Abstract
The aptamer database is designed to contain comprehensive sequence information on aptamers and unnatural ribozymes that have been generated by in vitro selection methods. Such data are not normally collected in ‘natural’ sequence databases, such as GenBank. Besides serving as a storehouse of sequences that may have diagnostic or therapeutic utility, the database serves as a valuable resource for theoretical biologists who describe and explore fitness landscapes. The database is updated monthly and is publicly available at http://aptamer.icmb.utexas.edu/.
INTRODUCTION
Functional nucleic acids can be selected from random sequence libraries. In general, in vitro selection mimics natural selection, in that a pool of heritable diversity is generated (typically by chemical synthesis), the pool is sieved for binding or catalysis, and successful variants are preferentially amplified by some combination of reverse transcription, PCR and in vitro transcription (1–5) This process has also been described as the Systematic Evolution of Ligands by EXponential enrichment or SELEX (6). Nucleic acid binding species generated by in vitro selection have been referred to as aptamers (7). Aptamers can be RNA, modified RNA, single-stranded DNA or double-stranded DNA, and have been selected to bind targets ranging from small organic molecules to entire organisms. Novel nucleic acid catalysts can also be selected, in general by modifying selection schemes so that variants that make or break covalent (rather than non-covalent) bonds are selectively retained in the population (8–11). Since its introduction over 10 years ago, in vitro selection has been widely adopted as a tool for the development of research reagents, and shows promise for the generation of diagnostic and therapeutic agents (2,12–14).
The Aptamer Database is not only extremely useful both for identifying what aptamers and unnatural ribozymes already exist, but also for garnering information about in vitro selection experiments as a whole and for better understanding the distribution of functional nucleic acids in sequence space and the topographies of fitness landscapes. We have collaborated with theoretical biologists for several years on analyses of the Aptamer Database, and now wish to make this resource much more widely available. In addition, comparative sequence analysis tools should facilitate mappings between natural and unnatural sequence spaces, ultimately providing insights into both. For example, selected transcription factor binding sites have proven to be similar to and predictive of natural transcription factor binding sites (15–18).
Like other types of sequence data, the amount of sequence data generated by in vitro selection experiments has been accumulating exponentially. The sheer number and diversity of selection experiments has risen to the point where it is now essential to gather all the sequence data into a comprehensive, continuously updated database. Unfortunately, GenBank and other sequence databases do not have extensive collections of non-natural sequences, and journals do not typically require the entry of non-natural sequences into these databases. We have now privately maintained the Aptamer Database for 2 years, and expand its content on a monthly basis.
Another database, the SELEX_DB, also contains some information from in vitro selection experiments (19,20). However, the SELEX_DB focuses on in vitro selection experiments that have helped to define natural DNA and RNA recognition sites for proteins, rather than including the entire repertoire of in vitro selection experiments. While there is some overlap between the two sites, the Aptamer Database is in general more complete, and contains entries from 239 published in vitro selection experiments; in contrast SELEX_DB has entries from only 116 publications. The focus of the SELEX_DB on known binding sites ultimately limits its utility for exploring connections between selection experiments and the natural world. For example, natural aptamers that can bind ligands and regulate gene expression (so-called ‘riboswitches’) have been discovered by Breaker and his co-workers (21–23), and the sequences of both riboswitches and aptamers that bind cyanocobalamin can be readily compared using the Aptamer Database.
DATABASE CONTENT
At present, the Aptamer Database contains sequences drawn from 239 published in vitro selection experiments. Each entry is described by the following fields: Author (last name, first name and middle initial for each author); Title; Medline (accession number and a direct link to the PubMed record); Target name (the name of the ligand that was used for the selection of an aptamer); Target type (the classifications we have chosen are proteins, peptides, nucleic acids, organic molecules, inorganic molecules or other); Journal (year, volume, issue, pages); Pool category (DNA or RNA); Modified (Y or N) (indicates whether the nucleic acid pool used in a selection was natural or modified); Buffer conditions; Template description (describes the length of the random region); Template sequence (describes the primer binding sites); and Sequences (the list of all the sequences isolated from the selection).
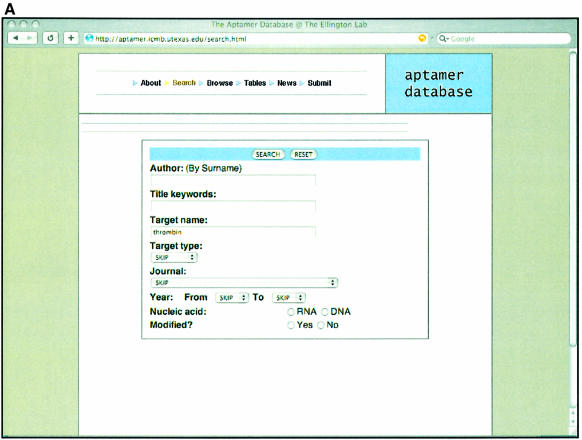
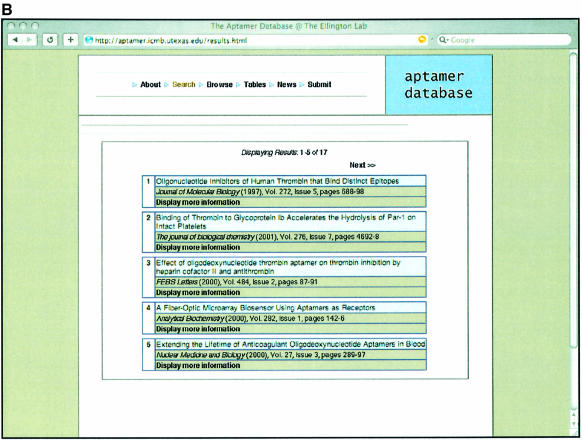
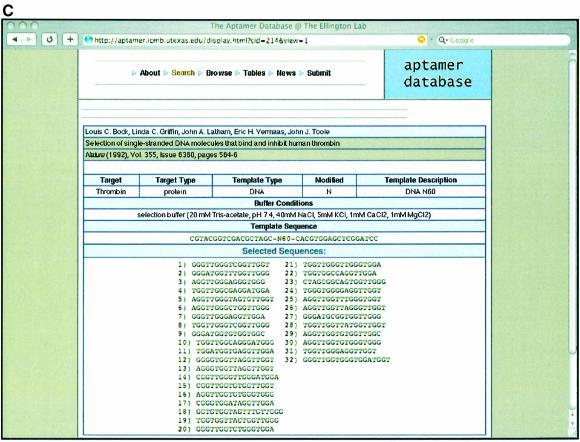
References to and sequences from in vitro selection experiments can be searched by providing specific queries relevant to one of the fields such as author name, target name, type of target, type of pool and so forth, as shown in Figure 1A. In this example the database is being searched for a given Target, thrombin (24). Once the user makes a selection, the results will show all those papers that match the criteria supplied as shown in Figure 1B. The user can then hone in on the information in one or more particular publications, as shown in Figure 1C.
Figure 1.
Examples of Aptamer Database pages. (A) Search page. Users can search the database by supplying a broad range of terms such as Author’s last name, Title keywords or Target name. Records in the database can also be searched based on combination of different criteria [such as the Target type, the journal or year a particular record was published and the type of pool (RNA or DNA) that was used to carry out the selection]. In the example shown, the term ‘thrombin’ is supplied in the Target name dialogue box. (B) Results of the search. A display of the multiple records that were found for the target ‘thrombin’. (C) Sequences from the search. Any one of the recovered records has an associated set of aptamer or ribozyme sequences. For the ‘thrombin’ example, the sequences from the original single-stranded DNA selection that targeted thrombin (24) are shown. There is also more detailed information about the selection that produced these sequences, such as the nature of the pool and buffer conditions used for selection.
The database is updated monthly as new papers on the in vitro selection of aptamers or unnatural ribozymes become available. Initially, older papers were chosen for entry based on using the keywords ‘aptamer’ and ‘SELEX’ with the PubMed search engine. Additional searches with the same keywords were also conducted using the SciFinder search engine (available at http://www.cas.org/SCIFINDER/scicover2.html). Pubcrawler (25) is utilized for monthly updates using the keywords ‘aptamer’ and ‘SELEX.’ At present, data are entered manually. Each sequence is entered in the database template twice and the entries are compared to ensure accuracy. While we have attempted to use optical character recognition (OCR) software, it has proven both inefficient and inaccurate, since the formats of published data are very different from one another.
While we have attempted to make the database as complete as possible, we recognize that some selection papers have been published without appropriate or standard keywords, and thus that some literature may have been overlooked. We include older references as they are brought to our attention. In addition, one of our prime considerations in publishing the Aptamer Database is to facilitate the entry of data by authors and users, and have provided a template for data entry available for download at http://aptamer.icmb.utexas.edu/submit.html. Only data that have been peer-reviewed in conjunction with a publication will be accepted in the database.
DATABASE ANALYSIS
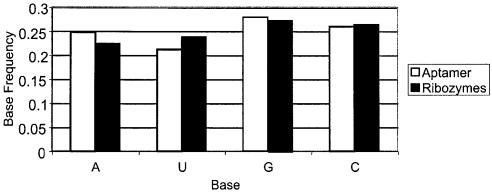
Given that aptamers and unnatural ribozymes were derived from random sequence libraries, it is interesting to consider whether functional sequences globally resemble unselected sequences, or whether there are particular qualities that functional sequences have in common. This question is more than just academic, in that a bias in sequence composition or function could help to inform genomic searches for natural, functional RNA molecules (26–28). In this regard, a base composition analysis of all the sequences from the database reveals that there is a slight statistical skewing towards G and C (p value < 0.01) in the base compositions of RNA aptamers and ribozymes relative to completely random sequences (equimolar A, C, G and U; Fig. 2). As Schultes et al. (29) have observed previously, functional sequences, whether natural or unnatural, appear to have a slight preponderance of guanosine and cytidine. Interestingly, while the content of (G+C) appears to be similar for aptamers and ribozymes, ribozymes contain proportionately more U (and less A) than aptamers (p value < 0.01). Beyond demonstrating that functional nucleic acid sequences of all sorts have particular sequence characteristics, these broad sequence analyses may inform the design of random sequence pools for selections; for example, it may be useful to skew the composition of a pool for the selection of ribozymes to a G:A:U:C ratio of 0.28:0.22:0.24:0.26. Since many selected ribozymes differ completely in sequence and function and cannot be aligned, this sort of analysis could be made much easier with the Aptamer Database.
Figure 2.
Sequence analyses based on the Aptamer Database. The random regions of all RNA aptamer and ribozyme sequences from the Aptamer Database were extracted, and aggregate base compositions were determined. White bars represent the nucleotide frequencies calculated for RNA aptamers. Black bars represent the nucleotide frequencies calculated for ribozymes. The individual residues (A, G, C or U) are shown on the x-axis, while the y-axis represents the frequency of each residue.
The collected availability of sequences in the database also facilitates other global analyses. For example, we exported all of the RNA aptamer and ribozyme sequences and analyzed their potential for forming secondary structures using the program RNAfold from the Vienna RNA package (30). The minimum free energy algorithm in RNAfold is based on the dynamic programming algorithm developed by Zuker et al. (31). The Zuker algorithm only generates minimum free energy structures. Another alternative would have been to look at the ensemble of lowest energy structures. Lawrence and his co-workers have conducted a study that compared the reliability of different RNA structure prediction algorithms (32). Probability models that generate ensembles, such as McCaskill’s algorithm (33), did prove to be a better predictor of the structures of longer sequences. However, for shorter (<120 nucleotides) sequences, the improvements in structure prediction were modest and could primarily be attributed to the inability of the Zuker algorithm to take into account pseudoknots, a motif that is generally difficult to predict for any algorithm. In the end, the comparative analysis (32) confirmed that the Zuker minimum free energy method was very reliable for structure prediction. This method should be particularly appropriate for obtaining a general overview of the structural characteristics of the short selected sequences found in the database.
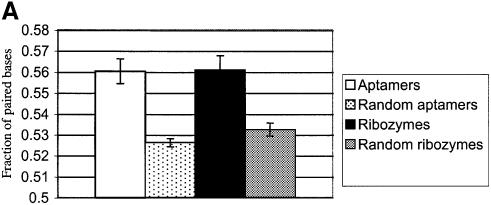
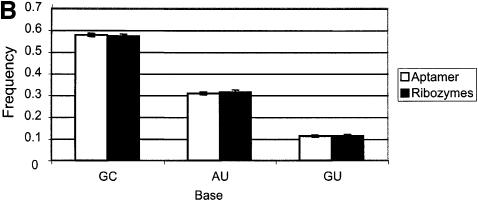
We counted the fraction of paired nucleotides in folded aptamer and ribozyme structures, and analyzed whether they were significantly different from the fraction of paired nucleotides in folded randomized structures. In this instance, the fraction of base pairs found in selected aptamer and ribozyme sequences is statistically greater than the fraction found in randomized versions of the same sequences as shown in Figure 3A (p value < 0.01). In addition, in selected sequences, G:C pairings are more abundant than the A:U or G:U pairings, as shown in Figure 3B (p value < 0.01). The large excess of G:C pairings is not a result of the only slightly higher concentrations of G and C in selected nucleic acids. Taken together, these results reveal that selection for binding or catalytic function of necessity results in selection for secondary structural stability. Schultes et al. (29) have also shown that there is a preponderance of G:C pairings in the stem regions of natural, functional RNA molecules relative to unpaired regions such as loops. Similarly, examinations of natural sequences have suggested that G:C pairings are required to stabilize and present protein binding sites (34–36).
Figure 3.
(A) Base-pairing in selected sequences. Overall base-pairing in RNA aptamer and ribozyme sequences. Secondary structures of the random regions of RNA aptamer and ribozyme sequences were obtained using the program RNAfold, implemented in the Vienna RNA Package (30). The minimum free energy algorithm was based on the dynamic programming algorithm developed by Zuker et al. (31). The fractions of base-paired residues were determined for RNA aptamers (white bar) or ribozymes (black bar). The base-pair fractions for all individual aptamers or all individual ribozymes were averaged; the error bars provide an indication of the spread of these values. To determine whether the number of base pairs that were formed was significantly influenced by selection, the random regions were randomized and re-folded for the RNA aptamers (dotted bar) and ribozymes (gray bar). (B) Distribution of individual base pairings. The data from (A) were re-tabulated in terms of the types of base pairs formed. The white bar represents RNA aptamer sequences, and the black bar represents ribozyme sequences. The y-axis represents the fraction of base pairs in each class, normalized to the total number of base pairings found in (A).
The sequence and structural comparisons we have carried out were relatively simplistic, they justify the contention that nucleic acids selected in vitro possess attributes similar to those of sequences found in nature. The Aptamer Database should continue to be a well-utilized source for such comparisons; for example, as new sequence or structural motifs are found, such as the well-characterized tetraloop sequences that have been found to stabilize natural RNA sequences (37–39), their prevalence and utility can be further confirmed by an unbiased search of the Aptamer Database.
DATABASE AVAILABILITY
The current version of our database can be viewed at http://aptamer.icmb.utexas.edu. Inquiries concerning the database should be directed to aptamer@ellingtonlab.org.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Rob Knight (Yarus Lab, University of Colorado, Boulder), Erik Schultes (Bartel Lab, MIT) for useful discussions. We would also like to thank members of the Ellington Lab for their suggestions and comments especially Scott Knudsen and Colin Cox. This research is funded by the National Science Foundation’s Integrative Graduate Education and Research Traineeship (IGERT) in computational phylogenetics, an Army Research Office grant (MURI DAAD19-99-1-0207) and a NIH grant (1R01 GM61789-01).
REFERENCES
- 1.Burgstaller P., Jenne,A. and Blind,M. (2002) Aptamers and aptazymes: accelerating small molecule drug discovery. Curr. Opin. Drug Discov. Dev., 5, 690–700. [PubMed] [Google Scholar]
- 2.Brody E.N. and Gold,L. (2000) Aptamers as therapeutic and diagnostic agents. J. Biotechnol., 74, 5–13. [DOI] [PubMed] [Google Scholar]
- 3.Eaton B.E. (1997) The joys of in vitro selection: chemically dressing oligonucleotides to satiate protein targets. Curr. Opin. Chem. Biol., 1, 10–16. [DOI] [PubMed] [Google Scholar]
- 4.Famulok M. and Mayer,G. (1999) Aptamers as tools in molecular biology and immunology. Curr. Top. Microbiol. Immunol., 243, 123–136. [DOI] [PubMed] [Google Scholar]
- 5.Feigon J., Dieckmann,T. and Smith,F.W. (1996) Aptamer structures from A to zeta. Chem. Biol., 3, 611–617. [DOI] [PubMed] [Google Scholar]
- 6.Klug S.J. and Famulok,M. (1994) All you wanted to know about SELEX. Mol. Biol. Rep., 20, 97–107. [DOI] [PubMed] [Google Scholar]
- 7.Ellington A.D. and Szostak,J.W. (1990) In vitro selection of RNA molecules that bind specific ligands. Nature, 346, 818–822. [DOI] [PubMed] [Google Scholar]
- 8.Illangasekare M. and Yarus,M. (1999) A tiny RNA that catalyzes both aminoacyl-RNA and peptidyl-RNA synthesis. RNA, 5, 1482–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang F., Yang,Z. and Yarus,M. (1999) Self-capping RNA catalysts derived from selection–amplification. Biol. Bull., 196, 320–321. [DOI] [PubMed] [Google Scholar]
- 10.Unrau P.J. and Bartel,D.P. (1998) RNA-catalysed nucleotide synthesis. Nature, 395, 260–263. [DOI] [PubMed] [Google Scholar]
- 11.Ekland E.H., Szostak,J.W. and Bartel,D.P. (1995) Structurally complex and highly active RNA ligases derived from random RNA sequences. Science, 269, 364–370. [DOI] [PubMed] [Google Scholar]
- 12.Lupold S.E., Hicke,B.J., Lin,Y. and Coffey,D.S. (2002) Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res., 62, 4029–4033. [PubMed] [Google Scholar]
- 13.Martell R.E., Nevins,J.R. and Sullenger,B.A. (2002) Optimizing aptamer activity for gene therapy applications using expression cassette SELEX. Mol. Ther., 6, 30–34. [DOI] [PubMed] [Google Scholar]
- 14.Jayasena S.D. (1999) Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem., 45, 1628–1650. [PubMed] [Google Scholar]
- 15.Roulet E., Busso,S., Camargo,A.A., Simpson,A.J., Mermod,N. and Bucher,P. (2002) High-throughput SELEX SAGE method for quantitative modeling of transcription-factor binding sites. Nat. Biotechnol., 20, 831–835. [DOI] [PubMed] [Google Scholar]
- 16.Kunsch C., Ruben,S.M. and Rosen,C.A. (1992) Selection of optimal κ B/Rel DNA-binding motifs: interaction of both subunits of NF-κB with DNA is required for transcriptional activation. Mol. Cell. Biol., 12, 4412–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lebruska L.L. and Maher,L.J.,3rd (1999) Selection and characterization of an RNA decoy for transcription factor NF-κB. Biochemistry, 38, 3168–3174. [DOI] [PubMed] [Google Scholar]
- 18.Wright W.E. and Funk,W.D. (1993) CASTing for multicomponent DNA-binding complexes. Trends Biochem. Sci., 18, 77–80. [DOI] [PubMed] [Google Scholar]
- 19.Ponomarenko J.V., Orlova,G.V., Frolov,A.S., Gelfand,M.S. and Ponomarenko,M.P. (2002) SELEX_DB: a database on in vitro selected oligomers adapted for recognizing natural sites and for analyzing both SNPs and site-directed mutagenesis data. Nucleic Acids Res., 30, 195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ponomarenko J.V., Orlova,G.V., Ponomarenko,M.P., Lavryushev,S.V., Frolov,A.S., Zybova,S.V. and Kolchanov,N.A. (2000) SELEX_DB: an activated database on selected randomized DNA/RNA sequences addressed to genomic sequence annotation. Nucleic Acids Res., 28, 205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winkler W.C., Nahvi,A., Sudarsan,N., Barrick,J.E. and Breaker,R.R. (2003) An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nature Struct. Biol. 10, 701–707. [DOI] [PubMed] [Google Scholar]
- 22.Winkler W.C., Cohen-Chalamish,S. and Breaker,R.R. (2002) An mRNA structure that controls gene expression by binding FMN. Proc. Natl Acad. Sci. USA, 99, 15908–15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winkler W., Nahvi,A. and Breaker,R.R. (2002) Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature, 419, 952–956. [DOI] [PubMed] [Google Scholar]
- 24.Bock L.C., Griffin,L.C., Latham,J.A., Vermaas,E.H. and Toole,J.J. (1992) Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature, 355, 564–566. [DOI] [PubMed] [Google Scholar]
- 25.Hokamp K. and Wolfe,K. (1999) What’s new in the library? What’s new in GenBank? Let PubCrawler tell you. Trends Genet., 15, 471–472. [DOI] [PubMed] [Google Scholar]
- 26.Sudarsanam P., Pilpel,Y. and Church,G.M. (2002) Genome-wide co-occurrence of promoter elements reveals a cis-regulatory cassette of rRNA transcription motifs in Saccharomyces cerevisiae. Genome Res., 12, 1723–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes J.D., Estep,P.W., Tavazoie,S. and Church,G.M. (2000) Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol., 296, 1205–1214. [DOI] [PubMed] [Google Scholar]
- 28.Klein R.J., Misulovin,Z. and Eddy,S.R. (2002) Noncoding RNA genes identified in AT-rich hyperthermophiles. Proc. Natl Acad. Sci. USA, 99, 7542–7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schultes E.A. and Bartel,D.P. (2000) One sequence, two ribozymes: implications for the emergence of new ribozyme folds. Science, 289, 448–452. [DOI] [PubMed] [Google Scholar]
- 30.Hofacker I., Fontana,W., Stadler,P., Bonhoeffer,L., Tacker,M. and Schuster,P. (1994) Fast folding and comparison of RNA secondary structures. Monatsh. Chem., 125, 167–188. [Google Scholar]
- 31.Zuker M. and Stiegler,P. (1981) Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res., 9, 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding Y. and Lawrence,C.E. (2001) Statistical prediction of single-stranded regions in RNA secondary structure and application to predicting effective antisense target sites and beyond. Nucleic Acids Res., 29, 1034–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCaskill J.S. (1990) The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers, 29, 1105–1119. [DOI] [PubMed] [Google Scholar]
- 34.Varani G. (1995) Exceptionally stable nucleic acid hairpins. Annu. Rev. Biophys. Biomol. Struct., 24, 379–404. [DOI] [PubMed] [Google Scholar]
- 35.Morgan W.D., Bear,D.G. and von Hippel,P.H. (1983) Rho-dependent termination of transcription. I. Identification and characterization of termination sites for transcription from the bacteriophage lambda PR promoter. J. Biol. Chem., 258, 9553–9564. [PubMed] [Google Scholar]
- 36.Morgan W.D., Bear,D.G., Litchman,B.L. and von Hippel,P.H. (1985) RNA sequence and secondary structure requirements for rho-dependent transcription termination. Nucleic Acids Res., 13, 3739–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorin E.J., Engelhardt,M.A., Herschlag,D. and Pande,V.S. (2002) RNA simulations: probing hairpin unfolding and the dynamics of a GNRA tetraloop. J. Mol. Biol., 317, 493–506. [DOI] [PubMed] [Google Scholar]
- 38.Li W., Ma,B. and Shapiro,B. (2001) Molecular dynamics simulations of the denaturation and refolding of an RNA tetraloop. J. Biomol. Struct. Dyn., 19, 381–396. [DOI] [PubMed] [Google Scholar]
- 39.Williams D.J. and Hall,K.B. (2000) Experimental and computational studies of the G[UUCG]C RNA tetraloop. J. Mol. Biol., 297, 1045–1061. [DOI] [PubMed] [Google Scholar]