Abstract
Azole resistance is an increasing clinical problem for Aspergillus fumigatus, with the majority of published resistance arising from mutations in the azole target gene CYP51A. Previous structural studies of this protein have suffered from a nonorthologous, low-homology template for homology modeling. Here we present a new model based on the human CYP51A orthologue that provides a higher-quality model for A. fumigatus CYP51A.
INTRODUCTION
Fungi are the causative agents of increasingly common life-threatening diseases (10). Invasive aspergillosis is one of the most common of these conditions and is caused by Aspergillus fumigatus in over 95% of cases (9). The most important, efficacious, and widely used class of antifungal compounds is the azoles. These compounds target sterol 14 demethylase and interfere with ergosterol production in the fungus (9). Resistance to azoles is constantly arising; resistance occurs in an estimated 8% of patients within the course of their treatment, and azole resistance is commonly found in plant-pathogenic fungi (4–6, 15). Fifty to 80 percent of azole resistance in human pathogens arises from mutations in the sterol demethylase gene (CYP51A) (4–6, 15). The most common CYP51A mutations that lead to azole resistance in fungi are M220, G54, and L98H. Protein structure modeling is an important tool in the study of drug action and resistance. Attempts to obtain a crystal structure for the fungal CYP51A protein have not been successful, and therefore we are forced to rely on in silico homology modeling. Previous models of the A. fumigatus CYP51A protein have been based on a crystal structure of a cytochrome P450 from Mycobacterium tuberculosis (1, 12, 13, 15). This enzyme is probably not a true orthologue of lanosterol demethylase (1); the amino acid homology is low (29% across 414/515 amino acids), and the A. fumigatus protein is 101 amino acids longer than the M. tuberculosis protein, resulting in lack of homology data for regions of the A. fumigatus gene that have been shown to be involved in azole resistance. Recently, the structure of the human CYP51A protein has been determined (http://www.pdb.org/pdb/explore/explore.do?structureId = 3JUS). This protein is a biosynthetic lanosterol demethylase and is 41% identical to the A. fumigatus protein across its complete amino acid sequence.
Models of the CYP51A proteins from two strains of A. fumigatus, Af293 and A1163, were constructed de novo for wild-type A. fumigatus CYP51A (AfCYP51A) and for sequences containing substitutions at sites commonly associated with drug resistance, namely, G54R, L98H, M220I, M220K, M220R, M220V, M220W, and G448S (4–6, 12), using the human 3JUS (azole-conjugated) and 3LD6 (no-azole) templates. For comparison, we modeled the A. fumigatus protein using the same parameters with the M. tuberculosis 1EA1 template. Models were constructed using Modeler 9.8 (7), with 3JUS or 3LD6 as a single template. Briefly, proteins were aligned using the ALIGN2D function, and then models were generated by the internal program. Discrete optimized potential energy (DOPE)-based loop modeling was performed, followed by model optimization using the Beale restart conjugate-gradients method (20 iterations at 300°C) and molecular dynamics optimization using the Verlet algorithm (100 iterations at 300°C). Quality scores for each model were obtained using DOPE modeling and the molecular pdf energy. Finally, models were tested for quality using QMEAN (2, 3). Models were also assessed using Ramachandran plots for each model iteration. Results of quality scoring are shown in Table 1, and model coordinates are available at http://130.88.242.202/medicine/Aspergillus/cyp51a/. Positions of heme and azole ligands were inferred by comparison to the human crystal structure.
Table 1.
Summary of structures tested and quality scores for the models deriveda
| Model | Template | Molpdf score | DOPE score | %ID | QMEAN (all) | QMEAN (no signal) |
|---|---|---|---|---|---|---|
| CYP51A | JUS | 754.11322 | −56,059.07031 | 41.34 | 0.662 | 0.801 |
| CYP51A | EAE | 4,125.88184 | −54,123.19922 | 29.97 | 0.584 | 0.584 |
| G54R | JUS | 2,959.02418 | −50,929.875 | 41.124 | 0.625 | 0.731 |
| G448S | JUS | 1,281.91821 | −50,139.01563 | 41.124 | 0.702 | 0.837 |
| M220I | JUS | 1,733.19971 | −50,897.00781 | 41.124 | 0.633 | 0.798 |
| M220V | JUS | 9,602.02637 | −49,395.46094 | 41.348 | 0.559 | 0.659 |
| M220W | JUS | 1,467.80127 | −50,351.31641 | 41.124 | 0.609 | 0.715 |
| M220K | JUS | 1,374.15918 | −50,486.30059 | 41.348 | 0.657 | 0.746 |
| M220R | JUS | 831.92627 | −49,997.29688 | 41.124 | 0.64 | 0.781 |
| L98H | JUS | 784.87683 | −49,235.55839 | 41.348 | 0.676 | 0.827 |
Values are for the Af293 structures. Note that molecular probability density function (PDF) energy (Molpdf) and DOPE scores serve as internal quality metrics and are not externally comparable. QMEAN scores were calculated for full-length models (all) and models lacking the predicted 24-amino-acid secretion/ER retention signal (no signal). %ID, percent identity.
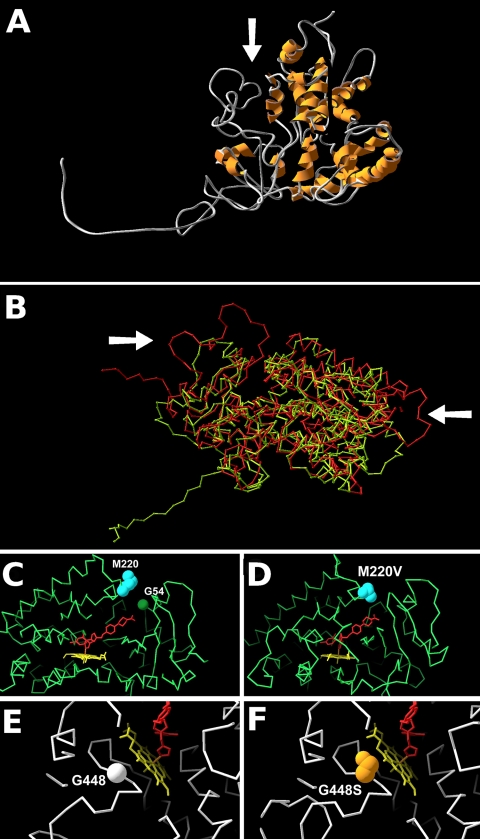
The wild-type Af293 A. fumigatus CYP51A model based on the human 3JUS structure (Af293CYP51A-3JUS) (Fig. 1 A) had a QMEAN score of 0.662; the model based on the M. tuberculosis 1EA1 structure had a QMEAN score of 0.584, suggesting that the new model was of higher quality than those previously constructed using this template (11, 13). Models from 3JUS and 1EAE are superimposed in Fig. 1 B, showing a number of clear structural differences, most notably at the entrance to the active-site cleft. Two poor-quality regions in the Af293CYP51A-3JUS model from QMEAN mapping and Ramachandran plots are in the 24 residues at the N terminus. This region is hydrophobic and predicted by PSORT2 analysis to function as an endoplasmic reticulum (ER)/Golgi retention signal. No mutations associated with azole resistance have been found in this region, and we do not expect it to play an important role in either enzyme function or drug binding (Table 2). When the predicted ER retention signal was omitted, the Af293CYP51A-3JUS model had a QMEAN score of 0.801. The 1EAE-based model lacked this sequence, and thus the comparable QMEAN score remained 0.584. The second area of poor quality in the model is the AfCYP51A position 415-to-440 loop. This sequence is not present in the human protein, and examination of the A. fumigatus gene raised the possibility that this was a readthrough intron, as it was precisely flanked by intron consensus sites and contained a fungal lariat consensus motif. Surprisingly, no full-length cDNA sequence has been published for this gene. We synthesized and sequenced the cDNA, but the region was present in the mRNA, suggesting the possibility that this region may have arisen through intronic decay. Multiple in-frame ATG sites occur within the promoter, so we also determined 5′ and 3′ mRNA boundaries (by rapid amplification of cDNA ends [RACE]) to confirm that the gene and protein predictions were correct. Transcriptional start sites were determined at positions −114 and −57 relative to the ATG, and termination was mapped to position +48 from the stop codon.
Fig. 1.
Models of CYP51A and azole resistance-associated amino acid variants. The protein CA (alpha carbon) backbone is shown in either green or white, heme is shown in yellow, and the inferred azole position for ketoconazole is shown in red. (A) Predicted structure of the Af293 CYP51A protein showing the active-site cleft (arrow). (B) Superimposed structures of Af293 CYP51A derived using human 3JUS (green CA trace) and m. tuberculosis 1eae (red ca trace). the arrows show regions of significant divergence between the models, including in the region of the access cleft (left arrow). (c) slab section (30 Å thick) showing the active-site cleft, with the ketoconazole position derived from 3jus. the wild-type positions of m220 and g54 are marked. (d) slab (30 Å) showing the active-site cleft, with the ketoconazole position derived from 3jus. the azole-resistant m220v mutation is marked. (e, f) slab (20 Å) showing the predicted position of the heme molecule. the wild-type g448 residue (e) is shown with the azole-resistant g448s residue (f).
Table 2.
Summary of azole resistance caused by mutations in the A. fumigatus CYP51A genea
| Azole | Resistance of strain with mutation: |
|||
|---|---|---|---|---|
| M220 | G54 | G448 | L98-TR* | |
| Itraconazole | R | R | R | R |
| Posaconazole | R | R | S | R |
| Voriconazole | I | S | R | R |
Data are a simplified version of Table 3 in the work of Howard et al. (8). R, resistance; S, susceptibility; I, intermediate or variable resistance or sensitivity; L98-TR*, L98 mutation associated with a tandem repeat in the gene promoter (it is unclear whether either mutation has an effect in isolation [11, 14]). The wild type was susceptible to all azoles.
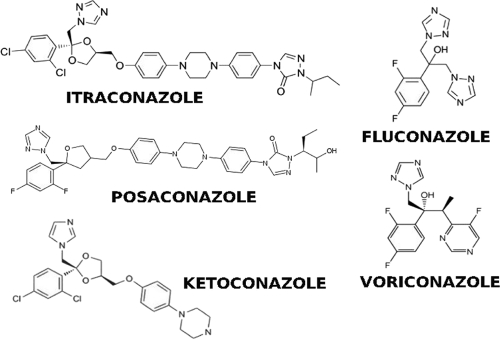
Models derived from the sequences of CYP51A genes from azole-resistant A. fumigatus isolates were examined to assess direct or indirect effects of mutations on azole access or azole binding. It is assumed that azoles act by insertion into the active-site cleft in the protein and prevent the access of lanosterol as well as sequester the heme. Mutations at M220 and G54 have clear potential to block access to the binding channel (Fig. 1C and D, with alternative images in Fig. S3C to G in the supplemental material). G448S is in an area on the opposite side of the protein adjacent to the heme position and appears to have the potential to disrupt the position of the heme within the protein (Fig. 1E and F). The heme molecule appears to be anchored by residues 290 to 297, S363, F459, I455, C454, H452, and F447. Mutation at G448 may act by reducing the ability of the azole to bind effectively to the heme, allowing replacement by the substrate. The model predicts that mutations at G54 or M220 (Fig. 1C and D) are likely to affect long-chain azole (itraconazole and posaconazole) (Fig. 2) binding more than compact azole binding (voriconazole) (Fig. 2) (4–6, 12). In particular, compared to ketoconazole, itraconazole and posaconazole have two further ring structures in the “tail” (Fig. 1C and D and 2) that might directly interact with M220 and G54. The L98H mutation is not adjacent to either heme or binding cleft and does not appear to alter the structure of the model. This mutation is unusual in that it appears to function only in the presence of a promoter duplication (11). Models based on 3JUS and 3LD6 show clear differences at the active-site cleft (Fig. S2 in the supplemental material). Alternatively, mutations in the active-site cleft have the potential to sterically interfere with azole binding.
Fig. 2.
Structures of azoles considered in this study.
The model of CYP51A presented here has clearly improved quality scores by several assessment methods, includes the full protein with a validated cDNA sequence, and provides a basis with which to address how mutations in this gene can affect drug resistance. It enables us to make testable predictions, for example, that mutations at S52 might lead to azole resistance, and we anticipate testing such hypotheses in the near future. Model coordinates are made freely available to the community for use or improvement.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 7 February 2011.
REFERENCES
- 1. Bellamine A., Mangla A. T., Nes W. D., Waterman M. R. 1999. Characterization and catalytic properties of the sterol 14alpha-demethylase from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 96:8937–8942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Benkert P., Tosatto S. C., Schomburg D. 2008. QMEAN: a comprehensive scoring function for model quality assessment. Proteins 71:261–277 [DOI] [PubMed] [Google Scholar]
- 3. Benkert P., Kunzli M., Schwede T. 2009. QMEAN server for protein model quality estimation. Nucleic Acids Res. 37:W510–W514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bueid A., et al. 2010. Azole antifungal resistance in Aspergillus fumigatus: 2008 and 2009. J. Antimicrob. Chemother. 65:2116–2118 [DOI] [PubMed] [Google Scholar]
- 5. Cools H. J., Fraaije B. A., Kim S. H., Lucas J. A. 2006. Impact of changes in the target P450 CYP51 enzyme associated with altered triazole-sensitivity in fungal pathogens of cereal crops. Biochem. Soc. Trans. 34:1219–1222 [DOI] [PubMed] [Google Scholar]
- 6. Cools H. J., Fraaije B. A. 2008. Are azole fungicides losing ground against Septoria wheat disease? Resistance mechanisms in Mycosphaerella graminicola. Pest Manag. Sci. 64:681–684 [DOI] [PubMed] [Google Scholar]
- 7. Eswar N., et al. 2007. Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 50:2.9.1-2.9.31 [DOI] [PubMed] [Google Scholar]
- 8. Howard S. J., et al. 2009. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg. Infect. Dis. 15:1068–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kelly S. L., Lamb D. C., Jackson C. J., Warrilow A. G., Kelly D. E. 2003. The biodiversity of microbial cytochromes P450. Adv. Microb. Physiol. 47:131–186 [DOI] [PubMed] [Google Scholar]
- 10. Lass-Florl C. 2009. The changing face of epidemiology of invasive fungal disease in Europe. Mycoses 52:197–205 [DOI] [PubMed] [Google Scholar]
- 11. Mellado E., et al. 2007. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob. Agents Chemother. 51:1897–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Podust L. M., Poulos T. L., Waterman M. R. 2001. Crystal structure of cytochrome P450 14alpha-sterol demethylase (CYP51) from Mycobacterium tuberculosis in complex with azole inhibitors. Proc. Natl. Acad. Sci. U. S. A. 98:3068–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Snelders E., Karawajczyk A., Schaftenaar G., Verweij P. E., Melchers W. J. 2010. Azole resistance profile of amino acid changes in Aspergillus fumigatus CYP51A based on protein homology modeling. Antimicrob. Agents Chemother. 54:2425–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Snelders E., et al. 2008. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med. 5:e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xiao L., et al. 2004. Three-dimensional models of wild-type and mutated forms of cytochrome P450 14alpha-sterol demethylases from Aspergillus fumigatus and Candida albicans provide insights into posaconazole binding. Antimicrob. Agents Chemother. 48:568–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.