7.2, 3.7, H-1′), 6.23 (v br s, 2H, NH2), 5.95 (dd, J = 17.4, 10.9 Hz, 1H, CH CHH), 5.48 (dd, J = 17.4, 1.7 Hz, 1H, CH
CHH), 5.48 (dd, J = 17.4, 1.7 Hz, 1H, CH CHH), 5.24 (dd, J = 10.8, 1.6 Hz, 1H, CH
CHH), 5.24 (dd, J = 10.8, 1.6 Hz, 1H, CH CH2), 4.71 (irregular t, J ≈ 7 Hz, 1H, H-3′), 4.50 (ABq, J = 11.9 Hz, 2H, OCH2Ph), 3.68 (ABq, J = 11.5 Hz, 2H, CH2OSi), 2.46 to 2.60 (m, 2H, H-2′a,b), 0.98 [s, 9H, SiC(CH3)3]; 13C NMR (100 MHz, CDCl3) δ 155.56, 152.88, 149.26, 138.88, 137.70, 135.59, 135.64, 134.66, 132.72, 132.62, 129.87, 129.82, 128.46, 127.85, 127.8, 127.47, 120.00, 116.43, 88.81, 82.50, 82.44, 72.53, 65.77, 37.00, 26.91, 19.22; FAB-MS m/z 606.4 (30.0%) [M + H]+. Analysis calculated for C35H39N5O3Si: C, 69.39; H, 6.49; N, 11.46. Found: C, 69.23; H, 6.43; N, 11.39.
CH2), 4.71 (irregular t, J ≈ 7 Hz, 1H, H-3′), 4.50 (ABq, J = 11.9 Hz, 2H, OCH2Ph), 3.68 (ABq, J = 11.5 Hz, 2H, CH2OSi), 2.46 to 2.60 (m, 2H, H-2′a,b), 0.98 [s, 9H, SiC(CH3)3]; 13C NMR (100 MHz, CDCl3) δ 155.56, 152.88, 149.26, 138.88, 137.70, 135.59, 135.64, 134.66, 132.72, 132.62, 129.87, 129.82, 128.46, 127.85, 127.8, 127.47, 120.00, 116.43, 88.81, 82.50, 82.44, 72.53, 65.77, 37.00, 26.91, 19.22; FAB-MS m/z 606.4 (30.0%) [M + H]+. Analysis calculated for C35H39N5O3Si: C, 69.39; H, 6.49; N, 11.46. Found: C, 69.23; H, 6.43; N, 11.39.
(vi) [(3S,5R)-5-(6-Amino-9H-purin-9-yl)-3-(benzyloxy)-2-vinyltetrahydrofuran-2-yl]methanol (compound 11).
A solution of compound 10 (0.772 g, 1.19 mmol) in anhydrous tetrahydrofuran (THF; 30 ml) was treated with a 1 M solution of tetra-n-butylammonium fluoride (TBAF; 7.1 ml). The reaction mixture was stirred for 30 min at room temperature and then concentrated to dryness in vacuo. The residue was purified by flash column chromatography (silica gel) using 50% ethyl acetate in hexanes, followed by 100% ethyl acetate. A second chromatography using 2% to 10% methanol in dichloromethane afforded a solid material that was triturated with water (30 ml) and methanol (5 ml). The solid was removed by filtration, washed with ether (10 ml), and dried in vacuo to give 0.295 g of compound 11 as a white solid. Melting point, 185 to 186°C; 1H NMR (400 MHz, CDCl3) δ 8.20 (br 2, 2H, H-2, H-8), 7.52 (m, 4H, Ph), 7.22 to 7.37 (m, 16H, Ph), 6.35 (dd, J = 7.2, 3.7, H-1′), 6.23 (v br s, 2H, NH2), 5.95 (dd, J = 17.4, 10.9 Hz, 1H, CH CHH), 5.48 (dd, J = 17.4, 1.7 Hz, 1H, CH
CHH), 5.48 (dd, J = 17.4, 1.7 Hz, 1H, CH CHH), 5.24 (dd, J = 10.8, 1.6 Hz, 1H, CH
CHH), 5.24 (dd, J = 10.8, 1.6 Hz, 1H, CH CH2), 4.71 (irregular t, J ≈ 7 Hz, 1H, H-3′), 4.50 (ABq, J = 11.9 Hz, 2H, OCH2Ph), 3.68 (ABq, J = 11.5 Hz, 2H, CH2OSi), 2.46 to 2.60 (m, 2H, H-2′a,b), 0.98 [s, 9H, SiC(CH3)3]; 13C NMR (100 MHz, CDCl3) δ 156.55, 152.80, 149.14, 139.85, 138.81, 136.70, 128.66, 127.85, 127.79, 119.65, 115.00, 89.76, 82.98, 79.33, 71.69, 65.37, 36.95; FAB-MS m/z 368.2 (71.1%) [M + H]+. Analysis calculated for C19H21N5O3: C, 62.11; H, 5.76; N, 19.06. Found: C, 62.02; H, 5.72; N, 18.97.
CH2), 4.71 (irregular t, J ≈ 7 Hz, 1H, H-3′), 4.50 (ABq, J = 11.9 Hz, 2H, OCH2Ph), 3.68 (ABq, J = 11.5 Hz, 2H, CH2OSi), 2.46 to 2.60 (m, 2H, H-2′a,b), 0.98 [s, 9H, SiC(CH3)3]; 13C NMR (100 MHz, CDCl3) δ 156.55, 152.80, 149.14, 139.85, 138.81, 136.70, 128.66, 127.85, 127.79, 119.65, 115.00, 89.76, 82.98, 79.33, 71.69, 65.37, 36.95; FAB-MS m/z 368.2 (71.1%) [M + H]+. Analysis calculated for C19H21N5O3: C, 62.11; H, 5.76; N, 19.06. Found: C, 62.02; H, 5.72; N, 18.97.
(vii) (3S,5R)-5-(6-Amino-9H-purin-9-yl)-2-ethyl-2-(hydroxymethyl)tetrahydrofuran-3-ol (compound 2).
A room temperature solution of compound 11 (0.270 g, 0.74 mmol) in anhydrous methanol (75 ml) was circulated through a 10% Pd/C cartridge in an H-Cube (Thales) hydrogenator at 50 lb/in2 (flow rate, 2 ml/min) for 18 h. The solvent was removed in vacuo to give 0.237 g (87.8%) of crude intermediate. A total of 0.377 g (1.0 mmol) that was accumulated after this operation was repeated was dissolved in 3.4% formic acid in methanol (200 ml) and flushed with argon before palladium black (200 mg) was added. The reaction mixture was stirred at room temperature until thin-layer chromatography (silica gel; dichloromethane-methanol, 7:1) showed product, some starting material, and adenine from depurination (24 h). The reaction mixture was filtered, the solvent was removed in vacuo, and the crude product was purified by flash chromatography (silica gel) using dichloromethane-methanol (from 20:1 to 4:1) to give 0.133 g (35.3%) of starting material and 0.125 g of product. This product was further purified using a C18 packed column eluted with water and then water-methanol (from 20:1 to 3:1). The methanol was removed and the aqueous solution was lyophilized to give a fluffy solid that was triturated with ether-hexanes (1:1.6) to give 0.117 g (41%) of compound 2 as a white solid. Melting point, 98 to 99°C; 1H NMR (400 MHz, DMSO-d6) δ 8.28 (s, 1H, H-8), 8.07 (s, 1H, H-2), 7.23 (br s, 2H, NH2), 6.25 (dd, J = 7.8, 6.0 Hz, 1H, H-1′), 5.15 (dd, J = 6.6, 4.8 Hz, 1H, CH2OH), 5.11 (d, J = 4.7 Hz, 1H, OH), 4.37 (m, 1H, H-3′), 3.48 (dd, J = 11.5, 4.8 Hz, 1H, CHHOH), 3.37 (dd, J = 11.5, 6.6 Hz, 1H, CHHOH), 2.83 (m, 1H, H-2′a), 2.21 (2 times irregular q, J ≈ 3.0 Hz, 1H, H-2′b), 1.58 (m, 2H, CH2CH3), 0.83 (t, J = 7.41 Hz, 3H, CH2CH3); 13C NMR (100 MHz, DMSO-d6) δ 155.51, 152.72, 152.68, 149.28, 139.99, 119.68, 113.35, 90.21, 83.49, 83.37, 72.16, 64.27, 24.20, 8.71; FAB-MS m/z 280.1 (61.9%) [M + H]+. Analysis calculated for C12H17N5O3 · 0.5H2O: C, 50.01; H, 6.26; N, 24.30. Found: C, 49.90; H, 6.27; N, 24.01.
Cell-based assays.
The human embryonal kidney cell line 293T was obtained from the American Type Culture Collection (ATCC). The human osteosarcoma cell line HOS was obtained from Richard Schwartz (Michigan State University, East Lansing, MI). The HeLa cell-derived TZM-bl cell line was obtained from the NIH AIDS Research and Reference Reagent Program. Cell lines were maintained in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with 5% (vol/vol) fetal bovine serum, 5% newborn calf serum, and penicillin (50 units/ml) plus streptomycin (50 μg/ml) (Quality Biological, Gaithersburg, MD). Vesicular stomatitis virus G protein (VSV-G)-pseudotyped HIV vectors were produced by transfection of 293T cells. On the day prior to transfection, 293T cells were plated in 100-mm dishes at a density of 9 × 105 cells per plate. 293T cells were cotransfected with 10 μg of pNLNgoMIVR+ΔEnv.LUC (wild-type or NRTI resistance mutant) and 3 μg of pHCMV-G (obtained from Jane Burns, University of California San Diego) using calcium phosphate precipitation (24). After 48 h, virus-containing supernatants were harvested, clarified by low-speed centrifugation and filtration, and diluted 1 to 5 in preparation for infection assays. HOS (or TZM-bl) cells were plated in 96-well luminescence cell culture plates at a density of 4,000 cells in 100 μl per well on the day prior to infection. On the day of infection, cells were pretreated with the target compounds for 3 h. Infections were carried out by adding 100 μl of virus-containing supernatants to each well and incubating for 48 h. Infectivity was determined using a luciferase reporter assay (27). Luciferase activity was activated using a Steadylite plus regent kit (PerkinElmer, Waltham, MA); luminescence was measured using a microplate reader. Activity was normalized to infections done in the absence of target compounds for the appropriate NRTI variant. Regression analysis on the data was performed using a 4-parameter sigmoidal binding model, f(x) = a + b/[1 + (x/c)d], and 50% effective inhibitory concentrations (EC50s) were determined from the fit. For a sigmoidal hill binding curve, f(x) is the mathematical function represented by the equation, a represents the slope of the baseline, b represents the difference between the baseline and the maximum, c is the EC50 value, and d is the hill coefficient.
Cytotoxic effects were determined by measuring the effects of the compounds on the ATP levels in the cells. ATP levels were measured after incubation of HOS (or TZM-bl) cells with the target compounds for 48 h. The level of ATP was determined with a PerkinElmer ATPlite kit. Luminescence data were normalized to HOS cell data in the absence of target compound. The data were fit as described above, and 50% cytotoxic concentrations (CC50s) were determined from the fit.
Real-time PCR.
Real-time PCR was performed on DNA purified from infected cells at various times over the first 24 h after infection. Primers and probes used for the PCRs were as previously described and correspond to the following regions: RU5, minus-strand transfer, Gag, and plus-strand transfer (16).
Polymerase extension assay.
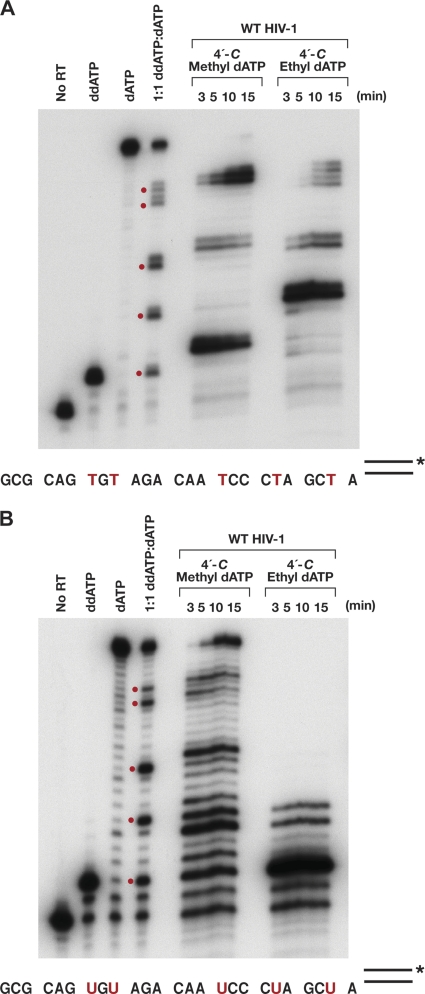
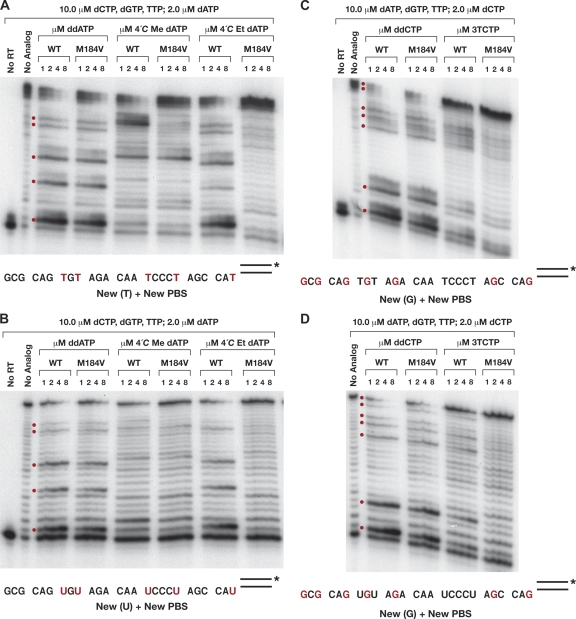
The expression and purification of HIV-1 RT and the polymerase assays were done as previously described. For the extension assays, 7.0 μl of a 2.0 μM stock of synthetic DNA oligonucleotide (5′-CAGGTCACTGTTCGAGCACCA-3′; Biosource, CA) was 5′ end labeled and then annealed to an excess of either a DNA template (5′-GCGCAGTGTAGACAATCCCTAGCTATGGTGCTCGAACAGTGACCTG-3′) or an RNA template (5′-GCGCAGUGUAGACAAUCCCUAGCUAUGGUGCUCGAACAGUGACCUG-3′). The underlined A residue is the first nucleotide past the 3′ end of the primer. The annealed template-primer (T/P) was suspended in a final concentration of 25.0 mM Tris (pH 8.0), 75 mM KCl, 8.0 mM MgCl2, 100.0 μg/ml bovine serum albumin (BSA), 10.0 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), and 10.0 μM (each) dCTP, dGTP, and TTP. For the RNA template reactions, the mixture was also supplemented with 1 U/μl of SuperAsin (Ambion). The reaction buffer was aliquoted into individual tubes and then supplemented with a total of 10.0 μM dATP and/or an ATP analog (as described in the legends to Fig. 6 and 8). The reactions were initiated by the addition of 1.0 μg RT, and the reaction mixtures were incubated at 37°C for various times, as indicated in Fig. 5. Reactions were stopped by the addition of EDTA, and the nucleic acids were precipitated by the addition of isopropyl alcohol. The pellets were resuspended in formamide gel loading buffer II (Ambion), heated at 70°C for 5 min to denature the reaction products, and fractionated on a 15% polyacrylamide sequencing gel.
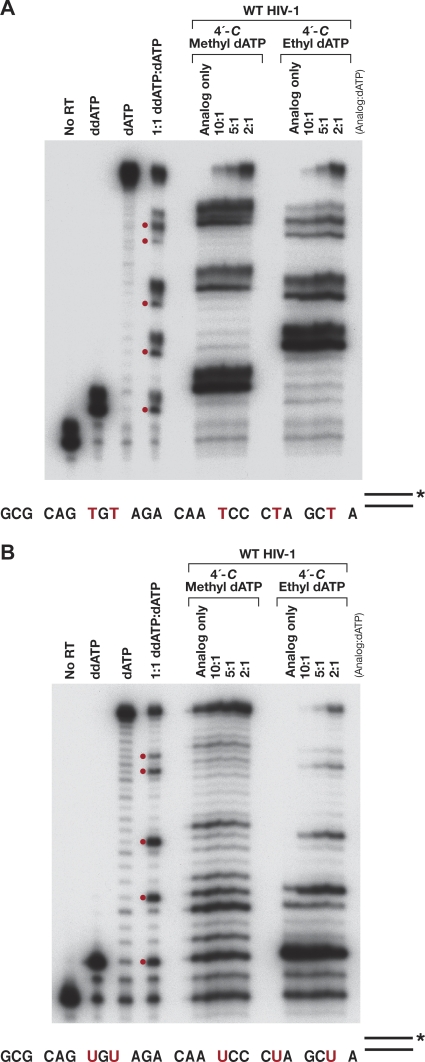
Fig. 6.
In vitro DNA polymerase assays using purified recombinant HIV-1 RT, various ratios of 4′-C-methyl- or 4′-C-ethyl-2′-dATP to dATP, a labeled DNA primer, and either a DNA template (A) or an RNA template (B). Polymerase assays done in the presence of ddATP show the expected DNA products resulting from immediate chain termination, while the assays done in the presence of dATP show the fully extended DNA products. Bands in the lane containing equimolar amounts of ddATP and dATP show a partial block to DNA synthesis caused by the incorporation of ddATP. The red dots correspond to positions where dATP or a dATP analog could be incorporated. The corresponding positions in the sequence are indicated by red Ts and red Us.
Fig. 5.
Time course of in vitro DNA polymerase assays performed using purified recombinant HIV-1 RT, 10.0 μM dCTP, dGTP, and TTP, 10.0 μM analog and/or dATP (ddATP, ddATP-dATP, 4′-C-methyl- or 4′-C-ethyl-2′-dATP), a labeled DNA primer, and either a DNA template (A) or an RNA template (B). Polymerase assays done in the presence of ddATP show the expected DNA products resulting from immediate chain termination, while the assays done in the presence of dATP lane show fully extended DNA products. Bands in the lane containing equimolar amounts of ddATP and dATP show a partial block to DNA synthesis caused by the incorporation of ddATP. Reactions were done at 37°C for the times indicated. The red dots correspond to positions where dATP or a dATP analog could be incorporated. The corresponding positions in the sequence are indicated by red T's and red U's.
Analog inhibition assay.
The analog inhibition assay is similar to that described above. For the extension assays, the DNA oligonucleotide (5′-CAGGTCACTGTTCGAGCACCA-3′; Biosource, CA) was 5′ end labeled and then annealed to an excess of either a DNA template (5′-GCGCAGTGTAGACAATCCCTAGCTATGGTGCTCGAACAGTGACCTG-3′) or an RNA template (5′-GCGCAGUGUAGACAAUCCCUAGCUAUGGUGCUCGAACAGUGACCUG-3′). The underlined A residue is the first nucleotide past the 3′ end of the primer. The annealed T/P was suspended in a final concentration of 25.0 mM Tris (pH 8.0), 75 mM KCl, 8.0 mM MgCl2, 100.0 μg/ml BSA, 10.0 mM CHAPS, and 10.0 μM (each) dCTP, dGTP, and TTP. The reaction buffer was aliquoted into individual tubes and then supplemented with 10.0 μM analog only or a mixture of analog and dATP (final concentration, 10 μM) at analog/dATP ratios of 10:1, 5:1, and 2:1. The reactions were initiated by the addition of 1.0 μg RT, and the reaction mixtures were incubated at 37°C for 10 min. Reactions were stopped by the addition of EDTA, and the nucleic acids were precipitated by the addition of isopropyl alcohol. The pellets were resuspended in formamide gel loading buffer II (Ambion), heated at 70°C for 5 min to denature the reaction products, and fractionated on a 15% polyacrylamide sequencing gel.
Analog inhibition assay (WT versus M184V).
The analog inhibition assay is similar to the assays described above. The DNA oligonucleotide (5′-CAGGTCACTGTTCGAGCACCA-3′; Biosource, CA) was 5′ end labeled and then annealed to an excess of either a DNA template (5′-GCGCAGTGTAGACAATCCCTAGCATTGGTGCTCGAACAGTGACCTG-3′) or an RNA template (5′-GCGCAGUGUAGACAAUCCCUAGCAUUGGUGCUCGAACAGUGACCUG-3′). The underlined T (or U) residue is the first nucleotide past the 3′ end of the primer. The annealed T/P was suspended in a final concentration of 25.0 mM Tris (pH 8.0), 75 mM KCl, 8.0 mM MgCl2, 100.0 μg/ml BSA, 10.0 mM CHAPS, 10.0 μM (each) dCTP, dGTP, and TTP, and 2.0 μM dATP. The reaction buffer was aliquoted into individual tubes and then supplemented with various concentrations of analog (1.0, 2.0, 4.0, and 8.0 μM). The reactions were initiated by the addition of 1.0 μg RT, and the reaction mixtures were incubated at 37°C for 5 min. Reactions were stopped by the addition of EDTA, and the nucleic acids were precipitated by the addition of isopropyl alcohol. The pellets were resuspended in formamide gel loading buffer II (Ambion), heated at 70°C for 5 min to denature the reaction products, and fractionated on a 15% polyacrylamide sequencing gel.
RESULTS
Synthesis of compounds 1 and 2.
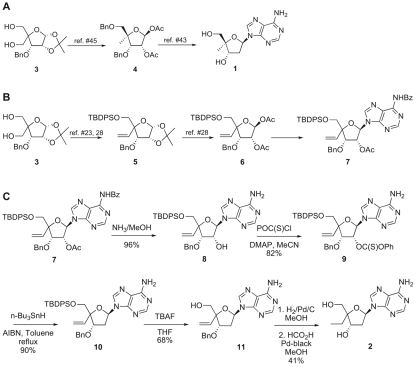
The key intermediate, compound 3, was prepared from commercially available 1,2:5,6-di-O-isopropylidene-α-d-allofuranose as reported by Youssefyeh et al. (45), After four steps, according to the method of Waga et al. (42), we obtained compound 4. Coupling with N6-benzoyladenine, followed by the removal of the 2′-OH via radical deoxygenation and final deprotection, gave target compound 1, as reported by Waga et al. (43) (Fig. 2A).
Fig. 2.
Schematic representation of the synthesis of 4′-C-methyl-2′-deoxyadenosine (compound 1) (A) and 4′-C-ethyl-2′-deoxyadenosine (compound 2) (B and C) from the common intermediate, compound 3. TBDPS, tertiarybutyldiphenylsilyl; Bz, benzoyl; MeOH, methanol; OC(S)CI, chlorocarbonothioyl; MeCN, acetonitrile.
Starting from the same intermediate, compound 3, selective protection of the less hindered primary alcohol as the tert-butyldiphenylsilyl ether, according to Obika et al. (23), allowed the selective oxidation of the remaining primary alcohol to the aldehyde, which, after Wittig olefination, as reported by Rangam et al. (28), yielded compound 5. After the exchange of protecting groups, as described by Rangram et al. (28), we obtained the desired sugar precursor, compound 6, which was readily condensed with N6-benzoyladenine to give the new compound, compound 7 (Fig. 2B). From compound 7, the rest of the sequence involved the removal of the 2′ functionality via radical deoxygenation, as shown in Fig. 2C, to give intermediate compound 10. The silyl ether was removed with TBAF, and the 4′-C-vinyl group was reduced to the desired ethyl group. Finally, removal of the 3′-O-benzyl ether protection by catalytic transfer hydrogenation afforded the second target compound, compound 2.
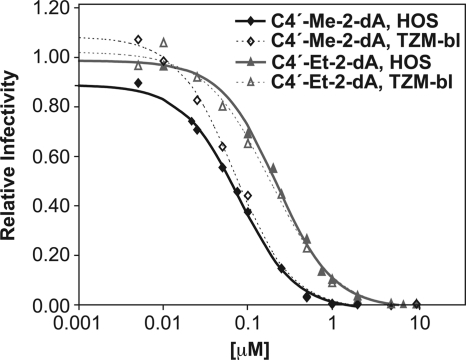
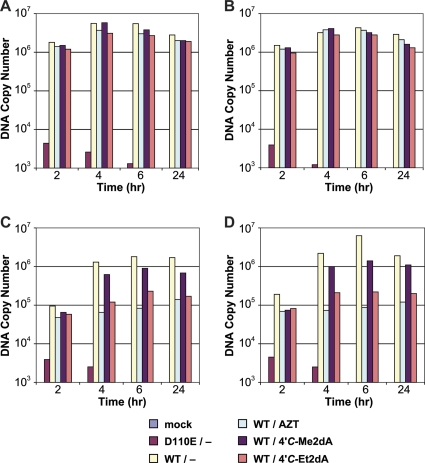
We wanted to compare the ability of 4′-C-methyl- and 4′-C-ethyl-2-deoxyadenosine to inhibit the replication of WT and NRTI-resistant HIV-1 to the corresponding 4′-C-methyl- and 4′-C-ethyl-2′-thymidines in HOS and TZM-bl cells using a single-cycle infectivity assay. We used an HIV-1-based vector, which had a deletion in the Env-coding region and a luciferase reporter in the Nef-coding region, pseudotyped with VSV-g (24). Luciferase is produced only if viral DNA is synthesized by RT and is then successfully integrated into the host genome. Both compounds inhibited HIV-1 replication in the cell lines tested (Fig. 3) with submicromolar EC50s (Table 1). Both 4′-C-methyl- and 4′-C-ethyl-2-deoxyadenosine have favorable EC50s (measured in a one-round assay in cultured cells) compared to those of our previous compound, d-carba thymidine, at 3.2 μM (6), and the approved NRTI tenofovir at 1 μM (data not shown), although they are less potent than AZT at 1.4 nM (6). In previous work, we found that related compounds were not effective in blocking HIV replication unless the cells also expressed the HSV TK. This suggests that the previous compounds were poorly phosphorylated by host cell kinases. We infer, from the fact that the compounds that we describe in this report effectively inhibited viral replication in cells that do not express HSV TK, that the new compounds are reasonably well phosphorylated in cultured cells. The proposal that the 4′-C-methyl- and 4′-C-ethyl-2-deoxyadenosine compounds are phosphorylated is supported by the fact that the 4′-C-ethynyl-2-fluoro-2′-deoxyadenosine (4′-C-E-2FdA) compound is efficiently phosphorylated (22). A previous report by Ohrui and Mitsuya indicated that 4′-C-methyl-2′-deoxyadenosine was cytotoxic in MT-4 cells and, for that reason, was not a good drug candidate (26); however, we found, using a relatively sensitive assay for cytotoxity, that these compounds showed good selectivity (Table 1). The fact that the compounds are active and are known to block reverse transcription reactions in vitro (described below) suggested that the compounds act by inhibiting viral DNA synthesis; however, we wanted to rule out the possibility that the inhibitors could block some other step in the viral life cycle. To determine the effect(s) of the inhibitors on HIV-1 reverse transcription in cell culture, we isolated DNA from drug-treated infected cells and used real-time PCR to determine the amounts of viral DNA corresponding to initiation (RU5), minus-strand transfer (U3), late-first strand products (Gag), and plus-strand transfer products present in the infected cells at various times after infection. The real-time PCR analysis shows that there is no significant difference in the amounts of the RU5 DNA in treated and untreated cells (Fig. 4). However, cells treated with 4′-C-ethyl-2′-deoxyadenosine produced smaller amounts of the later-stage viral DNA products than untreated infected cells. The decreases seen in the amounts of the viral DNAs present in the infected cells corresponded to the step in the reverse transcription process: the effects of the compound increased as viral DNA synthesis progressed. The results with the 4′-C-ethyl-2′-deoxyadenosine were quite similar to what was seen when the cells are treated with AZT and suggest that, like AZT, this compound blocks viral DNA synthesis in cells infected with the HIV-1 vectors. The ethyl derivative appears to have a greater effect on the synthesis of the minus-strand DNA (which is what is measured in this assay) than the methyl derivative. The methyl derivative still has a significant impact on minus-strand DNA synthesis (the data in Fig. 4 are shown on a log scale); however, it is possible that a portion of the effect of the methyl compound is on the synthesis of the second (plus) DNA strand. In order to show that incorporation of these compounds can block viral DNA synthesis, we performed DNA extension assays using purified recombinant HIV-1 RT and the triphosphate forms of 4′-C-methyl- and 4′-C-ethyl-deoxyadenosine.
Fig. 3.
Relative infectivity of a one-round HIV-1 vector that replicates using wild-type RT in HOS or TZM-bl cells pretreated with various concentrations of either 4′-C-methyl- or 4′-C-ethyl-2′-deoxyadenosine.
Table 1.
Anti-HIV-1 efficacy and cytotoxicity of 4′C-methyl- and -ethyl-2′-deoxyadenosine
| Compound and cell line | EC50 (nM) | CC50 (μM) | Selective indexa |
|---|---|---|---|
| 4′-C-Me-2′-dA | |||
| HOS | 63 ± 7 | 59 ± 5 | 936 |
| TZM-bl | 69 ± 4 | 169 ± 22 | 2,550 |
| 4′-C-Et-2′-dA | |||
| HOS | 221 ± 3 | 369 ± 22 | 1,669 |
| TZM-bl | 184 ± 20 | 204 ± 28 | 1,109 |
Selective index = CC50/EC50.
Fig. 4.
Viral DNA synthesis in HIV-1-infected cells, measured by real-time PCR. Infections were carried out using a vector that replicates using wild-type RT (the active-site mutant D110E was included as a negative control) in the presence or absence of an inhibitory compound. Results for infections carried out in the absence of an inhibitor are shown. The inhibitors tested were 4′-C-methyl-2′-deoxyadenosine, 4′-C-ethyl-2′-deoxyadenosine, and AZT. Mock transfections and the D110E RT mutant were used as DNA-negative controls. DNA products corresponding to initiation (A), minus-strand transfer (B), plus-strand transfer (C), and Gag (D) are shown.
Our previous work using 4′-C-alkylated thymidine analogs indicated that the methyl analog was neither an immediate nor a delayed chain terminator but was a kinetic chain terminator, whereas the ethyl analog acted, despite the presence of a 3′-OH, as a conventional (immediate) chain terminator (2). We examined the ability of recombinant HIV-1 RT to extend a DNA primer using a DNA or an RNA template in the presence of the triphosphate form of the analog 4′-C-Me-dATP either alone (Fig. 5) or in the presence of normal dATP (Fig. 6). Control reactions were done using an immediate chain terminator ddATP, normal dATP, and an equimolar mixture of both. The ddATP lane showed that the addition of a single nucleotide, which is the expected product when only an immediate chain terminator is present in the reaction mixture in the presence of the normal dATP HIV-1 RT, extended the DNA primer to the end of the template. The 1:1 mixture of ddATP and dATP caused a partial block to DNA synthesis at every site where the dideoxynucleotide could be incorporated. If 4′-C-Me-dATP was present, the template was DNA, and if no dATP was present, there was a pause during DNA synthesis immediately after the analog was incorporated; however, at longer times, the DNA could be extended (Fig. 5A). When 4′-C-Et-dATP was used instead of dATP, strong pausing was not observed at the first site where the analog was incorporated; however, a strong pause is observed at the second incorporation site, with longer DNA products produced at longer times. The in vitro incorporation assays showed that neither 4′-C-Me- nor 4′-C-Et-dATP behaves strictly as an immediate chain terminator and that RT is able to extend the DNA chain.
Polymerase assays performed using an RNA template showed greater differences in the activities of 4′-C-Me- and 4′-C-Et-dATP relative to those shown by assays done with a DNA template (Fig. 5B). When the substrate was 4′-C-Me-dATP, pausing was usually observed at points just after the analog was incorporated. The block to DNA synthesis at the first incorporation site was much less intense on the RNA template than the DNA template. In addition, there was a partial block to DNA synthesis just before the second incorporation site. The presence of this band on the gel suggests that the analog was added slowly at this position. However, once the analog was incorporated, there was a second pause after the analog was incorporated. There were additional pauses at the other positions where the analog was incorporated, indicating that RT has trouble adding a normal dNTP after the 4′-C-methyl analog has been incorporated. When the substrate was 4′-C-Et-dATP and the template was RNA, a very strong band was observed after the first site where the analog was incorporated. However, some of the DNAs were extended through to the second incorporation site, similar to what was seen with 4′-C-Me-dATP and the RNA template, but the second band was of lower intensity and there was no significant accumulation of full-length products; these results are not consistent with the properties expected for an immediate chain terminator. The data show that when 4′-C-Et-dATP is incorporated using an RNA template, the block to DNA synthesis was stronger than when the analog was incorporated using a DNA template, suggesting that the RNA template helps to create a configuration at the active site that is unfavorable for the addition of the next nucleotide triphosphate. In contrast, 4′-C-Me-dATP appears to block DNA synthesis more effectively with a DNA than an RNA template. Differences in the activities of the compounds on RNA versus DNA templates in vitro support real-time PCR data which show that the incorporation of 4′-C-Et-dA by HIV-1 RT causes a reduction in the amount of viral DNA produced but that the effect is smaller than the effect of 4′-C-Me-dA (and AZT) on viral DNA synthesis.
The data obtained with the HIV-1 vectors suggest that the 4′-C analogs can compete effectively with the normal substrate dATP. If 4′-C-Me- or 4′-C-Et-dATP is able to block the extension of a DNA primer by HIV-1 RT in the presence of normal dATP, then the pausing patterns seen in reactions that contain a mixture of normal dNTPs and the triphosphate forms of the analogs should be similar to what was seen when the inhibitors were used in the reactions done without any dATP. If, however, the analog is a relatively poor substrate, like south methanocarba-dATP, the pausing would be eliminated if normal dATP was present in the reaction mixture (3). Polymerase reactions were performed using various mixtures of dATP and 4′-C-Me-dATP or dATP and 4′-C-Et-dATP. The gel analyses show that 4′-C-Me-dATP and 4′-C-Et-dATP can compete with dATP in vitro (Fig. 6), and the data support the idea that the ability of the 4′-C-Me-dATP analog to block DNA synthesis depends on the nature of the template (DNA versus RNA). In the cultured cell assay, 4′-C-Et-dA effectively blocked first (minus)-strand DNA synthesis; this is the DNA strand that is copied from the viral genome's RNA. In vitro, 4′-C-Et-dATP can inhibit DNA polymerization when the template is either RNA or DNA. In contrast, 4′-C-Me-dATP is much more effective at blocking viral DNA synthesis in vitro when the template is DNA than RNA. This could explain the fact that the 4′-C-methyl derivative is also less potent in its ability to block minus-strand DNA synthesis in an infected cell and would suggest that the ability of this compound to block viral replication in infected cells may rest on its ability to block second (plus)-strand DNA synthesis.
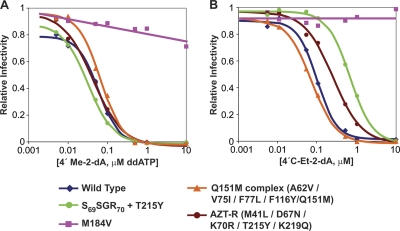
The gel data show that immediate chain termination does not occur with either 4′-C-Me- or 4′-C-Et-dATP. Because normal dNTPs are incorporated after the analogs, the analogs do not remain at the end of the primer strand, which should make it difficult for the virus to develop resistance to these compounds by the excision mechanism. This suggests that it is more likely that resistance would involve the exclusion mechanism. To test this conjecture, 4′-C-Me-2′-dA and 4′-C-Et-2′-dA were analyzed in infectivity assays using two excision mutants, SSGR-T125Y (T69K70/T215 → S69SGR70 + T215Y) and AZT-R (M41L/D67N/K70R/T215Y/K219Q), and two exclusion mutants, the Q151M complex (A62V/V75I/F77L/F116Y/Q151M) and M184V (Fig. 7). 4′-C-Methyl-2′-deoxyadenosine effectively inhibited vectors that replicate using WT RT or the excision-proficient RTs (Fig. 8; Table 2). Higher concentrations of 4′-C-ethyl-2′-deoxyadenosine were required to inhibit these mutant vectors. The Q151M complex caused an approximately 2-fold increase in EC50s for both compounds; however, neither compound effectively inhibited the M184V mutant (Table 2).
Fig. 7.
Relative infectivity of one-round HIV-1 vectors that replicate using wild-type or NRTI-resistant RTs in HOS cells in the presence of various concentrations of 4′-C-methyl-2′-deoxyadenosine (A) or 4′-C-ethyl-2′-deoxyadenosine (B). The compounds are effective against most of the NRTI-resistant mutants that we tested, with the exception of the M184V exclusion mutant.
Fig. 8.
In vitro DNA polymerase assays using purified recombinant HIV-1 RT (WT or the RT mutant M184V), 10.0 μM dCTP, dGTP, and TTP, 2.0 μM dATP, and increasing concentrations of ddATP, 4′-C-methyl-2′-dATP, or 4′-C-ethyl-2′-dATP (1.0, 2.0, 4.0, 8.0 μM) in the presence of a labeled DNA primer and either a DNA template (A) or an RNA template (B). PBS, phosphate-buffered saline. Polymerase assays done in the presence of ddATP show the expected DNA products resulting from immediate chain termination, while the assays done in the presence of dATP show the fully extended DNA products. Control reactions were done with both WT RT and the M184V mutant to compare the ability of ddCTP and 3TCTP to block DNA synthesis on either a DNA template (C) or an RNA template (D). As expected, both compounds interfered with DNA synthesis by WT RT; in contrast, the M184V mutant was resistant to 3TCTP. Reactions were done at 37°C for the times indicated. The red dots correspond to positions where dATP or a dATP analog could be incorporated. The corresponding positions of the sequence are indicated by red Ts and red Us.
Table 2.
Anti-HIV-1 efficacy of 4′-C-methyl- and 4′-C-ethyl-2′-deoxyadenosine against vectors with selected NRTI resistance mutations in HOS cells
| Strain | 4′-C-Me-2′-dA |
4′-C-Et-2′-dA |
||
|---|---|---|---|---|
| EC50 (nM) | Fold changea | EC50 (nM) | Fold changea | |
| Wild type | 63 ± 7 | 221 ± 3 | ||
| M184V | >10,000 | >150 | >10,000 | >45 |
| AZT-R | 64 ± 5 | 1.0 | 869 ± 55 | 3.9 |
| SSGR + T215Y | 25 ± 9 | 0.38 | 700 ± 100 | 3.2 |
| Q151M complex | 111 ± 8 | 1.7 | 520 ± 25 | 2.3 |
Versus wild type.
To better understand the resistance caused by the M184V mutation, we asked whether the analogs could be incorporated by either wild-type RT or the M184V mutant in reactions that contained the same concentration of normal dNTPs and increasing amounts of analog (Fig. 8). A control reaction using ddATP showed a decrease in the amount of the full-length product as the concentration of ddATP was increased. No difference was seen in reaction products synthesized in the presence of ddATP by wild-type RT and the M184V mutant. A control reaction using ddC triphosphate (ddCTP) also showed that the two enzymes made similar products. However, in the presence of increasing amounts of 3TC triphosphate (3TCTP), the M184V mutant, which is known to be 3TC resistant, made much larger amounts of the full-length product than wild-type RT (Fig. 8C and D). When a DNA template was used (Fig. 8A), 4′-C-Me-dATP competed with dATP in the reactions catalyzed by wild-type RT, blocking DNA synthesis. In the reaction catalyzed by M184V RT, there was less pausing, and an increased amount of the full-length product was made, indicating resistance. The M184V mutant was not inhibited by 4′-C-Et-dATP, even at high concentrations, consistent with high-level resistance. When an RNA template was used (Fig. 8B), 4′-C-Me-dATP was less able to block DNA synthesis in the presence of dATP in the reactions carried out by both the wild-type RT and the M184V mutant. On the other hand, 4′-C-Et-dATP still blocked DNA synthesis, even in the presence of dATP, in reactions catalyzed by wild-type RT on an RNA template, but 4′-C-Et-dATP was less effective when the M184V mutant was used in the reaction.
DISCUSSION
The simple interpretation of the in vitro data is that the 4′-modified dA analogs are efficiently incorporated at most sites, but once they are incorporated, the DNA is poorly extended, presumably because the presence of the 4′-alkyl group causes the 3′-OH to be out of position, thus making it difficult to add the next nucleotide. Ohrui and Mitsuya suggested that substitutions at the 4′-C position introduce a rotational constraint about the C-3′—C-4′ bond of the ribose which places the 3′-OH in a position that is unfavorable for reactivity (26). If the 4′-substituent is methyl, it would appear that this restriction is not sufficient to completely block DNA synthesis but, rather, serves as a kinetic barrier for DNA extension that is eventually bypassed. The gel data show that the addition of one normal nucleotide is sufficient to alleviate this distortion and restore the ability of RT to efficiently carry out efficient DNA polymerization.
The ability of DNA polymerases, including HIV-1 RT, to distinguish between normal dNTPs and dideoxynucleoside analogs depends on the sequence of the template. It is likely that this is due to subtle effects that the sequence context has on the structure of the nucleic acid at the polymerase active site. It should not be not surprising that the ability of delayed chain terminators to block DNA synthesis by HIV-1 RT is also sensitive to sequence context, a phenomenon that has previously been documented with the fixed-conformation analog north-methanocarba-dATP, which blocks DNA synthesis 2 nucleotides after it is incorporated (3). In those experiments, efficient read-through occurred when the U5-derived template strand contained the sequence CCCT. Altering the template sequence to CACT increased the amount of termination 2 nucleotides beyond the site at which the analog was incorporated. The addition of a second CCCT sequence elsewhere in the template also allowed HIV-1 RT to continue DNA synthesis, despite the incorporation of the analog. Because CCCT allowed read-through to occur with the north-methanocarba-dATP analog, we tested this sequence for its effect on termination by the 4′-C-methyl and 4′-C-ethyl analogs. Although the incorporation of the 4′-C-ethyl analog caused a block when CCCT was the template sequence, there was no corresponding block with the 4′-C-methyl analog (Fig. 5A).
Given these differences in the ability of the analogs to block DNA synthesis depending on the sequence of the template, it is not surprising that there are differences in the ability of the analogs to block DNA synthesis when a DNA template is compared to an RNA template with the same sequence. When the template strand was RNA, the incorporation of the 4′-C-ethyl analog caused a strong block to DNA synthesis at the first site where the analog was incorporated; however, there was no corresponding block on a DNA template. Although there are differences in the pattern of pausing seen with the 4′-C-methyl analog and the DNA and RNA templates, these differences are more subtle. The obvious difference is that in the presence of 4′-C-Me-dATP, no full-length product is made on the DNA template, although full-length product is made on the RNA template. These differences in the effects of incorporating the analog are presumably the result of the differences in the structure of the nucleic acid when the template strand was RNA or DNA (32). However, it is the ability of the compounds to cause a block to DNA synthesis that is the basis for their ability to inhibit HIV-1 replication in cell culture. The real-time PCR data obtained from experiments done in cell culture provide strong support for this interpretation and suggest that the 4′-C-ethyl compound is particularly potent in its ability to block minus-strand DNA synthesis, which matches its ability to block DNA synthesis from an RNA template in vitro. In contrast, the 4′-C-methyl compound, which is less potent in blocking DNA synthesis from an RNA template in vitro, also has a more modest effect on minus-strand DNA synthesis in an infected cell. It is possible that a significant portion of the impact of 4′-C-Me-dA on viral replication involves its ability to block the synthesis of the second (plus) strand.
The 4′-C-Me- and 4′-C-Et-2dA compounds are effective in blocking the synthesis of viruses that replicate using wild-type RT and RTs that carry several important NRTI resistance mutations in RT. As has already been discussed, this suggests that the compounds are efficiently phosphorylated in cultured cells. Inclusion of a 3′-OH group not only imparts diminished susceptibility to excision-proficient mutations in cell culture but also protects the compound from the exclusion mechanism mutations associated with the Q151M complex. However, the compounds failed to block the replication of vectors that have the M184V mutation. A 4′-C-substituted analog designed by Mitsuya and coworkers that maintains a 3′-OH group, 4′-C-ethynyl-2-fluoro-2′-deoxyadenosine, has favorable activity against viruses that replicate using WT RT and a number of NRTI-resistant mutants in cell culture (22, 25). In these assays, the RTs contained mutations that have been associated with both the excision and exclusion resistance mechanisms, including viruses that use RTs that contain the M184V mutation. We measured the ability of 4′-C-E-2FdA to inhibit HIV-1 infectivity in our single-cycle assay using HIV-1 vectors that replicate using either wild-type RT or an RT that carried the M184V mutation. The M184V mutation caused a 4-fold increase in the EC50 for 4′-C-E-2FdA relative to that for the wild-type virus (EC50s, 124 ± 5 and 31 ± 2 nM, respectively). This is consistent with what has been reported and shows that it is possible to develop a 4′-modified nucleoside that is relatively insensitive to the M184V mutation (22, 25) The CC50 of WT RT was 90 ± 25 μM, and the selective indexes (CC50/EC50) of WT RT and the RT that carried the M184V mutation were 2,903 and 725, respectively.
There is a concern that any nucleoside analog can be incorporated by a cellular DNA polymerase and that this might lead to toxicity. Both the 4′-C-methyl- and 4′-C-ethyl-dA compounds have reasonably good selectivity indexes in cultured cells. At least one nucleoside analog that retains a 3′-OH (entecavir) has been approved for the treatment of hepatitis B virus infections. It should be pointed out that administration of entecavir to hepatitis B virus-infected individuals who are coinfected with HIV leads to the selection of HIV-1 RT mutants; this suggests that entecavir is able to interfere with HIV replication (19).
This work describes the ability of the triphosphates of 4′-C-alkyl-dA compounds to inhibit RT polymerization in in vitro assays and the ability of the parental compounds to block the replication of HIV-1 vectors in cultured cells: 4′-C-methyl- and 4′-C-ethyl-dA show both efficacy and selectivity against HIV-1. The compounds are also effective against key viruses that replicate using RTs and that carry NRTI resistance mutations, with the exception of the M184V exclusion mutant. Analysis of viral DNA synthesis in infected cells showed that viral DNA synthesis is blocked by the incorporation of either 4′-C-methyl- or 4′-C-ethyl-2′-deoxyadenosine. In vitro experiments with purified HIV-1 RT showed that 4′-C-methyl- and 4′-C-ethyl-2′-dATP can compete with dATP and that incorporation of the analog causes pausing in DNA synthesis. Selection of resistance mutants using replication-competent HIV-1 in the presence of 4′-C-methyl- and 4′-C-ethyl-2′-deoxyadenosine is ongoing and will provide additional information on the mechanisms that can give rise to resistance to nucleotide analogs with 4′-C modifications.
ACKNOWLEDGMENTS
We thank Pat Clark for the preparation of purified enzymes and Hiroaki Mitsuya for the gift of the nucleotide analog 4′-C-ethynyl-2-fluoro-2′-deoxyadenosine.
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute. This research has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. N01-CO-12400. Research in S.H.H.'s laboratory was supported by the National Cancer Institute.
Footnotes
Published ahead of print on 22 February 2011.
REFERENCES
- 1. Arion D., Kaushik N., McCormick S., Borkow G., Parniak M. A. 1998. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry 37:15908–15917 [DOI] [PubMed] [Google Scholar]
- 2. Boyer P. L., et al. 2007. The nucleoside analogs 4′C-methyl thymidine and 4′C-ethyl thymidine block DNA synthesis by wild-type HIV-1 RT and excision proficient NRTI resistant RT variants. J. Mol. Biol. 371:873–882 [DOI] [PubMed] [Google Scholar]
- 3. Boyer P. L., Julias J. G., Marquez V. E., Hughes S. H. 2005. Fixed conformation nucleoside analogs effectively inhibit excision-proficient HIV-1 reverse transcriptases. J. Mol. Biol. 345:441–450 [DOI] [PubMed] [Google Scholar]
- 4. Boyer P. L., Sarafianos S. G., Arnold E., Hughes S. H. 2002. Nucleoside analog resistance caused by insertions in the fingers of human immunodeficiency virus type 1 reverse transcriptase involves ATP-mediated excision. J. Virol. 76:9143–9151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boyer P. L., Sarafianos S. G., Arnold E., Hughes S. H. 2001. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 75:4832–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boyer P. L., et al. 2009. The nucleoside analogue d-Carba T blocks HIV-1 reverse transcription. J. Med. Chem. 52:5356–5364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen C. H., Vazquez-Padua M., Cheng Y. C. 1991. Effect of anti-human immunodeficiency virus nucleoside analogs on mitochondrial DNA and its implication for delayed toxicity. Mol. Pharmacol. 39:625–628 [PubMed] [Google Scholar]
- 8. Choi Y., et al. 2003. A conformationally locked analogue of the anti-HIV agent stavudine. An important correlation between pseudorotation and maximum amplitude. J. Med. Chem. 46:3292–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Comin M. J., et al. 2008. d-(+)-Iso-methanocarbathymidine: a high-affinity substrate for herpes simplex virus 1 thymidine kinase. ChemMedChem 3:1129–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Das K., et al. 2009. Structural basis for the role of the K65R mutation in HIV-1 reverse transcriptase polymerization, excision antagonism, and tenofovir resistance. J. Biol. Chem. 284:35092–35100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao H. Q., Boyer P. L., Sarafianos S. G., Arnold E., Hughes S. H. 2000. The role of steric hindrance in 3TC resistance of human immunodeficiency virus type-1 reverse transcriptase. J. Mol. Biol. 300:403–418 [DOI] [PubMed] [Google Scholar]
- 12. Hayakawa H., et al. 2004. Potential of 4′-C-substituted nucleosides for the treatment of HIV-1. Antivir. Chem. Chemother. 15:169–187 [DOI] [PubMed] [Google Scholar]
- 13. Henry M., et al. 2006. Coexistence of the K65R/L74V and/or K65R/T215Y mutations on the same HIV-1 genome. J. Clin. Virol. 37:227–230 [DOI] [PubMed] [Google Scholar]
- 14. Huang H., Chopra R., Verdine G. L., Harrison S. C. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669–1675 [DOI] [PubMed] [Google Scholar]
- 15. Iversen A. K., et al. 1996. Multidrug-resistant human immunodeficiency virus type 1 strains resulting from combination antiretroviral therapy. J. Virol. 70:1086–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Julias J. G., Ferris A. L., Boyer P. L., Hughes S. H. 2001. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J. Virol. 75:6537–6546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Larder B. A., Kemp S. D. 1989. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT). Science 246:1155–1158 [DOI] [PubMed] [Google Scholar]
- 18. Marquez V. E., Hughes S. H., Sei S., Agbaria R. 2006. The history of N-methanocarbathymidine: the investigation of a conformational concept leads to the discovery of a potent and selective nucleoside antiviral agent. Antiviral Res. 71:268–275 [DOI] [PubMed] [Google Scholar]
- 19. McMahon M. A., et al. 2007. The HBV drug entecavir—effects on HIV-1 replication and resistance. N. Engl. J. Med. 356:2614–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meyer P. R., Lennerstrand J., Matsuura S. E., Larder B. A., Scott W. A. 2003. Effects of dipeptide insertions between codons 69 and 70 of human immunodeficiency virus type 1 reverse transcriptase on primer unblocking, deoxynucleoside triphosphate inhibition, and DNA chain elongation. J. Virol. 77:3871–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meyer P. R., Matsuura S. E., So A. G., Scott W. A. 1998. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl. Acad. Sci. U. S. A. 95:13471–13476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakata H., et al. 2007. Activity against human immunodeficiency virus type 1, intracellular metabolism, and effects on human DNA polymerases of 4′-ethynyl-2-fluoro-2′-deoxyadenosine. Antimicrob. Agents Chemother. 51:2701–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Obika S., et al. 2002. Synthesis and conformation of 3′,4′-BNA monomers, 3′-O-4′-C-methyleneribonucleosides. Tetrahedron 58:3039–3049 [Google Scholar]
- 24. Oh J., McWilliams M. J., Julias J. G., Hughes S. H. 2008. Mutations in the U5 region adjacent to the primer binding site affect tRNA cleavage by human immunodeficiency virus type 1 reverse transcriptase in vivo. J. Virol. 82:719–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ohrui H., et al. 2007. 2′-Deoxy-4′-C-ethynyl-2-fluoroadenosine: a nucleoside reverse transcriptase inhibitor with highly potent activity against wide spectrum of HIV-1 strains, favorable toxic profiles, and stability in plasma. Nucleosides Nucleotides Nucleic Acids 26:1543–1546 [DOI] [PubMed] [Google Scholar]
- 26. Ohrui H., Mitsuya H. 2001. 4′-C-substituted-2′-deoxynucleosides: a family of antiretroviral agents which are potent against drug-resistant HIV variants. Curr. Drug Targets Infect. Disord. 1:1–10 [DOI] [PubMed] [Google Scholar]
- 27. Petropoulos C. J., et al. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rangam G., Rudinger N. Z., Müller H. M., Marx A. 2005. Synthesis and application of 4′-C-alkylated uridines as probes for uracil-DNA glycosylase. Synthesis 9:1467–1472 [Google Scholar]
- 29. Ray A. S., et al. 2003. Probing the molecular mechanisms of AZT drug resistance mediated by HIV-1 reverse transcriptase using a transient kinetic analysis. Biochemistry 42:8831–8841 [DOI] [PubMed] [Google Scholar]
- 30. Sarafianos S. G., et al. 2002. Structures of HIV-1 reverse transcriptase with pre- and post-translocation AZTMP-terminated DNA. EMBO J. 21:6614–6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sarafianos S. G., et al. 1999. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids. Proc. Natl. Acad. Sci. U. S. A. 96:10027–10032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sarafianos S. G., et al. 2001. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 20:1449–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sarafianos S. G., Hughes S. H., Arnold E. 2004. Designing anti-AIDS drugs targeting the major mechanism of HIV-1 RT resistance to nucleoside analog drugs. Int. J. Biochem. Cell Biol. 36:1706–1715 [DOI] [PubMed] [Google Scholar]
- 34. Sarafianos S. G., et al. 2009. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 385:693–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sarafianos S. G., Pandey V. N., Kaushik N., Modak M. J. 1995. Glutamine 151 participates in the substrate dNTP binding function of HIV-1 reverse transcriptase. Biochemistry 34:7207–7216 [DOI] [PubMed] [Google Scholar]
- 36. Schinazi R. F., Larder B. A., Mellors J. W. 2000. Mutations in retroviral genes associated with drug resistance: 2000-2001 update. Int. Antiviral News 8:65–91 [Google Scholar]
- 37. Schuurman R., et al. 1995. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC). J. Infect. Dis. 171:1411–1419 [DOI] [PubMed] [Google Scholar]
- 38. Shirasaka T., et al. 1995. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc. Natl. Acad. Sci. U. S. A. 92:2398–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Takebe Y., Telesnitsky A. 2006. Evidence for the acquisition of multi-drug resistance in an HIV-1 clinical isolate via human sequence transduction. Virology 351:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tu X., et al. 2010. Structural basis of HIV-1 resistance to AZT by excision. Nat. Struct. Mol. Biol. 17:1202–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tuske S., et al. 2004. Structures of HIV-1 RT-DNA complexes before and after incorporation of the anti-AIDS drug tenofovir. Nat. Struct. Mol. Biol. 11:469–474 [DOI] [PubMed] [Google Scholar]
- 42. Waga T., Nishizaki T., Miyakawa I., Ohrui H., Meguro H. 1993. Synthesis of 4′-C-methylnucleosides. Biosci. Biotechnol. Biochem. 57:1433–1438 [DOI] [PubMed] [Google Scholar]
- 43. Waga T., Ohrui H., Meguro H. 1996. Synthesis and biological evaluation of 4′-C-methyl nucleosides. Nucleosides Nucleotides 15:287–304 [Google Scholar]
- 44. Winters M. A., et al. 1997. Human immunodeficiency virus type 1 reverse transcriptase genotype and drug susceptibility changes in infected individuals receiving dideoxyinosine monotherapy for 1 to 2 years. Antimicrob. Agents Chemother. 41:757–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Youssefyeh R. D., Verheyden J. P. H., Moffatt J. G. 1979. 4′-Substituted nucleosides. 4. Synthesis of some 4′-hydroxymethyl nucleosides. J. Org. Chem. 44:1301–1309 [Google Scholar]