Abstract
MODBASE (http://salilab.org/modbase) is a relational database of annotated comparative protein structure models for all available protein sequences matched to at least one known protein structure. The models are calculated by MODPIPE, an automated modeling pipeline that relies on the MODELLER package for fold assignment, sequence–structure alignment, model building and model assessment (http:/salilab.org/modeller). MODBASE uses the MySQL relational database management system for flexible querying and CHIMERA for viewing the sequences and structures (http://www.cgl.ucsf.edu/chimera/). MODBASE is updated regularly to reflect the growth in protein sequence and structure databases, as well as improvements in the software for calculating the models. For ease of access, MODBASE is organized into different data sets. The largest data set contains 1 262 629 models for domains in 659 495 out of 1 182 126 unique protein sequences in the complete Swiss-Prot/TrEMBL database (August 25, 2003); only models based on alignments with significant similarity scores and models assessed to have the correct fold despite insignificant alignments are included. Another model data set supports target selection and structure-based annotation by the New York Structural Genomics Research Consortium; e.g. the 53 new structures produced by the consortium allowed us to characterize structurally 24 113 sequences. MODBASE also contains binding site predictions for small ligands and a set of predicted interactions between pairs of modeled sequences from the same genome. Our other resources associated with MODBASE include a comprehensive database of multiple protein structure alignments (DBALI, http://salilab.org/dbali) as well as web servers for automated comparative modeling with MODPIPE (MODWEB, http://salilab.org/modweb), modeling of loops in protein structures (MODLOOP, http://salilab.org/modloop) and predicting functional consequences of single nucleotide polymorphisms (SNPWEB, http://salilab.org/snpweb).
INTRODUCTION
Genome sequencing efforts are providing us with complete genetic blueprints for hundreds of organisms, including humans. We are now faced with assigning, understanding and modifying the functions of proteins encoded by these genomes. This task is generally facilitated by protein 3D structures (1), which are best determined by experimental methods such as X-ray crystallography and NMR spectroscopy.
Over the past 2 years, the number of sequences in the comprehensive public sequence databases, such as Swiss-Prot/TrEMBL (2) and GenPept (3), have increased by a factor of 2.3 from 522 959 to 1 208 659 on August 15, 2003. In contrast, despite structural genomics, the number of experimentally determined structures deposited in the Protein Data Bank (PDB) increased by a factor of only 1.3 over the same period, from 17 443 to 23 096 (4). Thus, the gap between the numbers of known sequences and structures continues to grow.
Protein structure prediction methods are attempting to bridge this gap (5). The most accurate models are generally obtained by homology or comparative modeling (6). Comparative modeling is carried out in four sequential steps: finding known structures (templates) related to the sequence to be modeled (target), aligning the target sequence with the templates, building the model and assessing the model. Therefore, comparative modeling is only applicable when the target sequence is detectably related to a known protein structure. Using automated comparative modeling, the fraction of sequences with comparative models for at least one domain has remained at ∼57% over the past 2 years (7).
The utility of comparative protein structure models depends on their accuracy. The accuracy of comparative modeling is correlated with sequence identity between the template structure and the modeled sequence (5). Protein structure models with high accuracy can be obtained when template structures with >50% sequence identity to the modeled sequence are available. At this level of similarity, the errors usually include some incorrectly packed side chains, small shifts or distortions in the main chain and a few incorrectly modeled loops. Comparative models with medium accuracy are based on 30–50% sequence identity between the target and template sequences. Such models tend to have additional errors in some loop regions and occasional alignment errors. Below 30% sequence identity, alignment and fold assignment errors become the most significant sources of mistakes in comparative modeling. The accuracy of automated comparative protein structure modeling has been quantified by the CAFASP effort (8) as well as automated web servers EVA (9) and LIVEBENCH (10).
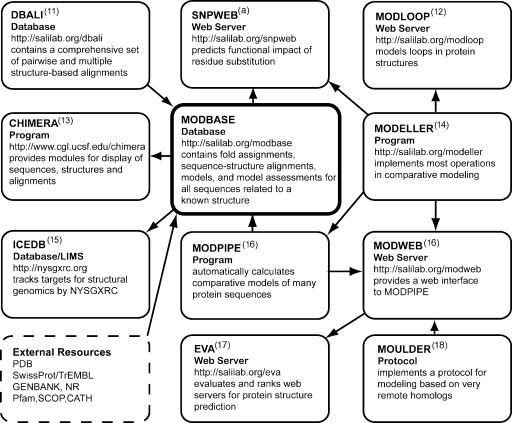
The process of comparative protein structure modeling usually requires the use of a number of programs to identify template structures, to generate sequence–structure alignments, to build the models and to evaluate them. In addition, various sequence and structure databases that are accessed by these programs are needed. Once an initial model is calculated, it is generally refined and ultimately analyzed in the context of many other related proteins and their functional annotations. To facilitate these tasks for both expert and novice users, we have developed several programs, servers and databases (Fig. 1).
Figure 1.
The relationships between MODBASE and associated resources. References are indicated by superscript numbers. aN. Mirkovic, M. A. Marti-Renom, A. Sali and A. N. A. Monteiro, submitted.
In this paper, we highlight the improvements of MODBASE that were implemented since the previous reports (7,19,20). These improvements include more sensitive and accurate software for calculating comparative models, an updated interface that relies on the CHIMERA package for viewing alignments and structures, integration of information about small ligand binding sites and protein–protein interactions with the model data sets, measurement of the contributions of structural genomics to the coverage of the sequence–structure space, and closer integration with a variety of other resources for deriving and using comparative models.
CONTENTS
MODBASE core
Models in MODBASE are calculated using MODPIPE, our entirely automated software pipeline for comparative modeling (16). MODPIPE can calculate comparative models for a large number of protein sequences, using many different template structures and sequence–structure alignments. MODPIPE relies on the various modules of MODELLER for its functionality and is streamlined for large-scale operation on a cluster of PCs using scripts written in PERL.
The templates used for model building consist of representative multiple structure alignments extracted from DBALI (11). These alignments were prepared by the SALIGN module of MODELLER (M. S. Madhusudan, M. A. Marti-Renom, A. Sali, in preparation), which implements a multiple structure alignment method similar to that in the program COMPARER (21). Sequence profiles are constructed for both the target sequences and the templates by scanning against the Swiss-Prot/TrEMBL database of sequences, relying on the BUILD_ PROFILE module of MODELLER (N. Eswar, M. S. Madhusudhan and A. Sali, in preparation). BUILD_ PROFILE is similar to PSI-BLAST (22), except that local dynamic programming is used instead of the BLAST heuristics. Sequence–structure matches are established by aligning the target sequence profile against the template profiles, using local dynamic programming in the SALIGN module and an assessment of statistical significance similar to that of PSI-BLAST (22) and COMPASS (23). Significant alignments covering distinct regions of the target sequence are chosen for modeling. Models are calculated for each of the sequence–structure matches using MODELLER (24). The resulting models are then evaluated by a composite model quality criterion that depends on the compactness of a model, the sequence identity of the sequence–structure match and statistical energy Z-scores (25).
The thoroughness of a search for the best model is modulated by a number of user parameters, including E-value thresholds for identifying useful sequence–structure relationships and the degree of conformational sampling given a sequence–structure alignment. The validity of sequence–structure relationships is not pre-judged at the fold detection stage, but is assessed after the construction of the model and its evaluation. This approach enables a thorough exploration of fold assignments, sequence–structure alignments and conformations, with the aim of finding the model with the best evaluation score.
The models in the version of MODBASE available until the end of 2003, however, were calculated using an earlier version of MODPIPE. These models were based on single template structures and built using sequence–structure matches generated by PSI-BLAST (22) and IMPALA (26).
Models in MODBASE are organized into data sets. The largest data set contains models of all sequences in the Swiss-Prot/TrEMBL database that are detectably related to at least one known structure in the PDB. Currently, there are 1 262 629 models for domains in 659 495 of the 1 182 126 sequences in the Swiss-Prot/TrEMBL database, with an average length of 235 residues per model. For example, there are models for 32 985 human sequences, 22 880 sequences from Arabidopsis thaliana, 15 195 sequences from Drosophila melanogaster and 9691 sequences from Escherichia coli. Because the sequence databases contain sequence information of different strains and mutations, the number of unique sequences for a given organism exceeds the number of genes in the genome. For example, there are about 16 700 unique E.coli sequences in Swiss-Prot/TrEMBL, compared with ∼4400 predicted genes in the E.coli genome.
Predicted interacting proteins
MODBASE links pairs of modeled sequences from the same organism that are predicted to interact with each other (H. Braberg, F. Davis, J. Espadaler, B. Oliva, A. Sali, M. S. Madhusudhan, in preparation). First, residue contacts between the two models are predicted based on a match of both modeled sequences to different parts of a single PDB file. Next, the residue contacts in a hypothetical interface are scored by their propensities to span an interface. These propensities were extracted from ∼8000 representative pairs of interacting domains. If the total score is sufficiently large, the two modeled sequences are predicted to interact with each other. The method is an extension of the Rosetta Stone approach, which was first applied to sequences (27) and is similar to several studies applied to structures (28,29). ∼10 000 modeled sequences in MODBASE are linked via ∼30 000 predicted pairwise interactions, with an estimated false positive ratio of 25%.
Predicted ligand binding sites
MODBASE contains a list of the binding sites of known structure for ∼50 000 ligands found in the PDB (30). The ligands include small molecules, such as metal ions, nucleotides and saccharides, but exclude water molecules, peptides and nucleic acids. Binding sites in the template structures are defined by residues with atoms within 5 Å of any ligand atom. In addition to the actual binding sites in the known structures, MODBASE also contains predicted binding sites on the template structures and models. The predicted binding sites on the template structures are inherited from any related known structure if at least 75% of the binding site residues are within 4 Å of the template residues in a global superposition of the two structures and if at least 75% of the binding site residue types are invariant. The structure superpositions are obtained from our comprehensive database of all pairwise structure superpositions, DBALI (11). The predicted binding sites on the model are defined by all the model residues that are aligned with either the actual or predicted binding site residues on the template. Forty-four percent of the models in MODBASE have at least one predicted binding site for a small ligand.
Application of MODBASE to structural genomics
MODBASE provides the basis for target selection and structure-based annotation by the New York Structural Genomics Research Consortium (NYSGXRC) (15), one of the nine pilot centers in the Protein Structure Initiative supported by the NIH (http://www.nigms.nih.gov/psi/). We highlight here the increased coverage of the sequence–structure space (31) by the NYSGXRC structures.
Relying on the 53 NYSGXRC structures, MODPIPE produced models for domains in 24 113 sequences in Swiss-Prot/TrEMBL (Table 1); the average target–template sequence identity was 18.9%. Only 10% of the sequences are modeled based on >30% sequence identity over more than 75 residues; 81% of the sequences have models that are predicted to have the correct fold based on the model score or the PSI-BLAST E-value (Table 1). The modeled sequences come from 1729 different organisms. Because the structures determined by NYSGXRC were selected by avoiding more than 30% sequence identity to any of the previously determined structures, most of the modeled sequences have been characterized structurally for the first time. The large number of models calculated based on the newly determined structures illustrates and justifies the premise of structural genomics.
Table 1. Summary of the automated modeling by MODPIPE for seven of the 53 structures determined by NYSGXRC.
| NYSGXRC X-ray structure | MODBASE models | |||||
|---|---|---|---|---|---|---|
| PDB code | Database accession number | Annotation | Total sequences | Fold and model | Fold | Model |
| 1b54 | P38197 | Hypothetical UPF0001 protein YBL036C | 151 | 132 | 2 | 17 |
| 1f89 | P49954 | Hypothetical 32.5 kDa protein YLR351C | 553 | 488 | 55 | 10 |
| 1njr | Q04299 | Hypothetical 32.1 kDa protein in ADH3-RCA1 intergenic region | 4 | 1 | 0 | 3 |
| 1nkq | P53889 | Hypothetical 28.8 kDa protein in PSD1-SKO1 intergenic region | 379 | 207 | 172 | 0 |
| 1jzt | P40165 | Hypothetical 27.5 kDa protein in SPX19-GCR2 intergenic region | 1058 | 39 | 1006 | 13 |
| 1jr7 | P76621 | Hypothetical protein ygaT | 11 | 10 | 0 | 1 |
| 1ku9 | 3025177 | YF63_METJA hypothetical protein MJ1563 | 598 | 131 | 214 | 253 |
The complete table is accessible at http://salilab.org/modbase/models_nysgxrc.html. The ‘PDB code’, ‘Database accession number’ and ‘Annotation’ columns define the template structure. ‘Total sequences’ is the number of sequences in SwissProt/TrEMBL could be modeled reliably using the NYSGXRC structure as a template. A sequence is modeled reliably if it has a reliable PSI-BLAST E-value of ≤10–4 (‘Fold’), a reliable model with model score ≥0.7 (‘Model’) or both (‘Fold and model’).
ACCESS AND INTERFACE
MODBASE is queryable through the web at http://salilab.org/modbase by PDB codes, Swiss-Prot/TrEMBL and GenPept accession numbers, annotation keywords, model reliability, model size, target–template sequence identity, alignment significance, and sequence similarity to the modeled sequences as detected by BLAST.
Models in MODBASE are organized into a number of data sets whose access by different users is regulated using a cookie mechanism (http://www.acm.org/crossroads/xrds7-1/cookies). The largest data set includes all modeled sequences from the Swiss-Prot/TrEMBL database and is freely accessible to all academic scientists. Other data sets include models calculated for NYSGXRC, MODWEB data sets from anonymous users and data sets associated with our other modeling projects.
The output of a search is displayed on pages with varying amounts of information about the modeled sequences, template structures, alignments and functional annotations. These tables also contain links to other sequence, structure and function annotation databases, such as PDB (4), GenBank (3), Swiss-Prot/TrEMBL (2), CATH (32), Pfam (33), ProDom (34), and UCSC Genome Browser (35). In addition, MODBASE models are directly accessible from the Swiss-Prot/TrEMBL sequence pages at http://www.expasy.org and UCSC Genome Browser at http://genome.ucsc.edu.
Visualization of sequences, structures and alignments with CHIMERA
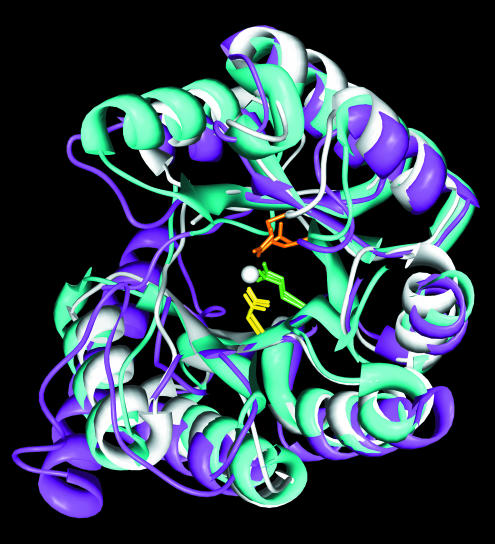
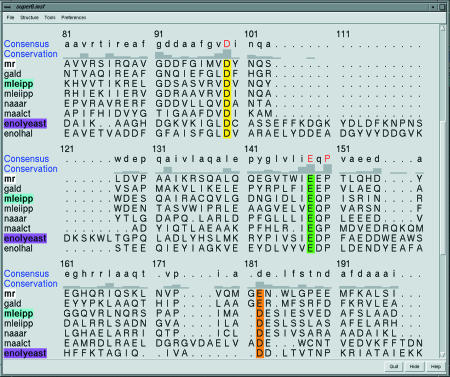
To simplify the process of visualizing the models contained in MODBASE, we created an extension to the CHIMERA Molecular Modeling System, which was developed by the researchers in the Resource for Biocomputing, Visualization, and Informatics at UCSF (Fig. 2) (http://www.cgl.ucsf.edu/chimera) (30). The data contained in a MODBASE entry are divided among three different files: a template file, a model file and an alignment file. Manually downloading and opening these files with visualization tools can be a cumbersome process. The new CHIMERA extension enables a web browser to communicate directly with CHIMERA. By clicking on a single link associated with each MODBASE model, information related to the model is transmitted to CHIMERA, which then displays the structures of the template and the model; their alignment is also displayed using CHIMERA’s multiple sequence alignment viewer, MultAlign Viewer. The user can then apply CHIMERA’s rich set of visualization and analysis tools to further study the model. CHIMERA runs on a local computer and is available for Linux, Windows, Mac OS 10.2, IRIX and COMPAQ Tru64 UNIX operating systems.
Figure 2.
CHIMERA and the MultAlign Viewer extension. The barrel domains of selected enolase superfamily members are shown, with sidechains displayed for active site metal-binding residues. The multiple sequence alignment contains the corresponding sequences with the metal-binding residues colored in the same way. The CHIMERA interface allows user selections within the sequences, to highlight the corresponding regions of the structures and vice versa.
FUTURE DIRECTIONS
MODBASE will be updated at least monthly to reflect the growth of the sequence and structure databases, as well as improvements in the methods and software used for calculating the models. We also plan to integrate access to the sequence profiles in the web-based interface and to include tools for target selection for structural genomics. Furthermore, we plan to improve the flexibility of searching for ligand binding sites. And finally, we will include additional search options to support associating structure and function.
CITATION
Users of MODBASE are requested to cite this article in their publications.
Acknowledgments
ACKNOWLEDGEMENTS
We are especially grateful to Roberto Sánchez for constructing the first version of MODBASE. We also thank Valentin Ilyin, Bino John, William Lane, Maria Sammut and Edward Wittenstein for their contributions to MODBASE, and Tom Goddard for his contribution to CHIMERA. We thank Elaine Meng for her assistance with preparing Figure 2. The project has been supported by NIH/NIGMS R01 GM 54762, NIH/NIGMS P50 GM62529, NIH/NCI R33 CA84699, NIH/NCRR P41 RR01081 (TF), Sun Academic Equipment Grant EDUD-7824-020257-US, an IBM SUR grant and an Intel computer hardware gift.
REFERENCES
- 1.Brenner S.E. and Levitt,M. (2000) Expectations from structural genomics. Protein Sci., 9, 197–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O’Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J., Rapp,B.A. and Wheeler,D.L. (2002) GenBank. Nucleic Acids Res., 30, 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berman H.M., Battistuz,T., Bhat,T.N., Bluhm,W.F., Bourne,P.E., Burkhardt,K., Feng,Z., Gilliland,G.L., Iype,L., Jain,S. et al. (2002) The Protein Data Bank. Acta Crystallogr. D, 58, 899–907. [DOI] [PubMed] [Google Scholar]
- 5.Baker D. and Sali,A. (2001) Protein structure prediction and structural genomics. Science, 294, 93–96. [DOI] [PubMed] [Google Scholar]
- 6.Marti-Renom M.A., Stuart,A.C., Fiser,A., Sanchez,R., Melo,F. and Sali,A. (2000) Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct., 29, 291–325. [DOI] [PubMed] [Google Scholar]
- 7.Pieper U., Eswar,N., Stuart,A.C., Ilyin,V.A. and Sali,A. (2002) MODBASE, a database of annotated comparative protein structure models. Nucleic Acids Res., 30, 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer D., Elofsson,A., Rychlewski,L., Pazos,F., Valencia,A., Rost,B., Ortiz,A.R. and Dunbrack,R.L.,Jr (2001) CAFASP2: the second critical assessment of fully automated structure prediction methods. Proteins (Suppl. 5), 171–183. [DOI] [PubMed] [Google Scholar]
- 9.Koh I.Y.Y., Eyrich,V.A., Marti-Renom,M.A., Przybylski,D., Madhusudhan,M.S., Eswar,N., Grana,O., Pazos,F., Valencia,A., Sali,A. et al. (2003) EVA: evaluation of protein structure prediction servers. Nucleic Acids Res., 31, 3311–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bujnicki J.M., Elofsson,A., Fischer,D. and Rychlewski,L. (2001) LiveBench-2: large-scale automated evaluation of protein structure prediction servers. Proteins (Suppl. 5), 184–191. [DOI] [PubMed] [Google Scholar]
- 11.Marti-Renom M.A., Ilyin,V.A. and Sali,A. (2001) DBAli: a database of protein structure alignments. Bioinformatics, 17, 746–747. [DOI] [PubMed] [Google Scholar]
- 12.Fiser A. and Sali,A. (2003) ModLoop: Automated modeling of loops in protein structures. Bioinformatics, in press. [DOI] [PubMed] [Google Scholar]
- 13.Huang C.C., Couch,G.S., Pettersen,E.F. and Ferring,T.E. (1996) CHIMERA: An extensible molecular modeling application constructed using standard components. Pacific Symp. Biocomput., 1, 724. [Google Scholar]
- 14.Sali A. (1995) MODELLER: Implementing 3D Protein Modeling. mc2. Molecular Simulations Inc., Vol. 2, p. 5.
- 15.Chance M.R., Bresnick,A.R., Burley,S.K., Jiang,J.S., Lima,C.D.S.A., Almo,S.C., Bonanno,J.B., Buglino,J.A., Boulton,S., Chen,H. et al. (2002) Structural Genomics: A pipeline for providing structures for the biologist. Protein Sci., 11, 723–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eswar N., John,B., Mirkovic,N., Fiser,A., Ilyin,V., Pieper,U., Stuart,A.C., Marti-Renom,M.A., Madhusudhan,M.S., Yerkovich,B. et al. (2003) Tools for comparative protein structure modeling and analysis. Nucleic Acids Res., 31, 3375–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eyrich V.A., Marti-Renom,M.A., Przybylski,D., Madhusudhan,M.S., Fiser,A., Pazos,F., Valencia,A., Sali,A. and Rost,B. (2001) EVA: continuous automatic evaluation of protein structure prediction servers. Bioinformatics, 17, 1242–1243. [DOI] [PubMed] [Google Scholar]
- 18.John B. and Sali,A. (2003) Comparative protein structure modeling by iterative alignment, model building and model assessment. Nucleic Acids Res., 31, 3982–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez R. and Sali,A. (1999) ModBase: a database of comparative protein structure models. Bioinformatics, 15, 1060–1061. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez R., Pieper,U., Mirkovic,N., de Bakker,P.I., Wittenstein,E. and Sali,A. (2000) MODBASE, a database of annotated comparative protein structure models. Nucleic Acids Res., 28, 250–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sali A. and Blundell,T.L. (1990) Definition of general topological equivalence in protein structures. A procedure involving comparison of properties and relationships through simulated annealing and dynamic programming. J. Mol. Biol., 212, 403–428. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadreyev R. and Grishin,N. (2003) COMPASS: a tool for comparison of multiple protein alignments with assessment of statistical significance. J. Mol. Biol., 326, 317–336. [DOI] [PubMed] [Google Scholar]
- 24.Sali A. and Blundell,T.L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol., 234, 779–815. [DOI] [PubMed] [Google Scholar]
- 25.Melo F., Sanchez,R. and Sali,A. (2002) Statistical potentials for fold assessment. Protein Sci., 11, 430–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaffer A.A., Wolf,Y.I., Ponting,C.P., Koonin,E.V., Aravind,L. and Altschul,S.F. (1999) IMPALA: matching a protein sequence against a collection of PSI-BLAST- constructed position-specific score matrices. Bioinformatics, 15, 1000–1011. [DOI] [PubMed] [Google Scholar]
- 27.Marcotte E.M., Pellegrini,M., Thompson,M.J., Yeates,T.O. and Eisenberg,D. (1999) A combined algorithm for genome-wide prediction of protein function. Nature, 402, 83–86. [DOI] [PubMed] [Google Scholar]
- 28.Aloy P. and Russell,R.B. (2003) InterPreTS: protein interaction prediction through tertiary structure. Bioinformatics, 19, 161–162. [DOI] [PubMed] [Google Scholar]
- 29.Lu L., Lu,H. and Skolnick,J. (2002) MULTIPROSPECTOR: an algorithm for the prediction of protein–protein interactions by multimeric threading. Proteins, 49, 350–364. [DOI] [PubMed] [Google Scholar]
- 30.Stuart A.C., Ilyin,V.A. and Sali,A. (2002) LigBase: a database of families of aligned ligand binding sites in known protein sequences and structures. Bioinformatics, 18, 200–201. [DOI] [PubMed] [Google Scholar]
- 31.Vitkup D., Melamud,E., Moult,J. and Sander,C. (2001) Completeness in structural genomics. Nature Struct. Biol., 8, 559–566. [DOI] [PubMed] [Google Scholar]
- 32.Orengo C.A., Pearl,F.M. and Thornton,J.M. (2003) The CATH domain structure database. Methods Biochem. Anal., 44, 249–271. [DOI] [PubMed] [Google Scholar]
- 33.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Servant F., Bru,C., Carrere,S., Courcelle,E., Gouzy,J., Peyruc,D. and Kahn,D. (2002) ProDom: automated clustering of homologous domains. Brief. Bioinform., 3, 246–251. [DOI] [PubMed] [Google Scholar]
- 35.Karolchik D., Baertsch,R., Diekhans,M., Furey,T.S., Hinrichs,A., Lu,Y.T., Roskin,K.M., Schwartz,M., Sugnet,C.W., Thomas,D.J. et al. (2003) The UCSC Genome Browser Database. Nucleic Acids Res., 31, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]