Abstract
Seventeen Klebsiella pneumoniae isolates producing the OXA-48 carbapenemase, obtained from 10 patients hospitalized from April to June 2010, mostly in the medical intensive care unit of the Villeneuve-Saint-Georges Hospital in a suburb of Paris, France, were analyzed. Seven patients were infected, of whom five were treated at least with a carbapenem, and five patients died. Molecular analysis showed that the isolates belonged to a single clone that harbored a 70-kb plasmid carrying the blaOXA-48 gene and coproduced CTX-M-15 and TEM-1 β-lactamases. This is the first reported outbreak of OXA-48-producing K. pneumoniae isolates in France.
INTRODUCTION
Carbapenems possess the most consistent in vitro activity against extended-spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae bacteria. Resistance to carbapenems, while still rare in Enterobacteriaceae, is increasing and represents a significant threat in the management of multidrug-resistant isolates (22, 25). It is mediated mostly by two main mechanisms. The first involves the production of a β-lactamase (a cephalosporinase or an ESBL) with a very low level of carbapenem-hydrolyzing activity combined with decreased permeability due to porin loss or alteration (13, 21). The second mechanism is related to carbapenem-hydrolyzing β-lactamases. The carbapenemases identified in Klebsiella pneumoniae isolates are metallo-β-lactamases (IMP, VIM, NDM) (6), plasmid-mediated clavulanic acid-inhibited β-lactamases (NmcA, IMI, SME, GES, and KPC) (22, 25), and the expanded-spectrum oxacillinase OXA-48 (23, 24).
The Ambler class D β-lactamase OXA-48, initially identified from a carbapenem-resistant K. pneumoniae isolate from Istanbul, Turkey, hydrolyzes penicillins and imipenem, sparing expanded-spectrum cephalosporins (23). The blaOXA-48 gene is located on Tn1999, a composite transposon made of two copies of the insertion sequence IS1999 (2). Outbreaks of OXA-48-producing K. pneumoniae and other enterobacterial isolates have been described in several cities in Turkey (1, 8, 9, 17) and once in the United Kingdom (27). Subsequently, single isolates of OXA-48-producing K. pneumoniae have been reported from Lebanon (20), Belgium (11), the United Kingdom (19), Tunisia (14), Israel (16), Morocco (4), Argentina (5), and India (3).
We describe here a nosocomial outbreak of carbapenem-resistant K. pneumoniae strains expressing OXA-48 associated with a CTX-M-15 ESBL in France.
In April 2010, three patients hospitalized in the medical intensive care unit (ICU) at the hospital of Villeneuve-Saint-Georges (VSG), a suburb of Paris, France, were infected by a multidrug-resistant K. pneumoniae with resistance to expanded-spectrum cephalosporins and carbapenems. During the subsequent period of time, all patients who had contact with these three patients were screened for fecal carriage of multidrug-resistant bacteria, using a medium designed to select for ESBL producers, BLSE agar (AES, Bruz, France) (26). From April 2010 to June 2010, a total of 260 patients hospitalized in the ICU and the internal medicine unit were screened. Of those 260 patients, 7 were infected and 3 were only colonized by multidrug-resistant K. pneumoniae isolates with the same susceptibility pattern (Table 1), indicating probable nosocomial transmission. Five patients had nosocomial pneumonia and three patients developed a catheter-related bloodstream infection, one of them having both types of infection. Infected patients were treated with carbapenems prior to and/or after the determination of antibiotic susceptibility. Colistin and/or amikacin were added in most of the infection cases. Despite treatment, five of the seven infected patients died; two of them improved. Until now, OXA-48 producers in France have been identified from patients transferred from a country in the Mediterranean area (12, 14). None of the VSG patients had a recent history of travel to a country known for having widespread OXA-48 producers, such as Turkey. The first patient infected with the OXA-48-producing K. pneumoniae bacteria had never traveled abroad and was retrospectively found to have been colonized at the end of March 2010, suggesting a possible lack of detection of the true index case and subsequent nosocomial transmission. Cohorting of patients and reinforced hygiene measures have been implemented, but several patients were already infected or colonized and may have contributed to the spread of OXA-48 producers.
Table 1.
Clinical features and MICs associated with isolation of carbapenem-resistant K. pneumonia isolates
| Patient | Date of hospitalization (mo/day/yr) | Hospital unit | Underlying disease | Isolate | Date of isolation (mo/day/yr) | Site of isolation | Infection(s)/colonization | Treatment | Patient outcome | MIC (μg/ml) ofa: |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IP | ETP | MP | DP | AN | CS | TGC | ||||||||||
| 1 | 03/30/10 | ICU | Legionnaire disease | VSG1 | 03/30/10 | Rectal swab | Colonization | None | ||||||||
| VSG2 | 04/16/10 | Catheter blood culture | Catheter-related bloodstream infection + thrombophlebitis | Doripenem + amikacin + colistin | Improved | 0.75 | 3 | 0.75 | 0.50 | <4 | 0.25 | 3 | ||||
| VSG3 | 04/17/10 | Endotracheal aspirate | Nosocomial pneumonia | |||||||||||||
| 2 | 04/14/10 | ICU | Severe pneumonia | VSG4 | 04/25/10 | Endotracheal aspirate | Nosocomial pneumonia | Imipenem | Deceased | 2 | 3 | 0.50 | 0.50 | <4 | 0.75 | 3 |
| 3 | 04/20/10 | ICU | Chronic bronchopneumonia | VSG5 | 05/12/10 | Endotracheal aspirate | Nosocomial pneumonia | Imipenem + amikacin + colistin (aerosol) | Deceased | 4 | 3 | 0.75 | 0.50 | <4 | 0.50 | 3 |
| VSG6 | 05/24/10 | Catheter | Colonization | None | ||||||||||||
| 4 | 05/10/10 | ICU | Pyelonephritis | VSG7 | 05/30/10 | Endotracheal aspirate | Nosocomial pneumonia | Meropenem + colistin | Improved | 1 | 4 | 1 | 0.75 | <4 | 0.75 | 2 |
| 5 | 05/14/10 | ICU | Chronic bronchopneumonia | VSG8 | 05/17/10 | Rectal swab | Colonization | None | ||||||||
| VSG9 | 05/27/10 | Catheter blood culture | Catheter-related bloodstream infection | Colistin + ciprofloxacin + ceftazidime | Deceased | 0.75 | 4 | 0.75 | 0.50 | <4 | 0.25 | 2 | ||||
| 6 | 05/14/10 | ICU | Lung cancer | VSG10 | 05/26/10 | Rectal swab | Colonization | None | ||||||||
| VSG11 | 06/04/10 | Endotracheal aspirate | Nosocomial pneumonia | None | Deceased | 0.75 | 4 | 1 | 0.75 | <4 | 0.19 | 2 | ||||
| 7 | 05/21/10 | ICU | Biliary duct adenocarcinoma | |||||||||||||
| 06/01/10 | Internal medicine | VSG12 | 06/07/10 | Rectal swab | Colonization | None | Improved | 0.75 | 3 | 0.75 | 0.75 | <4 | 0.25 | 3 | ||
| 8 | 05/31/10 | Internal medicine | Acute pulmonary edema | |||||||||||||
| 06/03/10 | ICU | VSG13 | 06/03/10 | Rectal swab | Colonization | None | Deceased | 1 | 4 | 0.50 | 0.50 | <4 | 0.75 | 2 | ||
| 9 | 06/07/10 | Internal medicine | Severe pneumonia | |||||||||||||
| 06/11/10 | ICU | VSG14 | 06/21/10 | Rectal swab | Colonization | None | ||||||||||
| VSG15 | 06/24/10 | Catheter | Catheter-related bloodstream infection | Meropenem + colistin | Deceased | 1 | 4 | 1 | 0.50 | <4 | 0.75 | 2 | ||||
| 10 | 06/14/10 | ICU | Brain stroke | VSG16 | 06/21/10 | Rectal swab | Colonization | None | Improved | 1 | 3 | 0.75 | 0.25 | <4 | 0.50 | 3 |
IP, imipenem; ETP, ertapenem; MP, meropenem; DP, doripenem; AN, amikacin; CS, colistin; TGC, tigecycline.
The antibiotic susceptibilities of the isolates was first determined with the Vitek 2 system (bioMérieux, Marcy l'Etoile, France) (AST-N103 card, software version 04.02), which identified resistance to carbapenems. As routinely performed, the antibiogram was confirmed by the disk diffusion method according to Clinical and Laboratory Standards Institute guidelines (10). Additionally, the MICs of several antibiotics were determined using Etest strips (bioMérieux) and the Vitek 2 system (Table 1). All isolates were resistant to all penicillins and expanded-spectrum cephalosporins, and they exhibited heterogeneous decreased susceptibilities to carbapenems, with ertapenem resistance, imipenem being in the intermediate or susceptible range, and meropenem and doripenem being in the susceptible range according to Clinical and Laboratory Standards Institute guidelines updated in June 2010 (10). Moreover, these isolates were resistant to aminoglycosides, except for amikacin, fluoroquinolones, cotrimoxazole, and tigecycline, and remained susceptible to colistin, according to guidelines from the Committé Antibiogramme—Société Française de Microbiologie (CA-SFM) (http://www.sfm-microbiologie.org/UserFiles/file/casfm_2010.pdf).
Specific primers were used for the detection of carbapenemases genes and for β-lactamase-encoding genes that had been previously identified in K. pneumoniae isolates that produce OXA-48, namely, blaTEM, blaSHV, blaCTX-M, blaOXA-1, and blaOXA-47 (9). All isolates were positive for the blaOXA-48, blaCTX-M-15, blaSHV-1, blaTEM-1, and blaOXA-1 genes (Table 2).
Table 2.
Structure of Tn1999, other β-lactamases, plasmid analysis, pulsotypes, and ST types of different OXA-48-producing Klebsiella pneumoniae isolates from various geographic origins
| Isolate | Origin | Reference | PCR and sequencing characterization of: |
Characterization of: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tn1999 |
Other β-lactamases |
blaOXA-48-carrying plasmid |
Bacterial isolate |
||||||||||
| OXA-48 | IS1999 | IS1 | SHV | TEM | CTX-M | OXA | Size (kb) | Inca | PFGE type | ST type | |||
| VSG1-16 | France | This study | + | + | + | SHV-11 | TEM-1 | CTX-M-15 | OXA-1 | 70 | P | A | ST-395 |
| UCL-1 | Belgium | 11 | + | + | + | SHV-11 | TEM-1 | 70 | P | B | ST-147 | ||
| KpE | Egypt | 12 | + | + | + | SHV-11 | TEM-1 | CTX-M-15 | OXA-1 | 70 | P | C | ST-152 |
| HPA-1 | Tunisia | 14 | + | + | + | SHV-11 | TEM-1 | CTX-M-15 | OXA-1 | 70 | P | D | ST-101 |
| KpL | Lebanon | 20 | + | + | + | SHV-11 | TEM-1 | CTX-M-15 | OXA-1 | 70 | P | D | ST-496 |
| 11978 | Turkey | 23 | + | + | − | SHV-11 | TEM-1 | OXA-1 | 70 | P | E | ST-14 | |
| SHV-2a | OXA-47 | ||||||||||||
Inc, plasmid incompatibility group.
The genetic relationship between the different isolates studied by pulsed-field gel electrophoresis (PFGE) revealed that the isolates were clonally related. These strains were compared with K. pneumoniae 11978 from Turkey (23) and with isolates identified in Lebanon (20), Belgium (11), Tunisia (14), and Egypt (12). This epidemic clone was genetically distinct from all the other isolates (Table 2). Multilocus sequence typing (MLST) with seven housekeeping genes (rpoB, gapA, mdh, pgi, phoE, infB, and tonB) was performed according to the method of Diancourt et al. (15). Allele sequences and sequence types (STs) were verified by using the PubMLST Klebsiella pneumoniae MLST Database (http://www.pasteur.fr/recherche/genopole/PF8/mlst/kpneumoniae.html). All isolates from VSG displayed ST-395 (allelic profile, 3-1-2-4-1-1-4) (Table 2). The analysis of STs using eBURST (http://pubmlst.org) showed that ST-395 is a single-locus variant of ST-134. The ST types of isolates from other geographical origins were also determined and were different, confirming the results of PFGE (Table 2).
Plasmid DNA extraction according to the Kieser technique (18) showed that in the VSG isolates, the blaOXA-48 gene was carried by a self-conjugative 70-kb plasmid, whereas the other β-lactamases genes were carried on larger plasmids (data not shown). Using specific primers as described by Carrër et al. (9), we identified the repP gene as the plasmid incompatibility group. Therefore, the plasmid content of the OXA-48-producing K. pneumoniae clone identified here was identical to that previously reported (9).
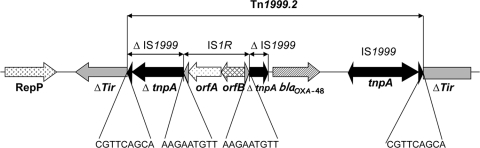
The genetic environment of the blaOXA-48 gene was determined by PCR using specific primers for the insertion sequence IS1999, located upstream and downstream of the blaOXA-48 gene in Tn1999 (2). A structure similar to Tn1999, Tn1999.2, was identified for all isolates. Tn1999.2 differs from Tn1999 by the presence of an IS1R element inserted in the IS1999 element located upstream of the blaOXA-48 gene (Fig. 1) (9).
Fig. 1.
Schematic representation of Tn1999.2, identified with the blaOXA-48 gene in the epidemic clone. Genes and their transcription orientations are represented as horizontal arrows. Black triangles represent inverted repeats of insertion sequence IS1999, and gray triangles represent inverted repeats of insertion sequence IS1R. Target site duplications are indicated below the sequence. Δ indicates that the gene/element is interrupted by an insertion element.
Here, we identify the first outbreak of OXA-48-positive, carbapenem-resistant K. pneumoniae isolates in France. During 2 months, 10 patients were found to be infected or colonized by similar multidrug-resistant isolates. Seven patients developed infection and, because of limited treatment options and comorbidities, five died. Two of the deceased patients were treated at least with imipenem (500 mg every 6 h), imipenem being reported in the intermediate susceptibility range according to the June 2010 updated guidelines of the CLSI. Although a valid comparison is difficult to establish, the death rate is comparable to that observed in another OXA-48-associated outbreak from Istanbul, where 10 of 15 infected patients died despite imipenem-containing treatment. Clinical laboratory detection of OXA-48-producing strains may be difficult because of a low level of resistance to carbapenems and, sometimes, lack of additional ESBLs (9, 11). In this study, detection of patients having gut carriage was facilitated by the additional resistance to cephalosporins due to the presence of the blaCTX-M-15 gene (7). The OXA-48 producers identified here were not clonally related to the previously identified OXA-48-positive K. pneumoniae isolates, but a very similar 70-kb plasmid harboring blaOXA-48 was identified (9). Therefore, spread of the blaOXA-48 gene may be very much associated with the spread of a single plasmid type. This study underlines that K. pneumoniae is an enterobacterial species prone to cause hospital-based outbreaks of strains carrying multidrug-resistant genes, such as ESBL and, now, carbapenemase genes. Outbreaks of OXA-48 K. pneumoniae producers are now identified in Western Europe after those producing other types of carbapenemases, in particular KPC-2 and VIM-1.
Acknowledgments
We thank A. Carrër for helpful discussions. We thank the platform Genotyping of Pathogens and Public Health (Institut Pasteur) for coding MLST alleles and profiles.
This work was funded by INSERM, France, by a grant in aid from the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539), Université Paris XI, Paris, and by a grant from the European Community (TEMPOtest-QC, HEALTH-2009-241742).
Footnotes
Published ahead of print on 22 February 2011.
REFERENCES
- 1. Aktas Z., et al. 2008. Carbapenem-hydrolyzing oxacillinase OXA-48 persists in Klebsiella pneumoniae in Istanbul, Turkey. Chemotherapy 54:101–106 [DOI] [PubMed] [Google Scholar]
- 2. Aubert D., Naas T., Héritier C., Poirel L., Nordmann P. 2006. Functional characterization of IS1999, an IS4 family element involved in mobilization and expression of β-lactam resistance genes. J. Bacteriol. 188:6506–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bell J. M., Mathai D., Jones R. N., Turnidge J. D. 2010. Emergence of OXA-48 carbapenemases among Klebsiella spp. from India, abstr. C2-650. Abstr. 50th Intersci. Conf. Antimicrob. Agents Chemother American Society for Microbiology, Washington, DC [Google Scholar]
- 4. Benouda A., Touzani O., Khairallah M. T., Araj G. F., Matar G. M. 2010. First detection of oxacillinase-mediated resistance to carbapenems in Klebsiella pneumoniae from Morocco. Ann. Trop. Med. Parasitol. 104:327–330 [DOI] [PubMed] [Google Scholar]
- 5. Castanheira M., Watters A. A., Smayevsky J., Casellas J. M., Gur D., Jones J. 2010. Emergence of blaOXA-48 among Enterobacter cloacae (ECL) in Argentina and its prevalence among carbapenem (CARB) non-susceptible (NS) Enterobacteriaceae, abstr. C2-647. Abstr. 50th Intersci. Conf. Antimicrob. Agents Chemother American Society for Microbiology, Washington, DC [Google Scholar]
- 6. Cagnacci S., et al. 2008. Bloodstream infections caused by multidrug-resistant Klebsiella pneumoniae producing the carbapenem-hydrolysing VIM-1 metallo-beta-lactamase: first Italian outbreak. J. Antimicrob. Chemother. 61:296–300 [DOI] [PubMed] [Google Scholar]
- 7. Carrër A., Fortineau N., Nordmann P. 2010. Use of ChromID extended-spectrum β-lactamase medium for detecting carbapenemase-producing Enterobacteriaceae. J. Clin. Microbiol. 48:1913–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carrër A., et al. 2008. Spread of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in Istanbul, Turkey. Antimicrob. Agents Chemother. 52:2950–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carrër A., et al. 2010. Spread of OXA-48-encoding plasmid in Turkey and beyond. Antimicrob. Agents Chemother. 54:1369–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial susceptibility testing; 20th informational supplement M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 11. Cuzon G., et al. 2008. Plasmid-encoded carbapenem-hydrolyzing beta-lactamase OXA-48 in an imipenem-susceptible Klebsiella pneumoniae strain from Belgium. Antimicrob. Agents Chemother. 52:3463–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cuzon G., et al. 2009. Plasmid-mediated carbapenem-hydrolyzing beta-lactamase OXA-48 in Klebsiella pneumoniae from Egypt, abstr. C2-649. Abstr. 49th Intersci. Conf. Antimicrob. Agents Chemother American Society for Microbiology, Washington, DC [Google Scholar]
- 13. Cuzon G., Naas T., Guibert M., Nordmann P. 2010. In vivo selection of imipenem-resistant Klebsiella pneumoniae producing extended-spectrum beta-lactamase CTX-M-15 and plasmid-encoded DHA-1 cephalosporinase. Int. J. Antimicrob. Agents 35:265–268 [DOI] [PubMed] [Google Scholar]
- 14. Cuzon G., Naas T., Lesenne A., Benhamou M., Nordmann P. 2010. Plasmid-mediated carbapenem-hydrolysing OXA-48 beta-lactamase in Klebsiella pneumoniae from Tunisia. Int. J. Antimicrob. Agents 36:91–93 [DOI] [PubMed] [Google Scholar]
- 15. Diancourt L., Passet V., Verhoef J., Grimont P. A., Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goren M. G., Chmelnitsky I., Carmeli Y., Navon-Venezia S. 2009. First report of plasmid-encoded OXA-48-like carbapenemase in E. coli from Israel, abstr. C1-1765. Abstr. 49th Intersci. Conf. Antimicrob. Agents Chemother American Society for Microbiology, Washington, DC [Google Scholar]
- 17. Gulmez D., et al. 2008. Carbapenem-resistant Escherichia coli and Klebsiella pneumoniae isolates from Turkey with OXA-48-like carbapenemases and outer membrane protein loss. Int. J. Antimicrob. Agents 31:523–526 [DOI] [PubMed] [Google Scholar]
- 18. Kieser T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19–36 [DOI] [PubMed] [Google Scholar]
- 19. Livermore D. M. 2009. Has the era of untreatable infections arrived? J. Antimicrob. Chemother. 64:29–36 [DOI] [PubMed] [Google Scholar]
- 20. Matar G. M., et al. 2008. Oxacillinase-mediated resistance to carbapenems in Klebsiella pneumoniae from Lebanon. Clin. Microbiol. Infect. 14:887–888 [DOI] [PubMed] [Google Scholar]
- 21. Mena A., et al. 2006. Characterization of a large outbreak by CTX-M-1-producing Klebsiella pneumoniae and mechanisms leading to in vivo carbapenem resistance development. J. Clin. Microbiol. 44:2831–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nordmann P., Cuzon G., Naas T. 2009. The real threat of KPC carbapenemase-producing bacteria. Lancet Infect. Dis. 9:321–331 [DOI] [PubMed] [Google Scholar]
- 23. Poirel L., Héritier C., Tolün V., Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Poirel L., Naas T., Nordmann P. 2010. Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob. Agents Chemother. 54:24–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Queenan A. M., Bush K. 2007. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 20:440–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Réglier-Poupet H., et al. 2008. Performance of chromID ESBL, a chromogenic medium for detection of Enterobacteriaceae producing extended-spectrum beta-lactamases. J. Med. Microbiol. 57:310–315 [DOI] [PubMed] [Google Scholar]
- 27. Thomas C. P., et al. 2009. Hospital outbreak of Klebsiella pneumoniae producing OXA-48 carbapenemase in the United Kingdom, abstr. C2-648. Abstr. 49th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC [Google Scholar]