Abstract
Lantibiotics such as nisin (NIS) are peptide antibiotics that may have a role in the chemotherapy of bacterial infections. A perceived benefit of lantibiotics for clinical use is their low propensity to select resistance, although detailed resistance studies with relevant bacterial pathogens are lacking. Here we examined the development of resistance to NIS in Staphylococcus aureus, establishing that mutants, including small-colony variants, exhibiting substantial (4- to 32-fold) reductions in NIS susceptibility could be selected readily. Comparative genome sequencing of a single NISr mutant exhibiting a 32-fold increase in NIS MIC revealed the presence of only two mutations, leading to the substitutions V229G in the purine operon repressor, PurR, and A208E in an uncharacterized protein encoded by SAOUHSC_02955. Independently selected NISr mutants also harbored mutations in the genes encoding these products. Reintroduction of these mutations into the S. aureus chromosome alone and in combination revealed that SAOUHSC_02955(A208E) made the primary contribution to the resistance phenotype, conferring up to a 16-fold decrease in NIS susceptibility. Bioinformatic analyses suggested that this gene encodes a sensor histidine kinase, leading us to designate it “nisin susceptibility-associated sensor (nsaS).” Doubling-time determinations and mixed-culture competition assays between NISr and NISs strains indicated that NIS resistance had little impact on bacterial fitness, and resistance was stable in the absence of selection. The apparent ease with which S. aureus can develop and maintain NIS resistance in vitro suggests that resistance to NIS and other lantibiotics with similar modes of action would arise in the clinic if these agents are employed as chemotherapeutic drugs.
INTRODUCTION
There is an urgent need for novel antibacterial agents to treat infections caused by antibiotic-resistant bacteria (44). One strategy for bringing new agents to the antibacterial pipeline involves developing compounds with known antibacterial activity which are currently unexploited for clinical use in humans. The lantibiotics may represent promising candidates in this regard (1, 43). Lantibiotics are a large, diverse group of ribosomally synthesized antimicrobial peptides, named to reflect the fact that they contain the posttranslationally modified amino acids lanthionine and 3-methyl-lanthionine (32). They are classified as type A(I), type A(II), or type B, depending on their structure, with the type A(I) peptide nisin (NIS) being the best-characterized lantibiotic (32). NIS has several properties which suggest its potential for use in antibacterial chemotherapy (1, 38, 43). First, it exhibits potent and rapid bactericidal activity against a variety of Gram-positive bacterial species in vitro, including multidrug-resistant clinical isolates (13, 37, 38). Second, it has a long history of safe use as a food preservative (8) and has also been employed successfully in veterinary medicine to treat mastitis (1). Third, the mode of action of NIS is relatively well defined: NIS binds to the pyrophosphate cage of the peptidoglycan precursor, lipid II, and subsequently inserts into the cytoplasmic membrane to form pores, which cause a loss of membrane potential and leakage of intracellular contents (5, 16, 17, 20, 23).
An additional perceived benefit of NIS and other type A(I) lantibiotics for clinical use is a low propensity to select resistance (7, 17, 43), implying that the activity of these agents would not become compromised rapidly by resistance following their introduction into the clinic (43). Although this idea has been expressed frequently in the scientific literature, it appears to be based predominantly on anecdotal evidence regarding the apparent lack of NIS resistance among food spoilage organisms despite decades of heavy NIS use in the food industry (7, 23, 28) and on theoretical considerations relating to the mode of action of NIS (17, 28, 43). With respect to the latter, it has been proposed that the cellular target of NIS is likely to limit the development of resistance, since it is not easy to envision how the well-conserved lipid II molecule (and, by implication, all of the enzymes involved in generating and/or processing this molecule) could undergo modification to prevent binding to NIS while still allowing peptidoglycan biosynthesis to occur (7, 17, 23).
However, the hypothesis that bacteria cannot readily develop resistance to NIS is challenged by in vitro studies with food-borne Gram-positive species such as Listeria monocytogenes (14, 15). Furthermore, some listerial NISr mutants are stable and apparently suffer little or no growth disadvantage compared to their NISs counterparts (14). Nonetheless, to accurately assess the resistance potential of lantibiotics such as NIS, further resistance studies with other relevant human pathogens are required. Here we report on studies that have examined the development, maintenance, and genetic basis of NIS resistance in Staphylococcus aureus.
MATERIALS AND METHODS
Bacterial strains, growth media, and antibacterial agents.
Bacterial strains used in this study are listed in Table 1. Luria-Bertani (LB) broth (Oxoid, Hampshire, United Kingdom) was used to culture Escherichia coli, while S. aureus strains were routinely propagated in tryptone soy broth (TSB; Oxoid). Iso-Sensitest broth (ISB; Oxoid) and Iso-Sensitest agar (ISA; Oxoid) were used for antibacterial susceptibility testing.
Table 1.
Bacterial strains used in this study
| Strain | Relevant genotype or description | Reference or source |
|---|---|---|
| Staphylococcus aureus | ||
| SH1000 | Standard laboratory strain | 21, 34 |
| 8325-4 | Standard laboratory strain (rsbU) | 34 |
| I10 | Engineered small-colony variant (8325-4 hemB::ermB) | 48 |
| DB24 | Engineered small-colony variant (8325-4 menD::ermC) | 4 |
| RN4220 | Restriction-deficient cloning host | 9 |
| MW2 | Community-acquired MRSA strain | 2 |
| Escherichia coli | ||
| DH5α | F− cloning strain | Invitrogen |
Pure NIS (6.6 mg/ml in 10 mM sodium citrate [pH 3.0]) was a gift from ImmuCell Corporation (ME), deoxyactagardin B and mersacidin were gifts from Novacta (Hertfordshire, United Kingdom), and daptomycin was a gift from Cubist (MA). Other antibacterial agents were from Sigma-Aldrich (Dorset, United Kingdom).
Selection of NISr mutants.
Several approaches were used to select NISr mutants. The serial subculture method involved propagating S. aureus SH1000 in ISB containing NIS at 1/4 the MIC, with daily transfer into fresh broth containing NIS. When the culture exhibited an increase in MIC, the selective NIS concentration used in the subculture was increased accordingly.
NISr mutants were also selected by plating saturated cultures of SH1000 onto ISA containing NIS at 4 times the MIC. In parallel, diluted cultures were plated onto nonselective ISA to achieve total counts, enabling calculation of the mutation frequency to NIS resistance (46). Mutation frequencies were determined from three independent cultures sampled in triplicate.
An approach intermediate between those described above was also used: after performing the subculture procedure for a single cycle, aliquots of the culture were plated onto agar containing NIS at 4 times the MIC.
Phenotypic characterization of NISr mutants.
Susceptibility testing was performed by agar dilution with ISA, using inocula of 104 CFU/spot. For daptomycin susceptibility determinations, agar was supplemented with 50 μg/ml Ca2+. The MIC was defined as the lowest concentration of antibiotic that prevented growth after 18 h at 37°C, and all determinations were performed at least in triplicate.
To determine doubling times, overnight cultures diluted 1/200 in ISB were incubated at 37°C with aeration, and the optical density at 600 nm (OD600) was monitored. OD600 values in the range of 0.1 to 0.3 were plotted against time, the gradient of the line was determined, and doubling times were calculated using the formula log10 2/gradient (27). Doubling-time results are means of three independent experiments. Competition assays to evaluate relative fitness were performed as previously described (47).
The stability of the NISr phenotype was evaluated by serial passage of NISr mutants in drug-free ISB. Briefly, cultures were grown overnight to saturation (ca. 3 × 109 CFU/ml), with daily transfer of aliquots (2 μl) into fresh ISB (9 ml). The number of bacterial generations (n) was determined using the formula n = log N − log N0 × 1/log 2, where N is the final cell number and N0 is the initial cell number (30). After several days of subculture, bacteria were plated onto drug-free ISA and individual colonies subjected to NIS susceptibility determinations.
Genotypic characterization of NISr mutants.
Chromosomal mutations in NISr mutant NR018 were identified by comparative genome sequencing (CGS) (18, 34) performed by Roche NimbleGen, using a custom CGS array based on the entire genome sequence of S. aureus NCTC 8325 (GenBank accession number CP000253). Genomic DNA was purified from NR018 and 8325 as described previously (34). The majority of mutations identified in NR018 by CGS represented established genetic differences between the 8325 and SH1000 genomes (34) and were disregarded. Mutations unique to NR018, identified in genes SAOUHSC_00467 and SAOUHSC_02955, were confirmed by PCR amplification and DNA sequencing using oligonucleotide primers 467U and 467L and primers 2955U and 2955L, respectively (Table 2).
Table 2.
Oligonucleotide primers used in this study
| Oligonucleotide | Sequence (5′–3′)a |
|---|---|
| 467U | AAATATTTATAATGCAGTTAACG |
| 467L | GACCAGAAGTATAAACCATACCA |
| 2955U | ACACATTAGCTGTTAACATGACG |
| 2955L | ATGTTTCACTTCTAGCAACATGG |
| 467attB1F1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTAACTGCACTATGTACTGGAAGAGGA |
| 467attB2R1 | GGGGACCACTTTGTACAAGAAAGCTGGGTGAAGCTTGATTAAACATATTAATCG |
| 2955attB1F1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTTAGATGAAGCAAAAGTCGTGTATC |
| 2955attB2R3 | GGGGACCACTTTGTACAAGAAAGCTGGGTATAAATTCATTTGATTAAGTGCTGT |
| 467_F1 | AAGAGTATTGGCATTTGAAATTGAT |
| 467_R1 | CCTTCTCAGCCTGACTAAGCAT |
| 467Sb | GATCCAAGACGTCGCGGTGAA |
| 467Sc | GAGTGTTGGGCATTATGTTCAATAC |
| 467Sd | CGAGTTTGGAAGAATTGGCTCTACT |
| 467Se | AACAGGCGAAGTCTTCAATAAGTGA |
| 467Sf | ACGTAGAACCTGGCAACAGTTTATC |
| 2955_F1 | TTCCATTTCAGACATAACGCCTGT |
| 2955_R1 | TGCATTAACCGAACCCTGTATATCA |
| 2955Sb | CACCGATGGCACCATTATATGTC |
| 2955Sc | GAAAGTTCAAATCAAGAGCCGTCA |
| 2955Sd | AAATGTTGGGAATCAGAAAACTTCA |
| 2955Se | CATTATGAACCTTGTCATCACGAAG |
| 2955Sf | CAAAGGGTACATCAGTTGGCAGTAC |
| mprFF1 | AAGAAGCACTCATAATCGGCTGTT |
| mprFR1 | GGCGACTTAACTTTAGCTCATTTCA |
| mprFS1 | TGTAGGTTTCGGTGGCTTTATTG |
| mprFS2 | CATCAGCTAAGAAGTACATTGAGGG |
| mprFS3 | GTATTGCGCTATTACTTCTGGCTTA |
| mprFS4 | TAACGCAATTTTCAACTTCAGGTAA |
Engineered attB sites for recombination of PCR amplicons into the pKOR1 vector are underlined.
The roles of putative NISr mutations in strain NR018 were determined by engineering them into the chromosome of S. aureus SH1000 by allelic replacement. Briefly, SAOUHSC_00467 and SAOUHSC_02955 containing putative NIS resistance mutations were PCR amplified using oligonucleotide primers 467attB1F1 and 467attB2R1 and primers 2955attB1F1 and 2955attB2R3, respectively (Table 2). The ca. 1,500-bp fragments were recombined into plasmid pKOR1 (3). The resulting constructs were passaged first in E. coli DH5α and then in S. aureus RN4220 and subsequently used to replace the wild-type allele from the chromosome of S. aureus SH1000 by homologous recombination (3). Successful allelic replacement was confirmed by PCR amplification of the loci and surrounding regions, using oligonucleotide primers 467_F1 and 467_R1 and primers 2955_F1 and 2955_R1, followed by DNA sequence analysis using oligonucleotide primers 467Sb-f and 2955Sb-f, respectively (Table 2). The allelic replacement strains were designated SH1000:PurR(V229G) and SH1000:NsaS(A208E). A strain harboring both mutations [SH1000:PurR(V229G), NsaS(A208E)] was generated by sequential allelic replacement.
Bioinformatic analysis and molecular modeling of NISr mutations.
The structure and function of NsaS were explored using bioinformatic tools that included Pfam (10) and InterProScan (49) to identify functional domains and DAS (6) and SOSUI (19) to predict transmembrane regions. STRING 8.2 (25) was used to predict physical and functional interactions between NsaS and other proteins. A predicted structure of the NsaS sensor histidine kinase was generated based on the X-ray crystal structure of a homologous sensor histidine kinase from Thermotoga maritima (Protein Data Bank [PDB] accession number 2C2A) (31) using I-TASSER (50) and rendered using PyMOL (W. L. De Lano [http://www.pymol.org]).
RESULTS
Selection and preliminary characterization of NISr mutants of S. aureus.
On the basis that it might require several genetic changes to confer appreciable NIS resistance, we initially attempted to select NISr mutants by serial subculture of S. aureus SH1000 in the presence of sub-MIC concentrations of NIS. In three independent subcultures, the NIS MIC rose from 4 μg/ml to 16 μg/ml after 1 day and reached 128 μg/ml (32-fold increase over that for SH1000) after 4 or 5 days. To identify the mutation(s) responsible for NIS resistance, we employed CGS to characterize one NISr mutant (NR018) for which the NIS MIC was 128 μg/ml. NR018 differed from SH1000 by only two nonsynonymous nucleotide substitutions, encoding the amino acid changes V229G in the purine operon repressor (PurR) and A208E in the putative sensor histidine kinase encoded by SAOUHSC_02955 (Table 3). Based on these findings and the results presented below, SAOUHSC_02955 was named nisin susceptibility-associated sensor histidine kinase (nsaS).
Table 3.
Properties of NISr mutants selected from S. aureus SH1000
| Strain | NIS MIC (μg/ml) | Doubling time (min) (mean ± SD) | Relative competitive fitness (W) | Amino acid substitution(s) |
|---|---|---|---|---|
| SH1000 | 4 | 26.43 ± 0.99 | 1.00 | None |
| NR006 | 16 | 28.08 ± 1.24 | 1.00 ± 0.12 | PurR(G228E) |
| NR017 | 128 | 27.80 ± 1.91 | 1.12 ± 0.11 | PurR(V229G), NsaS(A105T) |
| NR018 | 128 | 27.69 ± 1.72 | 1.18 ± 0.15 | PurR(V229G), NsaS(A208E) |
| NR111 | 128 | 33.76 ± 3.02 | 1.02 ± 0.12 | PurR(E184STOP), MprF(S295L) |
Since NIS resistance involved only a limited number of genetic changes, we subsequently examined whether NISr mutants could be recovered by a more direct route than subculture in the presence of sub-MIC levels of NIS. Plating saturated cultures of SH1000 directly onto agar containing NIS at 4 times the MIC recovered spontaneous NISr mutants at a mean frequency (± standard deviation) of 2.57 × 10−7 ± 0.45 × 10−7 after 48 h of incubation, and mutants typically exhibited an 8-fold increase in the NIS MIC, to 32 μg/ml. Mutants exhibiting higher-level NIS resistance (MIC of 128 μg/ml) could be selected by plating a culture grown to saturation in the presence of 1/4 the NIS MIC onto agar containing NIS at 4 times the MIC.
Approximately 20% of mutants isolated at 4 times the NIS MIC from plating of a NIS-naïve culture produced colonies on agar that were substantially smaller than those of the parent strain. To determine whether these represented classical small-colony variants (SCVs), all small colonies (n = 70) from a total of 332 colonies growing on agar containing NIS at 4 times the MIC were picked and their susceptibility to gentamicin determined. Of these, 16 displayed a 32-fold decrease in gentamicin susceptibility, strongly suggesting that they were SCVs (40). These strains were tested for both menadione and hemin auxotrophy, phenotypes commonly associated with SCVs (40). All were confirmed as menadione auxotrophs, as evidenced by restoration of colony size and gentamicin susceptibility in the presence of 1.5 μg menadione/ml of agar. Thus, 16/332 (∼5%) colonies selected on agar containing NIS at 4 times the MIC were shown to be menadione-auxotrophic SCVs. To confirm that SCVs which were not selected on agar containing NIS or that were hemin auxotrophs also exhibited reduced susceptibility to NIS, we performed NIS susceptibility determinations with the engineered SCV strains I10 and DB24 (Table 1). Both strains exhibited a 4-fold reduction in NIS susceptibility compared to the parental strain (MIC of 16 μg/ml versus 4 μg/ml).
To establish whether mutations in purR and nsaS are found in other in vitro-selected NISr mutants, we performed PCR amplification and DNA sequencing of these genes from three independently selected NISr mutants of SH1000 (NR006, NR017, and NR111). All three carried nonsynonymous mutations in purR; NR017 carried the same mutation as NR018, while the other strains carried different mutations within this gene (Table 3). NR017 also carried a mutation in nsaS encoding the substitution A105T, which differs from that found in this gene in NR018 (Table 3). In addition, NISr mutants were selected from a clinical S. aureus strain, community-acquired methicillin-resistant S. aureus (MRSA) strain MW2 (2). Two MW2 NISr mutants, for which NIS MICs were 32 μg/ml and 128 μg/ml, harbored mutations in nsaS encoding the substitutions R209I and G210D, respectively. However, neither of these NISr mutants carried mutations in purR.
Phenotypic characterization of NISr mutants.
To examine whether NIS resistance is associated with a fitness cost in vitro, doubling-time determinations and mixed-culture competition assays were performed on all four NISr strains derived from SH1000. Aside from NR111, which exhibited an increased doubling time, the NISr mutants showed no significant impairment in growth or competitive fitness (Table 3) when the results were compared by an unpaired t test. The NISr phenotype was stable in the absence of NIS selection in all strains for >50 generations, with no reduction in NIS susceptibility observed.
The possibility that NIS resistance might mediate cross-resistance to other antibacterial agents which act upon the cell envelope was explored by performing susceptibility determinations with the lantibiotics gallidermin, deoxyactagardin B, and mersacidin and the antibiotics daptomycin, oxacillin, and vancomycin. With the exception of strain NR111, which showed an 8-fold decrease in daptomycin susceptibility compared with SH1000, no cross-resistance was observed (data not shown). We speculated that the reduced susceptibility to daptomycin in strain NR111 was the result of a mutation(s) in the mprF gene (11). PCR amplification of mprF from NR111 using primers mprFF1 and mprFR1 and DNA sequencing using primers mprFS1 to -4 (Table 2) revealed a DNA mutation encoding amino acid substitution S295L, which has previously been associated with reduced susceptibility to daptomycin in a clinical isolate of S. aureus (11).
Further investigations into the molecular basis for NIS resistance.
To establish which of the mutated loci in the NISr mutant NR018 made the primary contribution to NIS resistance, we separately engineered the mutations into the chromosome of SH1000 by allelic replacement (3). Introducing the mutation encoding NsaS(A208E) into SH1000 conferred an increase of up to 16-fold in the NIS MIC, although introduction of the mutation encoding PurR(V229G) did not alter susceptibility to NIS (Table 4). The latter finding, though unexpected, is consistent with the observation that a strain recovered at an early stage of the NIS subculture experiment that ultimately generated NR018 contained the mutation encoding PurR(V229G) but did not display reduced susceptibility to NIS (data not shown). We subsequently examined whether the mutation encoding PurR(V229G), while not conferring NIS resistance detectable by standard susceptibility determination, might nonetheless offer the strain a selective growth advantage over SH1000 in the presence of NIS. However, the doubling times of SH1000:PurR(V229G) and SH1000 in the presence of a sub-MIC concentration of NIS (1 μg/ml) were not significantly different by unpaired t test (data not shown). To investigate the possibility that the PurR(V229G) substitution contributes to NIS resistance only in the NsaS(A208E) background, we generated a double mutant, SH1000:PurR(V229G), NsaS(A208E). However, the highest MIC of NIS observed for this strain was 64 μg/ml, the same as that seen for SH1000:NsaS(A208E) (Table 4).
Table 4.
Reintroduction into S. aureus SH1000 of putative NISr mutations identified in strain NR018 to evaluate their contributions to NIS resistance
| Strain | NIS MIC (μg/ml)a |
|---|---|
| SH1000 | 4 |
| NR018 [SH1000:PurR(V229G), NsaS(A208E)]b | 128 |
| SH1000:PurR(V229G)c | 4 |
| SH1000:NsaS(A208E)c | 32–64 |
| SH1000:PurR(V229G), NsaS(A208E)c | 32–64 |
MICs are determinations for a minimum of four independent experiments performed in triplicate.
Strain generated by serial subculture in the presence of NIS.
Strain generated by allelic replacement.
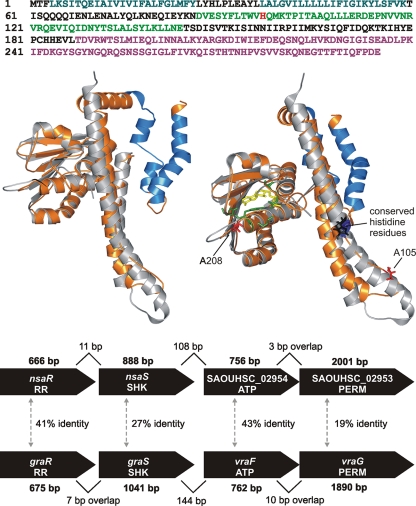
Consequently, at least in strain NR018, the mutation in NsaS makes the primary contribution to NIS resistance. NsaS has been annotated as a putative sensor histidine kinase but does not appear to have been subjected to further characterization. Pfam and InterProScan analyses corroborated this designation, revealing a conserved C-terminal catalytic and ATP-binding (HATPase_c) domain and a dimerization and histidine phosphotransfer domain (HisKA) (Fig. 1). SOSUI, DAS, and InterProScan all predicted two N-terminal transmembrane regions in NsaS (Fig. 1), a feature typical of membrane-associated histidine kinases (12).
Fig. 1.
Predicted functional domains and structure of the NsaS sensor histidine kinase and genetic context of the nsaS gene. (Top) Amino acid sequence of NsaS showing transmembrane domains (blue), the HisKA domain (green) containing the conserved histidine residue (bright red), and the HATPase_c domain (magenta), as predicted by InterProScan and SOSUI. (Middle left) Structural model of NsaS protein (orange) and X-ray crystal structure of the T. maritima sensor histidine kinase (gray) on which the model is based. Shown in blue are the transmembrane domains of NsaS predicted by SOSUI. (Middle right) Alternate view of the model on the left showing the ATP analogue ADPβN (yellow) bound into the T. maritima crystal structure and the residues with which it interacts (green). Also shown are NISr residues A208 and A105 in NsaS (red) and the conserved histidine residues in the T. maritima crystal structure (blue) and the NsaS model (black). (Bottom) Comparison of genetic architecture and protein products of the nsaRS and graRS operons of S. aureus SH1000. SHK, sensor histidine kinase; RR, response regulator; PERM, permease domain of ABC transporter; ATP, ATP-binding domain of ABC transporter. Gene sizes (bp) are indicated in bold, carets mark intergenic distances (bp), and % identity between paralogous protein products is shown.
To explore the potential impact of NISr mutations on the function of NsaS, a structural model of the protein was generated based on the published X-ray crystal structure of the cytoplasmic portion of a sensor histidine kinase from Thermotoga maritima (PDB accession number 2C2A) (31) (Fig. 1). Amino acid residues at positions 208 to 210 (mutated in NISr mutant NR018 and the two NISr derivatives of MW2) were found to lie in close proximity to residues that form hydrogen bonds with ATP within the ATP-binding site as defined in the T. maritima structure (31). In contrast, residue 105 (mutated in NISr mutant NR017) resides in the HisKA domain, carboxyl to the conserved histidine (Fig. 1).
STRING (25) was used to predict staphylococcal proteins that interact physically or functionally with NsaS. The top-ranked predicted functional partner was the putative cognate response regulator for NsaS, encoded by the neighboring gene SAOUHSC_02956 (which we consequently designated nsaR). The nsaRS genes reside in the same operon (45), upstream of the SAOUHSC_02953 and -4 genes, which encode a permease and the ATP-binding domain of a putative ABC transporter, respectively (Fig. 1, bottom panel). This genetic architecture closely resembles that of the two-operon cluster comprising graRS and vraFG, which determines susceptibility to vancomycin and the cationic antimicrobial peptide polymyxin B (33), and the protein counterparts encoded by these two systems are paralogous (Fig. 1).
DISCUSSION
Several parameters measured in vitro can be employed to predict the clinical resistance potential for an antibacterial agent (35). These include an assessment of the ease with which resistance arises, the impact that resistance exerts upon bacterial growth and competition, and the stability of resistance in the absence of selection (24, 46). Evaluation of these parameters for NIS suggests that this agent does not present a significant barrier to resistance development and maintenance in S. aureus. Thus, the frequency of mutation to low-level NIS resistance was high, with frequencies similar to those seen for antistaphylococcal agents which act upon one target and for which a single point mutation can confer clinically significant resistance, e.g., rifampin and fusidic acid (35). A single mutational event conferred ≤8-fold reduction in NIS susceptibility, while two mutations resulted in up to 32-fold reduction in NIS susceptibility. Furthermore, resistance was stable in the absence of selection and in most instances was not associated with a growth or competitive disadvantage in vitro.
In addition to NIS resistance developing in response to NIS selection, S. aureus genotypes which preexist in the clinic are capable of demonstrating cross-resistance to NIS. SCVs, which are found in patients with chronic staphylococcal infections (40), exhibit reduced susceptibility to NIS: this is presumably a consequence of the reduced membrane potential in these strains, leading to reduced affinity of cationic peptides for the bacterial membrane (39). Cross-resistance can also occur between NIS and daptomycin. Although NIS cross-resistance in daptomycin-resistant mutants has been reported previously (42), we have shown in the present study that selection for NIS resistance can in some cases result in cross-resistance to daptomycin, apparently as a consequence of mutation in the mprF gene.
Genetic characterization of in vitro-derived mutants resistant to an antibacterial agent can provide an indication of how resistance might arise in the clinical setting. Analysis of NR018 and other independently selected NISr mutants identified purR (encoding a purine operon repressor) and SAOUHSC_02955 (which we have designated nisin susceptibility-associated sensor [nsaS]), encoding a putative sensor histidine kinase, as important loci for acquired NIS resistance by S. aureus in vitro. Although PurR(V229G) from NR018 did not confer an increase in the NIS MIC when it was introduced into SH1000, the identification of distinct mutational changes in purR in three independently selected NISr mutants of SH1000 suggests that this locus is involved in NIS resistance in this strain background. One of these mutants (NR111) had a nonsense mutation in purR (E184STOP), suggesting that it is probably loss or impairment of PurR function that contributes to NIS resistance. However, the finding that spontaneous NISr mutants of strain MW2 did not have mutations in purR indicates that mutation of this gene is not essential for NIS resistance in S. aureus.
Allelic replacement of the mutation encoding NsaS(A208E) into SH1000 revealed that nsaS plays the major role in determining NIS resistance in NR018. It was anticipated that the double allelic replacement mutant SH1000:PurR(V229G), NsaS(A208E) would exhibit a NIS MIC equivalent to that of NR018, since it harbored the same two mutations detected in NR018. However, the NIS MIC was consistently at least 2-fold lower than that for NR018 and equivalent to that of the engineered strain SH1000:NsaS(A208E). It is conceivable that CGS failed to identify a third mutation present in NR018 which confers the additional increase in MIC from 64 μg/ml to 128 μg/ml, particularly since false-negative results (i.e., failure to detect all polymorphisms present in a genome) have previously been reported for this technique (18, 34).
The product of the nsaS gene has been annotated as a putative sensor histidine kinase, a designation further corroborated by the bioinformatic and structural modeling analyses presented here. The nsaS and nsaR genes, the latter of which is predicted to encode the cognate response regulator, lie in an operon a short distance upstream of a second operon which encodes a putative ABC transporter (45) (Fig. 1, bottom panel). This locus represents one of four in the S. aureus chromosome where genes encoding a sensor kinase/response regulator pair lie adjacent to those encoding an ABC transporter, and this appears to represent a common architecture for gene clusters determining resistance to antimicrobial peptides across the Firmicutes (26). In S. aureus, the best studied of these clusters comprises the operons encoding the GraRS two-component system and the VraFG ABC transporter (33) (Fig. 1, bottom panel). Both the architecture of the nsaRS operon and its downstream operon and the protein products which they encode are paralogous with the graRS/vraFG cluster (Fig. 1, bottom panel).
Thus, while further studies will be required to establish how nsaRS and the adjacent operon encoding a transporter participate in NIS resistance, it seems highly likely that this system is functionally similar to the graRS/vraFG system. In the latter system, mutations in the graRS regulatory circuit cause upregulation of VraFG, which is believed to participate in membrane detoxification and/or mediate export of components which alter the surface properties of the cell, thereby leading to shielding of lipid II by charge repulsion (29, 33). Furthermore, such regulatory mutations also trigger GraR-mediated upregulation of other genes that affect the cell surface (e.g., dlt and mprF) (36). By analogy, NIS resistance may result from NsaR-mediated upregulation of genes encoding the SAOUHSC_02953/4 ABC transporter and other proteins which affect the surface properties of the cell. Since loss of function of GraS results in increased susceptibility to antimicrobial peptides and putative gain-of-function mutations in this sensor are associated with increased resistance to vancomycin (22, 41), it seems reasonable to speculate that the NISr mutations we identified in nsaS also give rise to resistance owing to a gain of function.
Lantibiotics such as NIS exhibit potent antibacterial activity and may represent useful candidates for the development of novel antibacterial agents for human use (43). However, our results suggest that resistance to NIS would occur if it is employed clinically, and other lantibiotics may also be prone to resistance selection.
ACKNOWLEDGMENTS
Funding for this study was provided by the University of Leeds.
We thank J. Crabb (ImmuCell Corporation, ME) for providing pure nisin, G. Peters (University of Münster, Germany) for providing engineered SCV strains, and T. Bae (Indiana University School of Medicine—Northwest, IN) for advice concerning the use of pKOR1. We thank G. Cox for performing the I-TASSER structure prediction and K. Mariner and T. Tipton for technical assistance.
Footnotes
Published ahead of print on 7 February 2011.
REFERENCES
- 1. Asaduzzaman S. M., Sonomoto K. 2009. Lantibiotics: diverse activities and unique modes of action. J. Biosci. Bioeng. 107:475–487 [DOI] [PubMed] [Google Scholar]
- 2. Baba T., et al. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827 [DOI] [PubMed] [Google Scholar]
- 3. Bae T., Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63 [DOI] [PubMed] [Google Scholar]
- 4. Bates D. M., et al. 2003. Staphylococcus aureus menD and hemB mutants are as infective as the parent strains, but the menadione biosynthetic mutant persists within the kidney. J. Infect. Dis. 187:1654–1661 [DOI] [PubMed] [Google Scholar]
- 5. Breukink E., et al. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361–2364 [DOI] [PubMed] [Google Scholar]
- 6. Cserzo M., Wallin E., Simon I., von Heijne G., Elofsson A. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673–676 [DOI] [PubMed] [Google Scholar]
- 7. de Kruijff B., van Dam V., Breukink E. 2008. Lipid II: a central component in bacterial cell wall synthesis and a target for antibiotics. Prostaglandins Leukot. Essent. Fatty Acids 79:117–121 [DOI] [PubMed] [Google Scholar]
- 8. Delves-Broughton J., Blackburn P., Evans R. J., Hugenholtz J. 1996. Applications of the bacteriocin, nisin. Antonie Van Leeuwenhoek 69:193–202 [DOI] [PubMed] [Google Scholar]
- 9. Fairweather N., Kennedy S., Foster T. J., Kehoe M., Dougan G. 1983. Expression of a cloned Staphylococcus aureus alpha-hemolysin determinant in Bacillus subtilis and Staphylococcus aureus. Infect. Immun. 41:1112–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Finn R. D., et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Friedman L., Alder J. D., Silverman J. A. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:2137–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao R., Stock A. M. 2009. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63:133–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goldstein B. P., Wei J., Greenberg K., Novick R. 1998. Activity of nisin against Streptococcus pneumoniae, in vitro, and in a mouse infection model. J. Antimicrob. Chemother. 42:277–278 [PubMed] [Google Scholar]
- 14. Gravesen A., Jydegaard Axelsen A. M., Mendes da Silva J., Hansen T. B., Knochel S. 2002. Frequency of bacteriocin resistance development and associated fitness costs in Listeria monocytogenes. Appl. Environ. Microbiol. 68:756–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gravesen A., Sorensen K., Aarestrup F. M., Knochel S. 2001. Spontaneous nisin-resistant Listeria monocytogenes mutants with increased expression of a putative penicillin-binding protein and their sensitivity to various antibiotics. Microb. Drug Resist. 7:127–135 [DOI] [PubMed] [Google Scholar]
- 16. Hasper H. E., de Kruijff B., Breukink E. 2004. Assembly and stability of nisin-lipid II pores. Biochemistry 43:11567–11575 [DOI] [PubMed] [Google Scholar]
- 17. Hasper H. E., et al. 2006. An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 313:1636–1637 [DOI] [PubMed] [Google Scholar]
- 18. Herring C. D., Palsson B. O. 2007. An evaluation of comparative genome sequencing (CGS) by comparing two previously-sequenced bacterial genomes. BMC Genomics 8:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hirokawa T., Boon-Chieng S., Mitaku S. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379 [DOI] [PubMed] [Google Scholar]
- 20. Hobbs J. K., Miller K., O'Neill A. J., Chopra I. 2008. Consequences of daptomycin-mediated membrane damage in Staphylococcus aureus. J. Antimicrob. Chemother. 62:1003–1008 [DOI] [PubMed] [Google Scholar]
- 21. Horsburgh M. J., et al. 2002. sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Howden B. P., et al. 2008. Genomic analysis reveals a point mutation in the two-component sensor gene graS that leads to intermediate vancomycin resistance in clinical Staphylococcus aureus. Antimicrob. Agents Chemother. 52:3755–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hsu S.-T. D., et al. 2004. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat. Struct. Mol. Biol. 11:963–967 [DOI] [PubMed] [Google Scholar]
- 24. Hurdle J. G., O'Neill A. J., Ingham E., Fishwick C., Chopra I. 2004. Analysis of mupirocin resistance and fitness in Staphylococcus aureus by molecular genetic and structural modeling techniques. Antimicrob. Agents Chemother. 48:4366–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jensen L. J., et al. 2009. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37:D412–D416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jordan S., Hutchings M. I., Mascher T. 2008. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32:107–146 [DOI] [PubMed] [Google Scholar]
- 27. Koch A. L. 1994. Growth measurement, p. 248–292 In Gerhardt R., Murray R. G. E., Wood W. A., Krieg N. R. (ed.), Methods of general and molecular bacteriology. American Society for Microbiology, Washington, DC [Google Scholar]
- 28. Kramer N. E., et al. 2004. Resistance of Gram-positive bacteria to nisin is not determined by lipid II levels. FEMS Microbiol. Lett. 239:157–161 [DOI] [PubMed] [Google Scholar]
- 29. Li M., et al. 2007. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol. Microbiol. 66:1136–1147 [DOI] [PubMed] [Google Scholar]
- 30. Madigan M. T., Martinko J. M., Parker J. 2000. Brock biology of microorganisms, 9th ed. Prentice Hall International, Upper Saddle River, NJ [Google Scholar]
- 31. Marina A., Waldburger C. D., Hendrickson W. A. 2005. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 24:4247–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McAuliffe O., Ross R. P., Hill C. 2001. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 25:285–308 [DOI] [PubMed] [Google Scholar]
- 33. Meehl M., Herbert S., Gotz F., Cheung A. 2007. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 51:2679–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O'Neill A. J. 2010. Staphylococcus aureus SH1000 and 8325-4: comparative genome sequences of key laboratory strains in staphylococcal research. Lett. Appl. Microbiol. 51:358–361 [DOI] [PubMed] [Google Scholar]
- 35. O'Neill A. J., Chopra I. 2004. Preclinical evaluation of novel antibacterial agents by microbiological and molecular techniques. Expert Opin. Invest. Drugs 13:1045–1063 [DOI] [PubMed] [Google Scholar]
- 36. Otto M. 2009. Bacterial sensing of antimicrobial peptides. Contrib. Microbiol. 16:136–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Piper C., Cotter P. D., Ross R. P., Hill C. 2009. Discovery of medically significant lantibiotics. Curr. Drug Discov. Technol. 6:1–18 [DOI] [PubMed] [Google Scholar]
- 38. Piper C., Draper L. A., Cotter P. D., Ross R. P., Hill C. 2009. A comparison of the activities of lacticin 3147 and nisin against drug-resistant Staphylococcus aureus and Enterococcus species. J. Antimicrob. Chemother. 64:546–551 [DOI] [PubMed] [Google Scholar]
- 39. Proctor R. A., et al. 1998. Staphylococcal small colony variants have novel mechanisms for antibiotic resistance. Clin. Infect. Dis. 27(Suppl. 1):S68–S74 [DOI] [PubMed] [Google Scholar]
- 40. Proctor R. A., et al. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4:295–305 [DOI] [PubMed] [Google Scholar]
- 41. Sass P., Bierbaum G. 2009. Native graS mutation supports the susceptibility of Staphylococcus aureus strain SG511 to antimicrobial peptides. Int. J. Med. Microbiol. 299:313–322 [DOI] [PubMed] [Google Scholar]
- 42. Silverman J. A., Oliver N., Andrew T., Li T. 2001. Resistance studies with daptomycin. Antimicrob. Agents Chemother. 45:1799–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Smith L., Hillman J. D. 2008. Therapeutic potential of type A (I) lantibiotics, a group of cationic peptide antibiotics. Curr. Opin. Microbiol. 11:401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Spellberg B., et al. 2008. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin. Infect. Dis. 46:155–164 [DOI] [PubMed] [Google Scholar]
- 45. ten Broeke-Smits N. J. P., et al. 2010. Operon structure of Staphylococcus aureus. Nucleic Acids Res. 38:3263–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vickers A. A., O'Neill A. J., Chopra I. 2007. Emergence and maintenance of resistance to fluoroquinolones and coumarins in Staphylococcus aureus: predictions from in vitro studies. J. Antimicrob. Chemother. 60:269–273 [DOI] [PubMed] [Google Scholar]
- 47. Vickers A. A., Potter N. J., Fishwick C. W. G., Chopra I., O'Neill A. J. 2009. Analysis of mutational resistance to trimethoprim in Staphylococcus aureus by genetic and structural modelling techniques. J. Antimicrob. Chemother. 63:1112–1117 [DOI] [PubMed] [Google Scholar]
- 48. von Eiff C., et al. 1997. A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J. Bacteriol. 179:4706–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zdobnov E. M., Apweiler R. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848 [DOI] [PubMed] [Google Scholar]
- 50. Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]