Abstract
In Streptococcus pyogenes, inducible erythromycin (ERY) resistance is due to posttranscriptional methylation of an adenine residue in 23S rRNA that can be encoded either by the erm(B) gene or by the more recently described erm(TR) gene. Two erm(TR)-carrying genetic elements, showing extensive DNA identities, have thus far been sequenced: ICE10750-RD.2 (∼49 kb) and Tn1806 (∼54 kb), from tetracycline (TET)-susceptible strains of S. pyogenes and Streptococcus pneumoniae, respectively. However, TET resistance, commonly mediated by the tet(O) gene, is widespread in erm(TR)-positive S. pyogenes. In this study, 23 S. pyogenes clinical strains with erm(TR)-mediated ERY resistance—3 TET susceptible and 20 TET resistant—were investigated. Two erm(TR)-carrying elements sharing only a short, high-identity erm(TR)-containing core sequence were comprehensively characterized: ICESp1108 (45,456 bp) from the TET-susceptible strain C1 and ICESp2905 (65,575 bp) from the TET-resistant strain iB21. While ICESp1108 exhibited extensive identities to ICE10750-RD.2 and Tn1806, ICESp2905 showed a previously unreported genetic organization resulting from the insertion of separate erm(TR)- and tet(O)-containing fragments in a scaffold of clostridial origin. Transferability by conjugation of the erm(TR) elements from the same strains used in this study had been demonstrated in earlier investigations. Unlike ICE10750-RD.2 and Tn1806, which are integrated into an hsdM chromosomal gene, both ICESp1108 and ICESp2905 shared the chromosomal integration site at the 3′ end of the conserved rum gene, which is an integration hot spot for several mobile streptococcal elements. By using PCR-mapping assays, erm(TR)-carrying elements closely resembling ICESp1108 and ICESp2905 were shown in the other TET-susceptible and TET-resistant test strains, respectively.
INTRODUCTION
In Streptococcus pyogenes, erythromycin (ERY) resistance is due to two principal mechanisms: target site modification or active efflux (13, 29). The latter is normally mediated by the mef(A) gene, adjacent to an msr gene usually designated msr(D), and is associated with low-level resistance to 14- and 15-membered macrolides only (M phenotype). Conversely, target site modification generally consists in posttranscriptional methylation of an adenine residue in 23S rRNA caused by erm gene-encoded methylases and is associated with either constitutive (cMLS phenotype) or inducible (iMLS phenotype) coresistance to macrolide, lincosamide, and streptogramin B (MLS) antibiotics. While cMLS isolates are rather homogeneous in susceptibility patterns and their methylase gene is normally erm(B), iMLS isolates are more heterogeneous, and their methylase gene is either erm(B) or an erm(A) subclass commonly referred to as erm(TR) (26).
Until the present study, only two erm(TR)-carrying genetic elements had been completely sequenced: an integrative and conjugative element (ICE), designated ICE10750-RD.2 (∼49 kb), from the sequenced genome of a S. pyogenes strain (accession no. CP000262) (2), and a genetic element from Streptococcus pneumoniae, a species where erm(TR) is very uncommon, which was designated Tn1806 (∼54 kb, accession no. EF469826) (6). There are substantial identities between ICE10750-RD.2 and Tn1806, despite the occurrence of open reading frames (ORFs) unique to either element. Furthermore, both elements are integrated into an hsdM chromosomal gene.
Before ICE10750-RD.2 and Tn1806 were detected, we had demonstrated that the erm(TR) gene from inducibly resistant S. pyogenes donors could be transferred by conjugation to susceptible recipients of S. pyogenes and other Gram-positive species (9). Intraspecific transfer was associated with the insertion of a new DNA fragment whose size was dependent on the donor, suggesting that erm(TR) could be carried by different genetic elements. Partial sequencing of the transposable element from one of these donors (accession no. FM162351) enabled us to compare its erm(TR)-flanking region with those of ICE10750-RD.2 and Tn1806 and to document extensive similarities to both elements (29). Remarkably, other antibiotic (tetronasin and spectinomycin) resistance genes were found in the erm(TR)-flanking regions of the three elements.
The three available sequences of erm(TR)-carrying elements mentioned above are all from tetracycline (TET)-susceptible strains. However, TET resistance is widespread in erm(TR)-positive S. pyogenes (10, 11, 16, 19). In previous studies, we demonstrated the association of erm(TR) with tet(O) in ERY and TET coresistant S. pyogenes isolates and the cotransfer of the two resistance determinants, but an actual genetic linkage could not be proved (8).
In the present study, in addition to completing the sequencing, characterization, and comparative analysis of the erm(TR)-carrying element (designated ICESp1108) from our TET-susceptible strain, we demonstrate a completely different genetic element (designated ICESp2905), also carrying the tet(O) gene, in TET-resistant S. pyogenes isolates with erm(TR)-mediated ERY resistance. The two new elements were comprehensively characterized.
MATERIALS AND METHODS
Bacterial strains.
Twenty-three strains of S. pyogenes were used, all isolated from throat cultures of symptomatic patients and collected from Italian laboratories in the decade from 1998 to 2007. Strain identification was confirmed with bacitracin disks (Oxoid, Basingstoke, England) and by serogroup A agglutination (Streptex; Murex, Chatillon, France). The inclusion criterion was ERY resistance (MIC, ≥1 μg ml−1) mediated by the erm(TR) gene (determined by PCR using specific primers [12]). All isolates exhibited the iMLS phenotype. Twenty were TET resistant (MIC range, 64 to >128 μg ml−1), with resistance mediated by the tet(O) gene (determined by PCR using specific primers [17]), and three were TET susceptible (MIC range, ≤0.125 to 0.5 μg ml−1). Two test strains, both described in previous studies, were used for sequencing experiments: TET-susceptible C1 (9, 29) and TET-resistant iB21 (8), also called B2 in an earlier report (9).
PCR experiments.
The principal oligonucleotide primer pairs used in PCR experiments are listed in Table 1. Inverse PCR (23) was carried out to analyze unknown DNA regions. Genomic DNA digested with endonucleases MunI, HindIII (Roche Applied Science, Basel, Switzerland), BanI, Hpy188I, or AclI (New England Biolabs, Ipswich, MA) was ligated and used as the template in the PCR assays.
Table 1.
Principal oligonucleotide primer pairs used
| Procedure and genea | Primer | Sequence (5′–3′) | Source or reference | Product size (bp) |
|---|---|---|---|---|
| ICESp1108 mapping | ||||
| orf1 | ETR1 | GGGATAGGACTGATTGAA | This study | 10,110 |
| orf14 | ETR89 | GTGTTATACCTGTTGGAATACC | 6 | |
| orf14 | ETR99 | GGTATTCCAACAGGTATAACAC | 6 | 8,093 |
| orf20 | ETR94 | CTGTTCTTGGATATGTGATTAGC | 6 | |
| orf20 | ETR94-for | TTGGCTGGTAGGAATGAAT | This study | 6,991 |
| orf22 | ETR50-rev | TTTGAGTGGTAAGATGGTT | This study | |
| orf22 | ETR50 | TATGGTACAAGTAGAGTGAATGCC | 6 | 8,731 |
| erm(TR) | III8 | GCATGACATAAACCTTCA | 26 | |
| erm(TR) | TR1 | ATAGAAATTGGGTCAGGAAAAGG | 12 | 5,963 |
| orf34 | ETR13 | GCAAAAAAGCACGCAGAAAG | This study | |
| orf34 | ETR23 | GTCTTTCTGCTTGTAGTTCTGCC | This study | 5,369 |
| orf40 | ETR85 | GTTAGCAATAGTGTTTCTGTTTT | This study | |
| ICESp2905 mapping | ||||
| orf2 | CD1-for | AAAACGAACGGGAATATC | This study | 8,165 |
| erm(TR) | III8 | GCATGACATAAACCTTCA | 26 | |
| erm(TR) | TR1 | ATAGAAATTGGGTCAGGAAAAGG | 12 | 14,299 |
| orf29 | CD2-rev | TTGATGTAATGTAGGATAAAAG | This study | |
| orf29 | CD3-for | CAAAGCCAAATGTCCTAAATGAAA | This study | 13,452 |
| orf33 | CD3-rev | TCCGCAAAATCCGTCCTACA | This study | |
| orf34 | TnpV-for | GAATGGACAGGATGCCTCAA | This study | 14,319 |
| orf48 | CD4-rev | ACTCAAAATAATCGCTGTCCTT | This study | |
| orf49 | CD5-for | ATGTAGTTGAAAGAGAAAATAAGAA | This study | 12,981 |
| orf61 | CD5-rev | GGAGATTACCATACCGCCAACACC | This study | |
| ICESp1108 chromosomal integration site | ||||
| Spy1198* | LYT-for | TTACAGGGTCTGCGGCTATT | This study | 6,193 |
| orf5 | ETR49 | TCTTAGCTGGTATATACACTTACC | 6 | |
| orf37 | ETR45 | TCAAAAGCAGCATCTTTATCTTCG | 6 | 5,147 |
| Spy1195* | THIO-rev | TTCAAAGCCGTCTCAGTCAC | This study | |
| ICESp2905 chromosomal integration site | ||||
| Spy1099† | LYT-for | TTACAGGGTCTGCGGCTATT | This study | 2,043 |
| orf1 | TR-inv1 | CTAACTAAGAATCCAATAATCA | This study | |
| orf61 | CD6-for | TAATGTTGCGGTATTTTTTGA | This study | 2,445 |
| Spy1096† | THIO-rev | TTCAAAGCCGTCTCAGTCAC | This study | |
| Conserved core sequence | ||||
| erm(TR) | TR1 | ATAGAAATTGGGTCAGGAAAAGG | 12 | 1,497 |
| orf30‡ | Cyt-rev | ATGCCCTGAAGTTCCAAAG | This study | |
| Inverse PCR | ||||
| orf1‡ | C1LE-inv3 | TTCAATCAGTCCTATCCC | This study | |
| orf1‡ | C1LE-inv4 | AGTAAAGGGCAAAAGTCT | This study |
*, From the S. pyogenes MGAS10750 genome;
, from the S. pyogenes MGAS5005 genome;
, from ICESp1108.
DNA sequencing and sequence analysis.
Overlapping fragments of the erm(TR)-carrying elements were obtained by PCR assays and primer walking techniques using suitable primer pairs. Most oligonucleotides for long PCR experiments were designed from S. pyogenes ICE10750-RD.2 and ICE2096-RD.2 (accession numbers CP000262 and CP000261) (2) and from ICECd630 of Clostridium difficile (accession no. AM180355) (25). Amplicons were sequenced by ABI Prism (Perkin-Elmer Applied Biosystems, Foster City, CA) with dye-labeled terminators. ORF analysis was performed by using the online available software ORF Finder (http://www.ncbi.nlm.nih.gov/projects/gorf/). The criterion to designate a potential ORF was the existence of a start codon and a minimum coding size of 30 amino acids. Sequence similarity and conserved domain searches were carried out by using tools (BLAST and CDART) available online at the National Center for Biotechnology Information of the National Library of Medicine (Bethesda, MD) (http://www.ncbi.nlm.nih.gov/).
Nucleotide sequence accession numbers.
The complete sequences of two new erm(TR)-carrying genetic elements, ICESp1108 and ICESp2905, with their chromosomal junctions, have been submitted to the EMBL database under accession numbers FR691054 and FR691055, respectively.
RESULTS AND DISCUSSION
PCR mapping assays to detect ICE10750-RD.2.
The 23 test strains were PCR mapped using primer pairs designed from the sequence of ICE10750-RD.2, thus far the only completely sequenced S. pyogenes erm(TR) element. While the 3 TET-susceptible strains yielded positive PCR results with most primer pairs, none of the 20 TET-resistant isolates yielded PCR evidence of ICE10750-RD.2, except for a short region including erm(TR).
ICESp1108, the erm(TR)-carrying element from TET-susceptible strain C1.
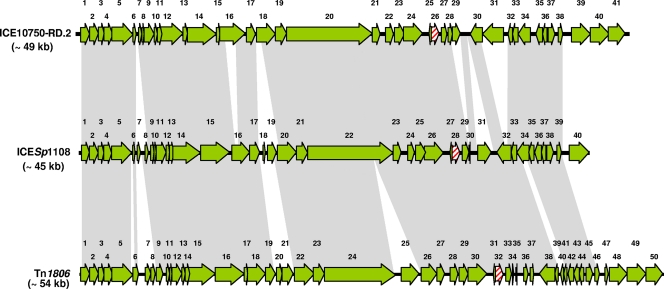
The erm(TR)-carrying element from S. pyogenes C1, one of the 3 TET-susceptible test strains, was sequenced (accession no. FR691054) and characterized. The new element, designated ICESp1108, was 45,456 bp in size. Its G+C content was 31%. Sequence analysis revealed 40 ORFs. erm(TR) was orf28 (100% identical to the genes of both ICE10750-RD.2 and Tn1806). orf29 encoded a spectinomycin phosphotransferase, 99.8% identical to the gene adjacent to erm(TR) in both ICE10750-RD.2 and Tn1806. In orf30 (120 bp, identical to the corresponding gene of Tn1806), only the first 85 bp matched (100%) the initial portion of the cytidine deaminase-encoding gene of ICE10750-RD.2 (390 bp). The ICESp1108 ORF map, aligned with the ORF maps of ICE10750-RD.2 and Tn1806, is shown in Fig. 1, while the major characteristics of the ORFs are detailed in the supplemental material (see Table S1 in the supplemental material).
Fig. 1.
ORF map of ICESp1108 from S. pyogenes strain C1 (accession no. FR691054) and its alignment with the ORF maps of S. pyogenes ICE10750-RD.2 (accession no. CP000262) and S. pneumoniae Tn1806 (accession no. EF469826). The ORFs, indicated as arrows pointing in the direction of transcription, are numbered consecutively (orf1 to orf40 in ICESp1108, with some predicted functions reported in Table S1 in the supplemental material; orf1 to orf41 in ICE10750-RD.2; and orf1 to orf50 in Tn1806). ORFs are depicted as green arrows except for erm(TR) (striped red). Gray areas between ORF maps denote >90% DNA identity.
ICESp1108 displayed close similarities to both ICE10750-RD.2 and Tn1806, as demonstrated by extensive DNA identities (>90%). However, a significant difference was noted at the right end: orf40, the last ORF of ICESp1108, encoding a recombinase, replaced the last three ORFs of ICE10750-RD.2 and Tn1806, which were >95% identical in the two elements and encoded recombinases totally unrelated to the one encoded by orf40.
ICESp2905, the erm(TR)- and tet(O)-carrying composite element from TET-resistant strain iB21.
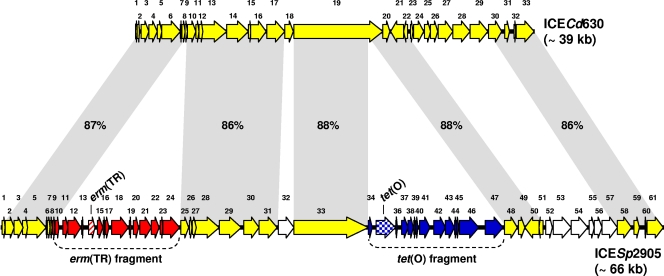
The erm(TR)-carrying element from S. pyogenes iB21—one of the 20 TET-resistant test strains, previously used as a donor in mating assays yielding cotransfer of erm(TR) and tet(O) (8)—was sequenced and characterized. High identities to different portions of a segment of the C. difficile 630 genome (accession no. AM180355) (25) were detected in both erm(TR)- and tet(O)-flanking regions, previously sequenced by inverse PCR analysis. We considered this segment (∼39 kb, 33 ORFs, G+C content 35% versus 29% of the chromosome), unmentioned in the genome analysis of C. difficile 630, as a putative ICE, and arbitrarily designated it ICECd630. These findings suggested that both erm(TR) and tet(O), located on separate fragments, were inserted in the same scaffold—ICECd630—to form a larger structure. The new composite element was designated ICESp2905 (accession no. FR691055). It was 65,575 bp in size, and its G+C content was 36%. Sequence analysis disclosed 61 ORFs. erm(TR) (orf14) and tet(O) (orf35) were far apart (almost 28 kb), explaining previous failures in demonstrating their linkage by PCR (8). The ICESp2905 ORF map, aligned with that of ICECd630, is shown in Fig. 2, while the major characteristics of the ORFs are detailed in the supplemental material (see Table S2 in the supplemental material). The organization of ICESp2905 is summarized below.
Fig. 2.
ORF map and genetic organization of ICESp2905 from S. pyogenes strain iB21 (accession no. FR691055), and its alignment with the ORF map of C. difficile ICECd630 (accession no. AM180355). The ORFs, indicated as arrows pointing in the direction of transcription, are numbered consecutively (orf1 to orf61 in ICESp2905, with some predicted functions reported in Table S2 in the supplemental material; and orf1 to orf33 in ICECd630). ICECd630 ORFs and related ORFs in ICESp2905 are depicted as yellow arrows. ICESp2905 ORFs of the erm(TR) fragment and the tet(O) fragment are depicted as red and blue arrows, respectively, except for erm(TR) (striped red) and tet(O) (checkered blue). Other ICESp2905 ORFs are depicted as white arrows. Gray areas between ORF maps denote significant DNA identities (>70%) as indicated.
(i) Initial region (bp 1 to 5074).
This region, spanning from orf1 to orf8, displayed high identity (87%) to a region of ICECd630 (bp 480415 to 485486 of the C. difficile 630 genome). The specific functions associated with some ORFs (orf2, orf3, and orf5) are presumably involved in the ICE conjugative transfer.
(ii) erm(TR) fragment (bp 5075 to 17690).
This erm(TR)-containing fragment (31% G+C), spanning orf9 to orf24, was inserted into orf8, close to its 3′ end. orf8 is 90% identical to the corresponding ORF of ICECd630. The insertion of the erm(TR) fragment did not interrupt the orf8 coding sequence, which was reconstituted by the first 7 nucleotides of the inserted fragment to produce an ORF shorter than in the wild type (159 versus 215 bp). erm(TR) was orf14 (99.9% identical to the gene of ICESp1108). orf15, encoding spectinomycin phosphotransferase, was 99.8% identical to the gene of ICESp1108). orf16 (141 bp) again displayed the same 85-bp segment mentioned above, matching the initial portion of the cytidine deaminase-encoding gene of ICE10750-RD.2. The region spanning from orf17 to orf22 was similar to a region of ICE6180-RD.1—an ∼11-kb element of S. pyogenes MGAS6180 (accession no. NC_007296)—spanning bases 1083646 to 1087913 of the MGAS6180 genome. orf24, the last ORF of the erm(TR) fragment, encoded a transposase indicated as tndX-like according to CDART analysis, but displayed no significant identity to the tndX gene of the Tn916 family transposon Tn5397 (14, 21).
(iii) Central region (bp 17691 to 36310).
This region is the portion of the ICECd630-like scaffold of ICESp2905 encompassed between the two insertions of the composite element, the one containing erm(TR) and the one containing tet(O). The region, spanning from orf25 to orf33, largely consisted of two portions (bp 17691 to 27418 and bp 29020 to 36310) displaying high identity (86 and 88%, respectively) to ICECd630; the only significant difference was orf32, which replaced an ORF encoding a different protein in the clostridial ICE.
(iv) tet(O) fragment (bp 36311 to 49746).
This fragment (44% G+C) was formed by an ∼11-kb tet(O)-containing portion, spanning orf34 to orf46, which was highly identical to a portion of ICE2096-RD.2 (an ∼63-kb element of S. pyogenes MGAS2096 [accession no. CP000261] [2]), plus orf47, the last ORF in the fragment, which was alien to ICE2096-RD.2. The tet(O) fragment was inserted into orf19 of ICECd630 (encoding a putative helicase), at base 503358 of the C. difficile 630 genome. In ICESp2905, this insertion split the original helicase gene into two ORFs: orf33 (the last in the central region, encoding a putative helicase) and orf48 (the first in the terminal region).
(v) Terminal region (bp 49747 to 65575).
This region spanned orf48 to orf61. High identities to ICECd630 were displayed by the two portions spanning from orf48 to orf51 (88% identity to the region from bp 503457 to 507442 of the C. difficile 630 genome) and from orf58 to orf61 (86% identity to the region from bp 515268 to 519796 of the same genome). It is worth noting that orf61, the last ORF in ICESp2905, and the last ORF (orf40) in ICESp1108, both encoding a putative recombinase, displayed 71% identity.
Chromosomal integration of ICESp1108 and ICESp2905.
DNAs from both strains C1 (harboring ICESp1108) and iB21 (harboring ICESp2905) yielded no PCR products using primers, designed from the MGAS10750 sequence, targeting hsdM, a chromosomal gene that is the integration site of erm(TR)-carrying elements ICE10750-RD.2 (2) and Tn1806 (6).
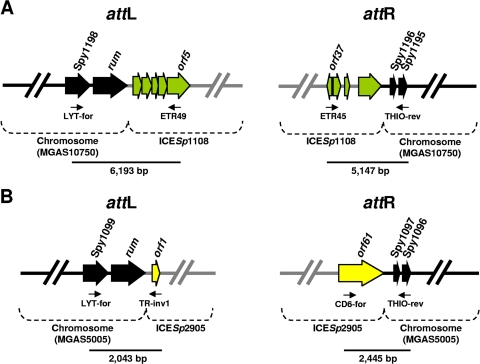
The left junction of ICESp1108 was identified by inverse PCR. In particular, Hpy188I-restricted genomic DNA from strain C1 was ligated and used as the template with primer pair C1LE-inv3/C1LE-inv4 (Table 1). Sequencing of the amplicon revealed significant DNA identity to a conserved RNA uracil methyltransferase (rum) gene detected in all S. pyogenes genomes sequenced thus far; the highest degree of identity was found to be with the rum gene from S. pyogenes MGAS10750 (99%). PCR experiments were carried out with two primer pairs, one for the left junction (attL) and one for the right junction (attR) (Fig. 3A). Sequencing of the resulting amplicons disclosed that ICESp1108 was integrated at the 3′ end of the rum gene at base 1142414 of the S. pyogenes MGAS10750 genome. The chromosomal insertion of ICESp1108 did not interrupt the orf40 coding sequence, which was reconstituted by the first 39 nucleotides of chromosomal origin.
Fig. 3.
Chromosomal integration of ICESp1108 (A) and ICESp2905 (B). (A) ICESp1108 was integrated into the chromosome of S. pyogenes C1 within the rum gene. This gene, detected in all S. pyogenes genomes sequenced to date, has the highest DNA identity with the corresponding gene (Spy1197) from S. pyogenes MGAS10750. Chromosomal ORF designations are thus from S. pyogenes MGAS10750. Chromosomal ORFs at the left (attL) and right (attR) junctions are indicated as black arrows, and ICESp1108 ORFs are indicated as green arrows. Amplicons obtained by pairing primers LYT-for/ETR49 (attL) and ETR45/THIO-rev (attR) are identified by bars. (B) ICESp2905 was integrated into the chromosome of S. pyogenes iB21 within the rum gene. This gene has the highest DNA identity with the corresponding gene (Spy1098) from S. pyogenes MGAS5005. Chromosomal ORF designations are thus from S. pyogenes MGAS5005. Chromosomal ORFs at the left (attL) and right (attR) junctions are indicated as black arrows, and ICESp2905 ORFs are indicated as yellow arrows. Amplicons obtained by pairing primers LYT-for/TR-inv1 (attL) and CD6-for/THIO-rev (attR) are identified by bars.
The junctions of ICESp2905 in the chromosome were also characterized by inverse PCR and direct sequencing. The highest degree of identity (99%) was with a rum gene from S. pyogenes MGAS5005 (accession no. NC_007297). Similar to ICESp1108, the integration site was at the 3′ end of the conserved rum gene at base 1070363 of the S. pyogenes MGAS5005 genome (Fig. 3B). The chromosomal insertion of ICESp2905 did not interrupt the orf61 coding sequence, which was reconstituted by the first 39 nucleotides of chromosomal origin.
Distribution of erm(TR)-carrying elements in all test strains.
After the complete sequence and the chromosomal integration of ICESp1108 from strain C1 and ICESp2905 from strain iB21 were established, the erm(TR) elements of all test strains were comparatively examined by PCR mapping using suitable primer pairs (Table 1).
The erm(TR) elements of the 2 TET-susceptible strains other than C1 showed an organization comparable to that of ICESp1108, which was also largely shared by ICE10750-RD.2 and Tn1806. Remarkably, the right ends of the elements of both strains reproduced the organization of the right end of ICESp1108: a single recombinase versus the three unrelated recombinases shared by ICE10750-RD.2 and Tn1806. Accordingly, the chromosomal integration site of both elements was at the 3′ end of the rum gene as for ICESp1108 versus the hsdM gene which is the integration site of ICE10750-RD.2 and Tn1806.
The erm(TR) elements of the 19 TET-resistant strains other than iB21 had an organization comparable to that of ICESp2905, with the tet(O) gene detected almost 28 kb downstream of erm(TR). Five strains yielded no amplification using the primer pair CD1-for/III8. All demonstrated a chromosomal integration site at the 3′ end of the conserved rum gene.
Twenty-two of the twenty-three test strains (the exception being a TET-resistant isolate) yielded PCR evidence of the conserved region including erm(TR), the spectinomycin phosphotransferase-encoding ORF, and the above-mentioned 85-bp sequence.
Conclusions.
We show here that two completely distinct categories of erm(TR)-carrying elements can be found in S. pyogenes: one in TET-susceptible strains, and another, where the erm(TR) gene is typically linked with the tet(O) gene, in TET-resistant strains. The former category, epitomized here by ICESp1108 (∼45 kb) from strain C1 and detected in the other TET-susceptible test strains, also includes the two previously sequenced erm(TR) elements ICE10750-RD.2 (∼49 kb) and Tn1806 (∼54 kb). The latter category, epitomized here by ICESp2905 (∼66 kb) from strain iB21 and detected in all of the other TET-resistant strains tested, is a totally new finding. We had documented in a previous study the presence of the tet(O) determinant in TET-resistant S. pyogenes isolates with erm(TR)-mediated ERY resistance (8). However, the genetic basis of the erm(TR)-tet(O) association was still unknown. Here we demonstrated an erm(TR)-tet(O) linkage in ICESp2905 and closely related elements, which turned out to be composite structures where separate erm(TR)- and tet(O)-containing fragments are inserted in a clostridial scaffold. Intriguingly, in S. pyogenes, also mef(A)-encoded ERY resistance has been shown to be mediated by different genetic elements in TET-susceptible and TET-resistant strains (4, 29): the closely related Tn1207.3 (24) and Φ10394.4 (1) in the former strains and Φm46.1 (3), with TET resistance again encoded by tet(O), in the latter.
Although the nomenclature of mobile genetic elements is evolving (5, 20), all of the erm(TR) genetic elements described in the present study can be considered ICEs (27, 30). In keeping with their exogenous origin, S. pyogenes ICEs differ by an average of 5% from endogenous S. pyogenes core genomes (2), whose G+C content is ca. 38.5% (2, 7). This is consistent with the detected G+C contents of ICESp1108 (31%) and ICESp2905 (36%). In the latter, however, the G+C contents of the two insertions—the erm(TR) fragment and the tet(O) fragment—are quite far apart (31 and 44%, respectively).
Despite broad differences between the two categories of erm(TR) elements, all ICEs from our S. pyogenes test strains, both TET-susceptible and TET-resistant, were integrated in the chromosome at the 3′ end of the conserved rum gene. This denotes a considerable divergence from the two previously sequenced erm(TR) elements—ICE10750-RD.2 and Tn1806—which share a different chromosomal integration site, namely, the hsdM gene. Interestingly, the 3′ end of the rum gene is an integration hot-spot for mobile streptococcal elements that is also shared by other S. pyogenes ICEs, such as ICE2096-RD.2 and ICE6180-RD.1 (2), and by prophages such as S. pyogenes Φm46.1 (3), Streptococcus agalactiae λSa04 (3, 28), and Streptococcus suis ΦSsUD.1 (18).
Moreover ICESp1108 and ICESp2905, as well as ICE10750-RD.2 and Tn1806, shared an almost identical conserved core sequence (2,062 bp, 99.8% identity) that included erm(TR), an adjacent ORF encoding a spectinomycin phosphotransferase, and a contiguous segment matching the first 85 bp of the cytidine deaminase-encoding gene of ICE10750-RD.2. PCR evidence of this conserved region was obtained in all but one of the 23 test strains. Since erm(TR) is an erm(A) subclass (22, 26), it is noteworthy, as underscored previously (6), that a different gene encoding spectinomycin resistance (designated spc) is found adjacent to erm(A) also in the Staphylococcus aureus transposon Tn554, where erm(A) was first detected and sequenced (15). Compared to the spectinomycin phosphotransferase-encoding gene detected in ICESp1108 (orf29) and ICESp2905 (orf15), the spc gene encodes a different enzyme (a spectinomycin adenyltransferase) and is transcribed in the opposite direction.
Supplementary Material
ACKNOWLEDGMENTS
This study was partly supported by the Italian Ministry of Education, University and Research. Andrea Brenciani was supported by an ESCMID Research Grant 2010.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 22 February 2011.
REFERENCES
- 1. Banks D. J., Porcella S. F., Barbian K. D., Martin J. M., Musser J. M. 2003. Structure and distribution of an unusual chimeric genetic element encoding macrolide resistance in phylogenetically diverse clones of group A streptococcus. J. Infect. Dis. 188:1898–1908 [DOI] [PubMed] [Google Scholar]
- 2. Beres S. B., Musser J. M. 2007. Contribution of exogenous genetic elements to the group A streptococcus metagenome. PLoS One 2:e800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brenciani A., et al. 2010. Characterization of Φm46.1, the main Streptococcus pyogenes element carrying mef(A) and tet(O) genes. Antimicrob. Agents Chemother. 54:221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brenciani A., et al. 2004. Distribution and molecular analysis of mef(A)-containing elements in tetracycline-susceptible and -resistant Streptococcus pyogenes clinical isolates with efflux-mediated erythromycin resistance. J. Antimicrob. Chemother. 54:991–998 [DOI] [PubMed] [Google Scholar]
- 5. Burrus V., Pavlovic G., Decaris B., Guédon G. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601–610 [DOI] [PubMed] [Google Scholar]
- 6. Camilli R., Del Grosso M., Iannelli F., Pantosti A. 2008. New genetic element carrying the erythromycin resistance determinant erm(TR) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 52:619–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ferretti J. J., Ajdic D., McShan W. M. 2004. Comparative genomics of streptococcal species. Indian J. Med. Res. 119(Suppl.):1–6 [PubMed] [Google Scholar]
- 8. Giovanetti E., Brenciani A., Lupidi R., Roberts M. C., Varaldo P. E. 2003. Presence of the tet(O) gene in erythromycin- and tetracycline-resistant strains of Streptococcus pyogenes and linkage with either the mef(A) or the erm(A) gene. Antimicrob. Agents Chemother. 47:1935–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Giovanetti E., et al. 2002. Conjugative transfer of the erm(A) gene from erythromycin-resistant Streptococcus pyogenes to macrolide-susceptible S. pyogenes, Enterococcus faecalis, and Listeria innocua. J. Antimicrob. Chemother. 50:249–252 [DOI] [PubMed] [Google Scholar]
- 10. Giovanetti E., Montanari M. P., Mingoia M., Varaldo P. E. 1999. Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob. Agents Chemother. 43:1935–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grivea I. N., Al-Lahham A., Katopodis G. D., Syrogiannopoulos G. A., Reinert R. R. 2006. Resistance to erythromycin and telithromycin in Streptococcus pyogenes isolates obtained between 1999 and 2002 from Greek children with tonsillopharyngitis: phenotypic and genotypic analysis. Antimicrob. Agents Chemother. 50:256–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kataja J., Seppälä H., Skurnik M., Sarkkinen H., Huovinen P. 1998. Different erythromycin resistance mechanisms in group C and group G streptococci. Antimicrob. Agents Chemother. 42:1493–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leclercq R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34:482–492 [DOI] [PubMed] [Google Scholar]
- 14. Mullany P., et al. 1990. Genetic analysis of a tetracycline resistance element from Clostridium difficile and its conjugal transfer to and from Bacillus subtilis. J. Gen. Microbiol. 136:1343–1349 [DOI] [PubMed] [Google Scholar]
- 15. Murphy E. 1985. Nucleotide sequence of ermA, a macrolide-lincosamide-streptogramin B determinant in Staphylococcus aureus. J. Bacteriol. 162:633–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nielsen H. U., et al. 2004. Tetracycline and macrolide co-resistance in Streptococcus pyogenes: co-selection as a reason for increase in macrolide-resistant S. pyogenes? Microb. Drug Resist. 10:231–238 [DOI] [PubMed] [Google Scholar]
- 17. Olsvik B., Olsen I., Tenover F. C. 1995. Detection of tet(M) and tet(O) using the polymerase chain reaction in bacteria isolated from patients with periodontal disease. Oral Microbiol. Immunol. 10:87–92 [DOI] [PubMed] [Google Scholar]
- 18. Palmieri C., Princivalli M. S., Brenciani A., Varaldo P. E., Facinelli B. 2011. Different genetic elements carrying the tet(W) gene in two human clinical isolates of Streptococcus suis. Antimicrob. Agents Chemother. 55:631–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ripa S., et al. 2001. SmaI macrorestriction analysis of Italian isolates of erythromycin-resistant Streptococcus pyogenes and correlations with macrolide-resistance phenotypes. Microb. Drug Resist. 7:65–71 [DOI] [PubMed] [Google Scholar]
- 20. Roberts A. P., et al. 2008. Revised nomenclature for transposable genetic elements. Plasmid 60:167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roberts A. P., Johanesen P. A., Lyras D., Mullany P., Rood J. I. 2001. Comparison of Tn5397 from Clostridium difficile, Tn916 from Enterococcus faecalis and the CW459tet(M) element from Clostridium perfringens shows that they have similar conjugation regions but different insertion and excision modules. Microbiology 147:1243–1251 [DOI] [PubMed] [Google Scholar]
- 22. Roberts M. C., et al. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sambrook J., Russell D. W. 2001. Inverse PCR, p. 8.81–8.85 In Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 24. Santagati M., et al. 2003. The novel conjugative transposon Tn1207.3 carries the macrolide efflux gene mef(A) in Streptococcus pyogenes. Microb. Drug Resist. 9:243–247 [DOI] [PubMed] [Google Scholar]
- 25. Sebaihia M., et al. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779–786 [DOI] [PubMed] [Google Scholar]
- 26. Seppälä H., Skurnik M., Soini H., Roberts M. C., Huovinen P. 1998. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob. Agents Chemother. 42:257–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Seth-Smith H., Croucher N. J. 2009. Genome watch: breaking the ICE. Nat. Rev. Microbiol. 7:328–329 [DOI] [PubMed] [Google Scholar]
- 28. Tettelin H., et al. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. U. S. A. 102:13950–13955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Varaldo P. E., Montanari M. P., Giovanetti E. 2009. Genetic elements responsible for erythromycin resistance in streptococci. Antimicrob. Agents Chemother. 53:343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wozniak R. A., Waldor M. K. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8:552–563 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.