Abstract
Single-dose nevirapine (NVP) is quite effective in preventing transmission of the human immunodeficiency virus (HIV) from mother to child; however, many women develop resistance to NVP in this setting. Comparing outcomes of clinical studies reveals an increased amount of resistance in subtype C relative to that in other subtypes. This study investigates how nonnucleoside reverse transcriptase inhibitor (NNRTI) drug resistance mutations of subtype C affect replication capacity. The 103N, 106A, 106M, 181C, 188C, 188L, and 190A drug resistance mutations were placed in a reverse transcriptase (RT) that matches the consensus subtype C sequence as well as the HXB2 RT, as a subtype B reference. The replicative fitness of each mutant was compared with that of the wild type in a head-to-head competition assay. The 106A mutant of subtype C would not grow in the competition assay, making it the weakest virus tested. The effect of the 106M mutation was weaker than those of the 181C and 188C mutations in the consensus C RT, but in subtype B, this difference was not seen. To see if the 106A mutation in a different subtype C background would have a different replicative profile, the same NNRTI resistance mutations were added to the MJ4 RT, a reference subtype C molecular clone. In the context of MJ4 RT, the 106A mutant was not the only mutant that showed poor replicative fitness; the 106M, 188C, and 190A mutants also failed to replicate. These results suggest that NNRTIs may be a cost-effective alternative for salvage therapy if deleterious mutations are present in a subtype C setting.
INTRODUCTION
The emergence of drug-resistant mutants is common among HIV-infected patients undergoing antiviral therapy. Clinically, the standard practice is to drop the antiviral to which the resistance has developed from the treatment regimen in such drug failure cases. However, a notable exception to this is the continuing use of lamivudine (3TC) as a salvage regimen because the 184V mutation linked to 3TC treatment has decreased replication ability relative to that of the wild type (WT) in subtype B (7). Mechanistically, Back et al. (2) found that the 184V mutant is less processive than wild-type RT under conditions of limiting deoxynucleoside triphosphates (dNTPs), and in primary lymphoid cells, the mutant is less fit than wild-type virus. These studies were extended further in vivo by Paredes et al. (35), who looked at multidrug-resistant virus from patients who were ending 3TC treatment. They used an allele-specific PCR assay to track the reversion of the M184V sequence and found that there was a fitness cost of 4 to 8% for the 184V mutation compared to that of the viral sequence that grew without 3TC drug pressure. Taken together, these studies illustrate how clinical management of HIV-infected patients can benefit from the study of the fitness cost of HIV drug-resistant mutants.
The use of antiretroviral drugs is increasing in the developing world. In particular, the use of single-dose nevirapine (NVP) has been shown to be an effective means of mitigating the transmission of HIV from mother to child (22, 40); however, the women exposed to this treatment have high levels of drug resistance in response to the slow decay of NVP (19, 40).
The literature on NVP resistance has been documenting an increased amount of resistance seen in subtype C HIV compared to that in other subtypes in response to single-dose NVP therapy. Toni et al. (42) discovered that after single-dose NVP, 20% of women infected with CRF02-AG had resistance mutations at 1 month postpartum. The HIVNET012 study found that 25% of women 6 to 8 weeks after single-dose NVP had resistance mutations in Uganda, a country with a subtype A and D epidemic (12). The same group compared results from the HIVNET012 trial with results from the NVAZ trial conducted in Malawi, where subtype C is the major subtype. They found that 69% of women with subtype C viruses harbored drug resistance mutations, versus 19% and 36% of women with subtype A and D viruses, respectively, 6 weeks after delivery (13). The Mashi trial found that 45% of women infected with a subtype C virus who received zidovudine (AZT) for 6 weeks before delivery and single-dose NVP during labor had resistance mutations at 1 month postpartum (40). A different study found similar numbers of subtype C resistance after administration of single-dose NVP, with 46.5% of infants having detectable resistance 2 months after birth (21). At least in the single-dose NVP setting, it seems that subtype C develops resistance more often than other subtypes that have been tested. This seems particularly important because HIV-1 C viruses account for the largest proportion of HIV infections, especially in developing world populations (33).
For subtype C, there are 1,295 sequences deposited in the Stanford University HIV Drug Resistance Database (37) that were isolated from individuals who have been treated with at least one nonnucleoside reverse transcriptase inhibitor (NNRTI). Of these, 23% had the 103N mutation, 11% had 181C, 11% had 106M, 10% had 190A, 4% had 188L, 1.5% had 106A, and less than 1% had 188C. A direct comparison between subtypes B and C is difficult because the average length of treatment and number of drugs received for the submitted subtype B viruses are greater than those for submitted subtype C viruses. There are 7,884 subtype B viruses that have been treated with any NNRTI, and the 103N mutation is the most common, at 33% prevalence, followed by 181C at 18%, 190A at 12%, 188L at 5%, 106A at 1.5%, and both 188C and 106M at less than 1%. Since people infected with subtype B viruses have been treated longer and placed on more drugs than those infected with subtype C, it is easy to explain the increased amounts of resistance seen in subtype B. However, it is interesting to note that there are some differences in relative frequency of mutations. In both subtypes, it is clear that the 103N mutation is the most common, but in subtype C, the 181C and 106M mutations occur at similar frequencies, while in subtype B, the 181C and 190A mutations have a clear advantage over any change at the 106 position.
In subtype B, the most prevalent drug resistance mutations seen clinically tend to be the most fit mutations in cell culture. Several groups have looked at the thymidine analog mutations (TAMs) that occur in response to treatment with nucleoside reverse transcriptase inhibitors (NRTIs). When the TAM-1 pathway, 41L/210W/215Y, is compared with the TAM-2 pathway, 67N/70R/215F, the TAM-1 mutant is found to be more fit than the TAM-2 mutant (17, 34). This follows the clinical data which show that the TAM-1 pathway occurs twice as often as the TAM-2 pathway in a heavily treated cohort (26). NNRTI mutations follow a similar pattern. Collins et al. inserted the NNRTI resistance mutations into a molecular clone of HIV-1B. They found that the most common NNRTI resistance mutation, 103N, was the most fit in a head-to-head fitness assay, and 181C, 190A, 188C, and 106A followed in order of fitness and prevalence (8).
There is some evidence that drug resistance mutations may have different patterns in different subtypes. The 106M mutation instead of the 106A mutation was found to occur in subtype C in response to efavirenz drug selection (5). The protease inhibitor resistance mutation 30N was found to be rare in subtype C in response to nelfinavir; this same mutation had a large impact on viral replication in subtype C in vitro (16). Brenner et al. (6) found that in cell culture, subtype C viruses developed resistance to tenofovir through the K65R mutation more than other subtypes. This finding has been validated in some clinical studies showing an increased prevalence of the K65R mutation in clinical samples (10, 32). A study from our laboratory found that in subtype C, a mixed TAM pathway, comprising 67N/70R/215Y, occurred most often in response to a dideoxyinosine (ddI)-containing regimen but that in subtype B this pathway rarely occurs (31). In vitro, the mixed-TAM-pathway mutation was found to be more fit than the TAM-2 mutation in subtype C, but for subtype B, the mixed-TAM-pathway mutant had no advantage over the TAM-2 mutant (1).
Thus far, the question of whether NVP resistance carries any fitness cost in subtype C has not been explored. Similarly, is it possible that differences in fitness cost may explain why NVP resistance is occurring more in one subtype than the other? To address these questions, we tested the viral fitness of the NNRTI resistance mutations in both subtype B and subtype C. The 103N, 106A, 106M, 181C, 188C, 188L, and 190A RT mutations were placed in the consensus C and HXB2 RTs by site-directed mutagenesis. The 106A mutation did not allow efficient replication in the consensus C background sequence. Relative to what was observed for subtype B, the 106M mutation appears to have a higher fitness cost in subtype C; however, the 103N and 188L mutants are the fittest mutants of both subtypes, with no replicative fitness cost relative to that of the consensus C sequence. In order to see if the 106A mutation may be tolerated better in the context of another subtype C RT, the NNRTI mutations were introduced into the RT from MJ4. In this RT background, not only was the replication of the 106A mutant not detected, but the 106M, 188C, and 190A mutants would not grow either. In contrast, the 188L mutant showed a high level of replicative fitness when compared to the MJ4 RT, indicating that the MJ4 RT is not an enzyme inherently refractory to any NNRTI resistance mutation. This study highlights that fitness costs of NNRTI drug resistance mutations can be quite high for some subtype C RTs, and there may be reason to use NNRTIs in salvage therapy for these sequences that cannot tolerate NNRTI resistance mutations.
MATERIALS AND METHODS
DNA clones and mutagenesis.
The HIV subtype C molecular clone MJ4 was used as the backbone virus for all clones (30). The subtype B sample in this study is the RT from HXB2 (36), replacing the RT in MJ4 so that the viruses are identical except for RT (1). The consensus C sequence for RT from the Los Alamos HIV database (23) was used as the template to create the consensus C RT used in this study. Ten amino acids were changed from MJ4 to match the consensus sequence: 36A, 39E, 166K, 173A, 207E, 248E, 286A, 292I, 296T, and 334H. The 334H mutation does not match the consensus sequence, which has 334Q, but the percentages are 28% and 33%, respectively. The 334H mutation matches the consensus sequence for Botswana, so we chose to use the 334H residue since the consensus sequence residue of 334Q had a similar frequency. The NNRTI resistance mutations 103N, 106A, 106M, 181C, 188C, 188L, and 190A were placed into each RT using a QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) in a subclone of each RT. The sequence of each mutant was verified by DNA sequencing. Once the RT sequence was verified, the RT was placed in MJ4, and plasmid DNA was isolated with a Qiafilter plasmid maxikit (Qiagen, Valencia, CA). Full-length clones were also sequenced after construction to verify that the correct drug resistance mutations and sequence tag were present.
Cells and cell culture.
293 cells were maintained in basal Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10 percent fetal bovine serum (FBS; Invitrogen), 100 U/ml penicillin G (Invitrogen), 100 U/ml streptomycin (Invitrogen), and 0.25 μg/ml amphotericin B (Invitrogen). Cell transfections were carried out with the Superfect transfection reagent (Qiagen) with a modified protocol. Briefly, 15 μg of plasmid DNA was diluted in 135 μl of DMEM; 110 μl of transfection reagent was added to DNA and incubated at room temperature for 10 min. DNA was diluted with 2 ml of 293 growth medium, and the mixture was added to 5.3 × 106 293 cells in a T75 flask. Cells were incubated at 37°C and 5% CO2 for 2 h. DNA was washed off, and 7.5 ml of fresh 293 growth medium was added to cells. After 3 to 4 days, viral titer was assessed with a p24 enzyme-linked immunosorbent assay (ELISA; Perkin-Elmer, Waltham, MA). Virus was harvested when there was at least 25 ng p24/ml. Cell-free viral stocks were stored at −80°C.
Human peripheral blood mononuclear cells (PBMCs) were isolated from anonymous donors by use of a Ficoll gradient (MP Biomedicals, Solon, OH). PBMCs were stimulated with phytohemagglutinin (PHA; Sigma-Aldrich, St. Louis, MO) at a concentration of 10 μg/ml in RPMI medium (Invitrogen) supplemented with 20% FBS, 5 half-maximal U/ml interleuken-2 (IL-2; ABi, Columbia, MD), 100 U/ml penicillin G, 100 U/ml streptomycin, and 0.25 μg/ml amphotericin B for 3 days. PBMCs were maintained in growth medium containing 20% FBS, 5 half-maximal U/ml IL-2, 100 U/ml penicillin G, 100 U/ml streptomycin, and 0.25 μg/ml amphotericin B in RPMI basal medium.
TCID50 and RT assay.
The 50% tissue culture infectious dose (TCID50) was determined according to the NIH-DAIDS protocol with a few modifications. Cultures grew for 11 days instead of 7 to account for the slow-growing nature of the mutant viruses and were fed on days 4 and 7. In addition, an extra row of viral dilutions without cells was maintained in parallel to distinguish between viral growth and dilution of the stock p24 value.
The colorimetric reverse transcriptase assay (Roche, Mannheim, Germany) was used to assess the viability of each RT. Virus was isolated from cell culture supernatant with a 30% polyethylene glycol (PEG; Sigma-Aldrich)-1.2 M NaCl (Mallinckrodt Baker, Philipsburg, NJ) precipitation. Viral supernatant was diluted to 10 ng p24/ml with Dulbecco's phosphate-buffered saline (dPBS; Invitrogen) or used full-strength if the results for the previous assays were negative. PEG solution was added to virus at a ratio of 1 part PEG to 2 parts virus; the mixture was incubated on ice overnight. To precipitate virus, the solution was centrifuged at 800 × g for 45 min at 4°C, and the viral pellet was used for the RT assay. The viral pellet was resuspended in 105 μl of lysis buffer, and 25 μl of the reaction mixture was added to 50 μl of the viral lysate. The reaction mixture was allowed to incubate at 37°C overnight for more-sensitive detection. The rest of the assay was done in accordance with the manufacturer's protocol, with the final substrate incubating for 12 to 15 min.
Competition assay.
PHA-stimulated PBMCs (5 × 105) were infected with two competing viruses at a total multiplicity of infection of less than 0.001. Cells were infected overnight, and the following day, the entire culture was spun at 400 × g for 5 min to pellet the cells. One hundred forty microliters of cell culture supernatant was saved for viral RNA isolation, and the rest was discarded. Cells were resuspended in 500 μl of fresh PBMC growth medium and incubated at 37°C at 5% CO2. The saved supernatant is the day zero sample. PBMCs (5 × 105) freshly stimulated with PHA were added on day 4, and cultures were fed on day 8. RNA was isolated on days 0, 4, 8, and 12 from cell-free cell culture supernatant using a QIAamp viral RNA minikit (Qiagen) including RNase-free DNase I (Qiagen) in the column digestion step. RNA was stored at −80°C until collection of all time points was complete.
Real-time PCR assay.
Two viruses were distinguished by a sequence tag in Nef as described by Boutwell et al. (4), consisting of four nucleotide changes in the wobble position of four consecutive codons. This gives two viruses identical in amino acid sequence and containing mutations of interest, but with a different nucleotide sequence that can be discriminated by an allele-specific quantitative PCR assay. Two unique forward primers distinguish the viral variants with a common reverse primer. The WT-nef forward primer sequence is 5′-CAACACAGCCGCCAATA-3′; the Mut-nef forward primer is CAACACTCCGGCGAACA-3′. The common reverse primer is 5′-CCCACAAATCAAGGATCT-3′. Quantifast SYBR green reverse transcription-PCR (RT-PCR) master mix (Qiagen) was used in the Applied Biosystems (Foster City, CA) 7900 Fast 96-well format system with the manufacturer's cycling conditions. Two microliters of RNA was added to triplicate wells containing 18 μl of the reaction mixture. Relative amounts of each variant were determined with a standard curve generated from a linearized plasmid with the approximate viral length, 9,078 bp. The curve extended from 1 × 102 to 1 × 107 for each tag, and the slopes for each curve are similar, signifying a similar reaction efficiency for each set of primers (4).
Calculations and statistics.
The relative fitness is calculated from the coefficient of selectivity (s) as defined by Maree et al. (3, 28):
where Mutday12% is the percentage of total virus on day 12 that is mutant, WTday12% is the percentage of total virus on day 12 that is WT, and WTday12 is the number of copies of WT virus on day 12. The relative fitness is calculated by adding 1 to s. Statistics were calculated using SigmaStat software (Systat, Chicago, IL). The Mann-Whitney rank sum test was used for comparisons between each group, and a P value of less than 0.05 was defined as significant. The average for each set (at least five replicates) of competitions is graphed, and the error bars represent 1 standard deviation.
RESULTS
NNRTI mutations in subtype C RT.
To investigate the effects of NNRTI drug resistance mutations in the subtype C setting, the NNRTI mutations 103N, 106A, 106M, 181C, 188C, 188L, and 190A were placed in an RT that matched the consensus C sequence from the Los Alamos database (23). The RT was placed in MJ4, a subtype C molecular clone (30).
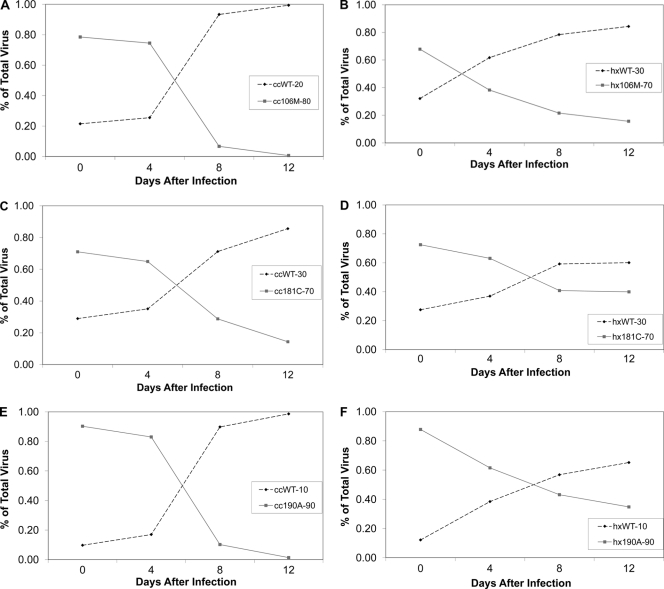
Each NNRTI drug-resistant mutant was competed with the consensus C RT; selected results are shown in Fig. 1. The 106A mutant did not grow in the competition assay; however, in the TCID50 test, the wells containing PBMCs held 25% more p24 than wells that did not contain cells at day 10 after infection (average of three wells; data not shown). While the 106A mutant could not be directly competed with the consensus C virus, the mutant was clearly the least fit mutant tested. The 103N and 188L mutations do not have a replication cost. The 106M mutant grew well but had a high fitness cost relative to that of the WT virus, as did the 181C, 188C (growth curve not shown), and 190A (Fig. 1A, C, and E) mutants.
Fig. 1.
Representative competitions of NNRTI drug resistance mutations in the consensus C RT and the subtype B RT competed with the WT. (A) Relative growth curve of the 106M mutant with the WT in the consensus C background, where the mutant makes up 80% of the inoculum and the WT is the remaining 20%. (B) Competition similar to that represented in panel A, except in the subtype B background, where the 106M mutant is at 70% and the WT starts at 30%. (C) Competition between the WT and the 181C mutant at an initial ratio of 30% WT to 70% mutation in the consensus C background. (D) Competition between the 181C mutant and the WT with an initial ratio of 30% WT to 70% mutation in the subtype B background RT sequence. (E) Consensus C competition between the 190A NNRTI mutant and the WT with an inoculum that is 10% WT and 90% mutant. (F) Competition between the 190A mutant and the WT in the subtype B background with an initial ratio of 10% WT to 90% mutant. “cc” signifies the consensus C RT, and “hx” indicates that the subtype B RT was used.
NNRTI resistance mutations in subtype B and comparison to the subtype C consensus sequence.
In order to compare the fitness costs of NNRTI resistance for subtype C to those for subtype B, the NNRTI resistance mutations were placed in the HXB2 RT (36). Selected results for competitions with the WT sequence are shown in Fig. 1. The 106A, 106M (growth curve not shown), 181C, and 190A mutations all had significant replication costs compared to that of the WT in subtype B (Fig. 1B, D, and F, respectively). Similar results were seen by Wang et al. (43) and Collins et al. (8).
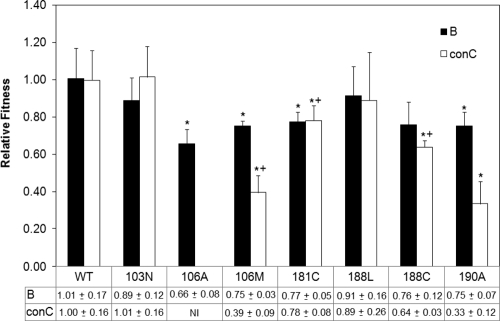
The results for five replicates for each competition are compared between subtypes in Fig. 2. While both the 103N and the 188L mutations appear to be the most fit in each subtype, with no significant difference between the mutants and the WT for both, the position 106 mutations have a higher fitness cost in subtype C than in subtype B. The 106A mutant would not grow in the competition assay in the consensus C RT, making it the least fit mutation tested. The effect of the 106M subtype C-specific mutation is still weak in the subtype C genetic background. While there was no difference between the 106M, 181C, and 188C mutations in subtype B, the 106M mutant was significantly less fit in the consensus C RT than the 181C and 188C mutants. The 190A mutant looks weaker than the 181C and 188C mutants but, with a P value of 0.096 (Mann-Whitney rank sum test), did not reach significance.
Fig. 2.
Comparison of subtype B and consensus C (conC) relative fitness costs of NNRTI drug resistance mutations. All bars represent the averages of results from five independent competitions, and the error bars represent 1 standard deviation. “NI” means that the virus was not infectious in this assay. An asterisk marks a significant difference between the mutant competition and the WT according to the Mann-Whitney rank sum test, where a P value of less than 0.05 is defined as significant. A plus sign marks a significant difference found between the 106M mutation and the 181C and 188C mutations when the competition results are compared using the Mann-Whitney rank sum test.
Comparison of the MJ4 background RT with NNRTI mutations and consensus C results.
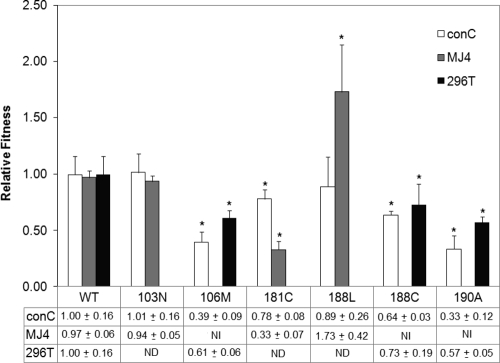
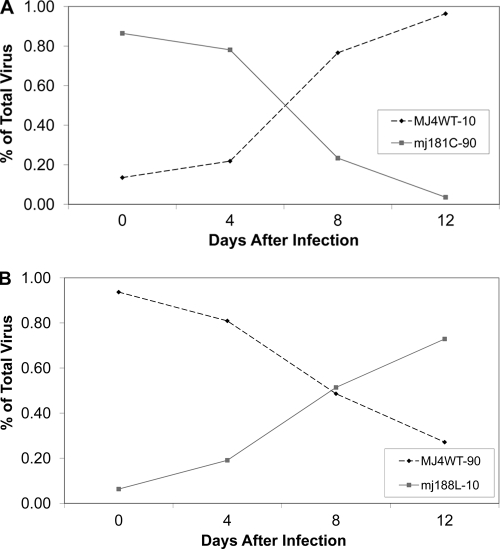
Given that there are several reports of the 106A mutation occurring clinically in patients for whom NNRTI-containing drug regimens fail or in response to single-dose NVP in subtype C (11, 12, 27, 29, 31, 38), we wanted to test a different subtype C RT to determine if the 106A mutant would grow in a clade C background. All of the NNRTI drug resistance mutations were added to the MJ4 RT by site-directed mutagenesis and competed with the MJ4 WT RT as shown in Fig. 4. Only three mutations produced a replication competent RT, 103N, 181C, and 188L. The 103N mutant had the same replication capacity as the WT, and the 181C mutant was much less fit than the WT (Fig. 3A). Curiously, the 188L mutation in the MJ4 background was more fit than the WT (Fig. 3B).
Fig. 4.
Comparison of relative fitness between the consensus C (conC) RT, the MJ4 RT, and the MJ4 RT containing the 296T point mutation. The bars represent the averages of results from five independent experiments, and the error bars represent 1 standard deviation. “ND” signifies that the experiment was not done, and “NI” means that the virus was not infectious in this assay. An asterisk marks a difference between the WT and the mutant relative fitnesses according to the Mann-Whitney rank sum test, where a P value of less than 0.05 is defined as significant.
Fig. 3.
Representative competitions of the NNRTI drug resistance mutations in the MJ4 RT. (A) Competition between the 181C mutant and the MJ4 WT with an inoculum viral ratio of 90% mutant to 10% WT. (B) 188L mutant competition with the WT where 90% of the viral inoculum is the WT and the remaining 10% is the mutant.
The results for the competition between the consensus C and the MJ4 RTs are compared in Fig. 4. The most striking feature of the comparison between the two subtype C RTs is that so many mutations are viable in only one sequence. In addition, the 188L mutation in MJ4 is the only mutation found to be fitter than the WT in any background tested.
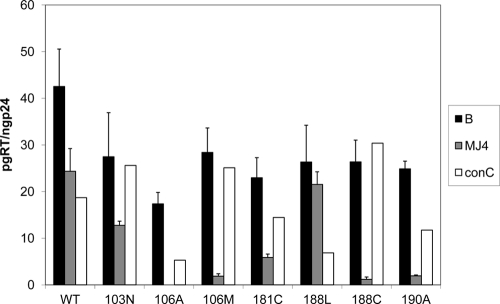
The 106A, 106 M, 188C, and 190A mutations are not viable in MJ4. The mutant RTs were placed in at least two different preparations of the backbone virus, but viable virus was never produced. In order to better understand where the block in infection is taking place, the viral stocks were used in an RT assay to assess RT activity (Fig. 5). It is clear that most mutants of the subtype C RT had reduced activity, but the noninfectious mutants had the smallest amount of active RT per standard sample. The 106A mutation in the MJ4 background did not produce a signal for RT, despite testing approximately 30 ng of p24 in each test; this result may mean that the 106A mutant RT was not packaged in the viral particle produced from transfection, that this RT may be degraded in the virion, or that this RT has no enzymatic activity.
Fig. 5.
RT activity of NNRTI drug resistance mutants for subtype B, MJ4, and consensus C (conC) RTs. The results are graphed in pg of RT, as determined by the RT assay, per ng of p24, as established by the p24 ELISA. The averages of results from three replicates are graphed, with the error bars representing 1 standard deviation. “B” represents the subtype B RT, and “MJ4” represents the MJ4 RT. “conC” represents the consensus C RT. The consensus C bars represent only one assay.
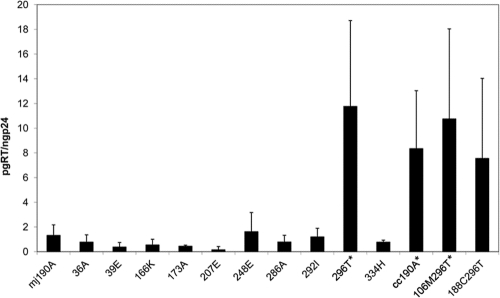
Single consensus C mutations in MJ4 RT containing the 190A resistance mutation.
In order to better understand the difference in viability between the consensus C RT and the MJ4 RT, we placed each of the 10 amino acid changes that differ between the consensus C RT and the MJ4 RT singly in the MJ4 RT containing the 190A NNRTI resistance mutation. We then screened each of these mutants with the RT assay to determine if viable RT was present (Fig. 6). Only one mutation, the N296T change, created an RT that met the viability cutoff (5 pg RT/ng p24). When the 296T mutation was added to the MJ4 RTs containing either the 106M or the 188C NNRTI mutation, these RTs were viable as well. The N296T amino acid was able to restore the replication ability of the NNRTI resistance mutations in the MJ4 background sequence.
Fig. 6.
RT assay showing the single-amino-acid differences between the MJ4 RT and the consensus C RT. The average of at least three replicates is shown, and the error bars are one standard deviation. mj signifies the MJ4 RT and cc indicates the consensus C RT. An asterisk signifies a P value of 0.05 or less (t test).
In order to ensure that the virus containing the 296T mutation acted similar to the consensus C RTs, we competed the 296T-containing NNRTI mutants with the consensus C WT virus. As Fig. 5 indicates, the 296T-containing mutants acted the same in cell culture as the consensus C mutants, and there was no statistical difference between the two sets of RTs containing the same NNRTI mutation.
DISCUSSION
This study looks at the replicative fitness costs to the NNRTI drug resistance mutations in subtype C HIV. We found that not only were the fitness costs of mutations subtype specific but a single background amino acid change rendered some NNRTI resistance mutations nonviable. Four mutations were not infectious in the RT isolated from MJ4, a subtype C molecular clone; however, when 106M or -A, 188C, and 190A are added to the consensus C RT, they are infectious. This change in phenotype was caused by changing one amino acid residue, N296T. In addition, one resistance mutant, 188L, has an increased replicative capacity compared to that of WT virus in MJ4, but in the consensus C backbone, there is no replicative benefit to this mutation (Fig. 4). The effect of the 106A mutation was weak in subtype C, with only a small amount of growth seen in the TCID50 assay for the consensus C backbone. While the 103N, 181C, and 188L mutations had similar fitness costs between subtype B and the consensus C sequence, the 106M and 188C mutants appeared to have a relatively higher replicative capacity in the subtype B RT. It is unclear how the change from N to T at position 296 can cause such a large change in phenotype. In the crystal structure, amino acid 296 lies in the thumb domain of p66 and makes contact with the minor groove of the DNA-RNA duplex (20). As N is still a polar amino acid, it seems unlikely that the change would drastically reduce the ability of the RT to bind the DNA-RNA hybrid. In addition, the resistance mutations 106M, 188C, and 190A do not seem to lie anywhere near the 296 residue in p66 or p51. Changes at the 296 residue appear to be quite rare, with not a single instance of an N residue occurring in the Stanford Drug Resistance Database among all submitted subtype C sequences (15, 39). The 334H substitution used in the consensus RT reflects the Botswana consensus sequence; however, the H residue is found in 28% of all subtype C sequences, and the Q amino acid is found in 33%, as well as being seen in MJ4. Neither of these amino acids at the 334 position seems to be important in NNRTI drug resistance mutation fitness. The other differences between the consensus C and MJ4 RT sequences appear to be common substitutions in subtype C RTs except for 39A and 248T. These two amino acid substitutions appear to be unique to MJ4, although there is some variability at these positions in the extended consensus sequence (15, 39).
A recent study by Iordanskiy et al. (18) found that differences in the replicative fitness of subtype B and C RTs is linked to the polymerase and the RNase H domain of RT. These findings cannot explain why some NNRTI resistance mutations are nonfunctional in the MJ4 background RT, since the RNase H and connective domains remained constant in all clones tested regardless of RT genotype. When the amino acid residues within these domains that have been implicated as accessory mutations for NNRTI resistance were examined, none of these residues in the background clone were found to be different from the consensus sequence. However, there could be inherent instabilities within the RNase H or connective domains that are stabilized by the subtype B RT, allowing for replication of all mutants tested in this context. This remains an interesting area for future inquiry.
Wang et al. (43) found that replicative fitness of NNRTI-resistant virions correlated with RT content of virions. These findings are quite similar to our findings with the RT assay. We found that level of fitness was associated with how much active RT was present in virions, with the smallest amount of RT present in the 106A and 106M mutants for the consensus C background. In addition, as the MJ4 background RT tolerated the mutations less well than the subtype B and consensus C RTs, these differences are also reflected in the amount of active RT present in purified virions. In the future, it will be interesting to determine the source of the instability in RT in the presence of these mutations.
It is interesting that the 106A mutation is so deleterious in subtype C. Brenner et al. noted the emergence of the 106M drug resistance mutation in patients being treated with efavirenz (EFV) who had subtype C HIV (5). Reports of the 106A mutation have been rare in subtype C, but they do occur (11, 12, 27, 29, 31, 38); however, the functionality of these subtype C 106A mutant RTs is unknown. The 106M mutation appears in the same studies at least three times as often as the 106A mutation in the same cohorts. In addition, according to the Stanford University Drug Resistance database, only 1.5% of all subtype C HIV sequences obtained from individuals who received at least one NNRTI contained the 106A mutation, while 11% had the 106M mutation (37). The fitness data can help explain why the 106M mutation is favored over the 106A mutation in the clinical setting. The consensus C RT showed a vastly reduced replicative capacity in the presence of the 106A mutation, and the mutation was completely nonviable in the MJ4 backbone. This implies that there are unknown compensatory mutations that allow the 106A mutation to be viable clinically in subtype C; however, there are several interesting residues. Comparing the sequences of subtype C viruses that contain the 106A mutation (37) with the consensus C sequence (23) reveals three noticeable differences. Nine out of 17 sequences contained a V60I change and a Q174K change in the RT sequence. A valine-to-isoleucine change is fairly conservative, but a glutamine-to-lysine change is a little more interesting, with a polar residue changed to a basic one. Half the sequences contained an arginine-to-lysine change at position 277, which is quite close to the 296T residue that we know from this work can play a role in the functionality of NNRTI-resistant RTs. Looking at the 3-D structure of the RT does not give a clear indication of which residue is likely to play a role in improving the functionality of the RT, and the signature amino acid remains to be identified. Given that the consensus nucleotide sequence for subtype C also favors a 1-nucleotide change to create a methionine codon (GTG g ATG) (5), it is understandable that the 106M mutation is favored in this setting.
These results do not explain why there seem to be more resistance mutations seen in subtype C in response to single-dose NVP than in other subtypes. Some studies use the LigAmp assay (41), which detects single mutations in a real-time PCR assay. These studies may be detecting more resistance in subtype C because they are only testing for the 103N mutation (14, 25). The 103N mutation is consistently the fittest mutation in subtype C, and some of the other common NNRTI resistance mutations, 106M and 190A, have a higher impact on replication in subtype C than they do in subtype B. These sensitive tests may be detecting an increased prevalence of the 103N mutation relative to the other resistance mutations and not increased resistance itself.
There are other reports of resistance determined using Viroseq, which sequences the entire RT and protease region (12, 21, 40, 42), where differences are seen between subtype C and other subtypes. These differences in mutation frequency are not as easily explained as they are when the 103N mutation is looked for specifically. We tested only two RTs from subtype C, and it is possible that there are other subtype C RTs that may be more predisposed to the development of NNRTI drug resistance mutations. Although, given that our fitness data for the consensus C RT mirror the prevalence of these mutations in subtype C, we believe that the fitness of the consensus C mutants is probably representative of the average subtype C RT.
In this study, we looked at subtype C in comparison with subtype B, but few women with a subtype B infection are treated with single-dose NVP, because subtype B HIV is rare in the developing world. Future studies looking at subtypes A and D may reveal differences that explain why subtype C develops more resistance than these clades under similar conditions; however, the studies are hampered by the lack of laboratory reagents that have been developed for non-B subtypes. Subtypes B and D are found to be the most similar of all subtypes when they are compared to each other genetically (9, 24), but further investigations are needed to determine why there is a difference in NVP resistance between subtypes C and D. While the fitness differences between subtypes B and C are clear, they do not seem to point to a reason for an increased prevalence of NNRTI resistance mutations in subtype C compared to the levels for other subtypes found in developing countries.
This study highlights the fact that phenotypic differences between subtypes are common, and these differences can lead to a difference in the ability to adapt to drug resistance mutations. It is interesting to note that several NNRTI mutations in subtype C affect the ability of the virus to grow more than they do in subtype B. If the sequence for patients in the developing world for whom all available regimens have failed is known, there may be situations where receiving an NNRTI as salvage therapy is warranted. Just as it is favorable to keep patients on 3TC or emtricitabine (FTC) in the presence of the M184V mutation, it may be favorable to keep patients on NVP or EFV in the presence of the 106A or 190A mutation. Given the cost effectiveness of NNRTIs and the expense of newly developed drugs, with further investigation there may be a viable alternative for salvage therapy in the developing world.
ACKNOWLEDGMENT
We thank M. McLane for technical assistance.
Footnotes
Published ahead of print on 14 March 2011.
REFERENCES
- 1. Armstrong K. L., Lee T. H., Essex M. 2009. Replicative capacity differences of thymidine analog resistance mutations in subtype B and C human immunodeficiency virus type 1. J. Virol. 83:4051–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Back N. K., et al. 1996. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 15:4040–4049 [PMC free article] [PubMed] [Google Scholar]
- 3. Bonhoeffer S., Barbour A. D., De Boer R. J. 2002. Procedures for reliable estimation of viral fitness from time-series data. Proc. R. Soc. Lond. B Biol. Sci. 269:1887–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boutwell C. L., Rowley C. F., Essex M. 2009. Reduced viral replication capacity of human immunodeficiency virus type 1 subtype C caused by cytotoxic-T-lymphocyte escape mutations in HLA-B57 epitopes of capsid protein. J. Virol. 83:2460–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brenner B., et al. 2003. A V106M mutation in HIV-1 clade C viruses exposed to efavirenz confers cross-resistance to non-nucleoside reverse transcriptase inhibitors. AIDS 17:F1–F5 [DOI] [PubMed] [Google Scholar]
- 6. Brenner B. G., et al. 2006. HIV-1 subtype C viruses rapidly develop K65R resistance to tenofovir in cell culture. AIDS 20:F9–F13 [DOI] [PubMed] [Google Scholar]
- 7. Campbell T. B., et al. 2005. Antiviral activity of lamivudine in salvage therapy for multidrug-resistant HIV-1 infection. Clin. Infect. Dis. 41:236–242 [DOI] [PubMed] [Google Scholar]
- 8. Collins J. A., et al. 2004. Competitive fitness of nevirapine-resistant human immunodeficiency virus type 1 mutants. J. Virol. 78:603–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cornelissen M., van den Burg R., Zorgdrager F., Lukashov V., Goudsmit J. 1997. pol gene diversity of five human immunodeficiency virus type 1 subtypes: evidence for naturally occurring mutations that contribute to drug resistance, limited recombination patterns, and common ancestry for subtypes B and D. J. Virol. 71:6348–6358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Doualla-Bell F., et al. 2006. High prevalence of the K65R mutation in human immunodeficiency virus type 1 subtype C isolates from infected patients in Botswana treated with didanosine-based regimens. Antimicrob. Agents Chemother. 50:4182–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Doualla-Bell F., et al. 2004. Mutations and polymorphisms associated with antiretroviral drugs in HIV-1C-infected African patients. Antivir. Chem. Chemother. 15:189–200 [DOI] [PubMed] [Google Scholar]
- 12. Eshleman S. H., et al. 2004. Comparison of nevirapine (NVP) resistance in Ugandan women 7 days vs. 6–8 weeks after single-dose nvp prophylaxis: HIVNET 012. AIDS Res. Hum. Retroviruses 20:595–599 [DOI] [PubMed] [Google Scholar]
- 13. Eshleman S. H., et al. 2005. Nevirapine (NVP) resistance in women with HIV-1 subtype C, compared with subtypes A and D, after the administration of single-dose NVP. J. Infect. Dis. 192:30–36 [DOI] [PubMed] [Google Scholar]
- 14. Flys T. S., et al. 2006. Quantitative analysis of HIV-1 variants with the K103N resistance mutation after single-dose nevirapine in women with HIV-1 subtypes A, C, and D. J. Acquir. Immune. Defic. Syndr. 42:610–613 [DOI] [PubMed] [Google Scholar]
- 15. Gifford R. J., et al. 2009. The calibrated population resistance tool: standardized genotypic estimation of transmitted HIV-1 drug resistance. Bioinformatics 25:1197–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grossman Z., et al. 2004. Mutation D30N is not preferentially selected by human immunodeficiency virus type 1 subtype C in the development of resistance to nelfinavir. Antimicrob. Agents Chemother. 48:2159–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu Z., et al. 2006. Fitness comparison of thymidine analog resistance pathways in human immunodeficiency virus type 1. J. Virol. 80:7020–7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iordanskiy S., Waltke M., Feng Y., Wood C. 2010. Subtype-associated differences in HIV-1 reverse transcription affect the viral replication. Retrovirology 7:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jourdain G., et al. 2004. Intrapartum exposure to nevirapine and subsequent maternal responses to nevirapine-based antiretroviral therapy. N. Engl. J. Med. 351:229–240 [DOI] [PubMed] [Google Scholar]
- 20. Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. 1992. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256:1783–1790 [DOI] [PubMed] [Google Scholar]
- 21. Kurle S. N., et al. 2007. Emergence of NNRTI drug resistance mutations after single-dose nevirapine exposure in HIV type 1 subtype C-infected infants in India. AIDS Res. Hum. Retroviruses 23:682–685 [DOI] [PubMed] [Google Scholar]
- 22. Lallemant M., et al. 2004. Single-dose perinatal nevirapine plus standard zidovudine to prevent mother-to-child transmission of HIV-1 in Thailand. N. Engl. J. Med. 351:217–228 [DOI] [PubMed] [Google Scholar]
- 23. Leitner T., et al. 2005. HIV sequence compendium 2005. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, NM [Google Scholar]
- 24. Leitner T., et al. 1995. Biological and molecular characterization of subtype D, G, and A/D recombinant HIV-1 transmissions in Sweden. Virology 209:136–146 [DOI] [PubMed] [Google Scholar]
- 25. Loubser S., et al. 2006. Decay of K103N mutants in cellular DNA and plasma RNA after single-dose nevirapine to reduce mother-to-child HIV transmission. AIDS 20:995–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marcelin A. G., et al. 2004. Thymidine analogue reverse transcriptase inhibitors resistance mutations profiles and association to other nucleoside reverse transcriptase inhibitors resistance mutations observed in the context of virological failure. J. Med. Virol. 72:162–165 [DOI] [PubMed] [Google Scholar]
- 27. Marconi V. C., et al. 2008. Prevalence of HIV-1 drug resistance after failure of a first highly active antiretroviral therapy regimen in KwaZulu Natal, South Africa. Clin. Infect. Dis. 46:1589–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maree A. F., Keulen W., Boucher C. A., De Boer R. J. 2000. Estimating relative fitness in viral competition experiments. J. Virol. 74:11067–11072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martinson N. A., et al. 2007. Selection and persistence of viral resistance in HIV-infected children after exposure to single-dose nevirapine. J. Acquir. Immune. Defic. Syndr. 44:148–153 [DOI] [PubMed] [Google Scholar]
- 30. Ndung'u T., et al. 2000. Molecular cloning and biological characterization of full-length HIV-1 subtype C from Botswana. Virology 278:390–399 [DOI] [PubMed] [Google Scholar]
- 31. Novitsky V., et al. 2007. The reverse transcriptase 67N 70R 215Y genotype is the predominant TAM pathway associated with virologic failure among HIV type 1C-infected adults treated with ZDV/ddI-containing HAART in southern Africa. AIDS Res. Hum. Retroviruses 23:868–878 [DOI] [PubMed] [Google Scholar]
- 32. Orrell C., et al. 2009. HIV type-1 clade C resistance genotypes in treatment-naive patients and after first virological failure in a large community antiretroviral therapy programme. Antivir. Ther. 14:523–531 [PMC free article] [PubMed] [Google Scholar]
- 33. Osmanov S., Pattou C., Walker N., Schwardlander B., Esparza J. 2002. Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J. Acquir. Immune. Defic. Syndr. 29:184–190 [DOI] [PubMed] [Google Scholar]
- 34. Paintsil E., Margolis A., Collins J. A., Alexander L. 2006. The contribution of HIV fitness to the evolution pattern of reverse transcriptase inhibitor resistance. J. Med. Virol. 78:425–430 [DOI] [PubMed] [Google Scholar]
- 35. Paredes R., et al. 2009. In vivo fitness cost of the M184V mutation in multidrug-resistant human immunodeficiency virus type 1 in the absence of lamivudine. J. Virol. 83:2038–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ratner L., et al. 1987. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res. Hum. Retroviruses 3:57–69 [DOI] [PubMed] [Google Scholar]
- 37. Rhee S. Y., et al. 2006. HIV-1 pol mutation frequency by subtype and treatment experience: extension of the HIVseq program to seven non-B subtypes. AIDS 20:643–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sen S., Tripathy S. P., Patil A. A., Chimanpure V. M., Paranjape R. S. 2007. High prevalence of human immunodeficiency virus type 1 drug resistance mutations in antiretroviral treatment-experienced patients from Pune, India. AIDS Res. Hum. Retroviruses 23:1303–1308 [DOI] [PubMed] [Google Scholar]
- 39. Shafer R. W., Stevenson D., Chan B. 1999. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 27:348–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shapiro R. L., et al. 2006. Maternal single-dose nevirapine versus placebo as part of an antiretroviral strategy to prevent mother-to-child HIV transmission in Botswana. AIDS 20:1281–1288 [DOI] [PubMed] [Google Scholar]
- 41. Shi C., et al. 2004. LigAmp for sensitive detection of single-nucleotide differences. Nat. Methods 1:141–147 [DOI] [PubMed] [Google Scholar]
- 42. Toni T. D., et al. 2005. Characterization of nevirapine (NVP) resistance mutations and HIV type 1 subtype in women from Abidjan (Cote d'Ivoire) after NVP single-dose prophylaxis of HIV type 1 mother-to-child transmission. AIDS Res. Hum. Retroviruses 21:1031–1034 [DOI] [PubMed] [Google Scholar]
- 43. Wang J., Bambara R. A., Demeter L. M., Dykes C. 2010. Reduced fitness in cell culture of HIV-1 with nonnucleoside reverse transcriptase inhibitor-resistant mutations correlates with relative levels of reverse transcriptase content and RNase H activity in virions. J. Virol. 84:9377–9389 [DOI] [PMC free article] [PubMed] [Google Scholar]