Abstract
In a number of higher plants, a substantial portion of the genome is composed of repetitive sequences that can hinder genome annotation and sequencing efforts. To better understand the nature of repetitive sequences in plants and provide a resource for identifying such sequences, we constructed databases of repetitive sequences for 12 plant genera: Arabidopsis, Brassica, Glycine, Hordeum, Lotus, Lycopersicon, Medicago, Oryza, Solanum, Sorghum, Triticum and Zea (www.tigr.org/tdb/e2k1/plant.repeats/index.shtml). The repetitive sequences within each database have been coded into super-classes, classes and sub-classes based on sequence and structure similarity. These databases are available for sequence similarity searches as well as downloadable files either as entire databases or subsets of each database. To further the utility for comparative studies and to provide a resource for searching for repetitive sequences in other genera within these families, repetitive sequences have been combined into four databases to represent the Brassicaceae, Fabaceae, Gramineae and Solanaceae families. Collectively, these databases provide a resource for the identification, classification and analysis of repetitive sequences in plants.
INTRODUCTION
Although plant genome size varies significantly due to ploidy differences, another contributor to genome size variation is the repetitive DNA content (1). For example, maize (Zea mays) has a genome size of 2.3–2.7 Gb (2), yet an estimated 50–80% of the genome is composed of repetitive sequences (1,3). A number of different repetitive sequences have been reported in plants and these can be classified into super-classes, classes and sub-classes based on structure and sequence composition. The transposable element super-class includes retrotransposons, transposons and miniature inverted-repeat transposable elements (MITEs) [for recent review see (4)]. Retrotransposons, which transpose through an RNA intermediate, include those with long terminal repeats (LTRs) as well as those without LTRs, which are termed long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs). Plant centromeres are composed of tracts of tandemly repeated sequence, also termed satellite repeats, that are interspersed with other repetitive sequences such as centromeric- and pericentromeric-specific retrotransposons (5,6). Telomeric sequences include the telomere repeat sequence and telomere-associated sequences [for review see (7)]. Another super-class of repetitive sequences are the 18S–5.8S–25S (45S) and 5S ribosomal DNAs (rDNAs), which encode the structural RNA components of ribosomes (8).
The high degree of similarity and duplication of repetitive sequences within certain plant genomes presents difficulties in annotation and genome analyses due to the false associations that can occur. To best identify and catalog the repetitive sequences within plant genomes as part of our overall genome annotation efforts in plants, we created a series of databases from 12 plant genera representing four plant families and made these available for searching and download.
CONSTRUCTION OF THE REPEAT DATABASES
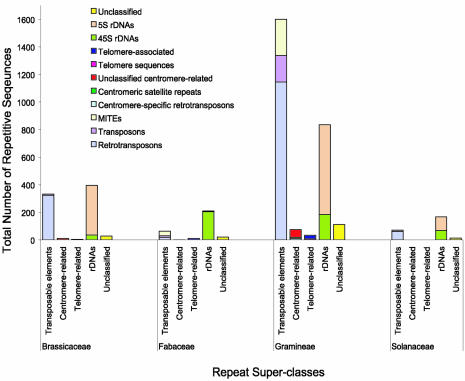
Repetitive DNA sequences of 12 plant genera (Arabidopsis, Brassica, Glycine, Hordeum, Lotus, Lycopersicon, Medicago, Oryza, Solanum, Sorghum, Triticum and Zea) were retrieved from GenBank and other published records based on their annotation. After elimination of duplicated sequences and trimming of contaminating vector sequences, the sequences were coded into five super-classes: transposable elements, centromere-related, telomere-related, rDNA and unclassified repetitive sequences. These super-classes were then broken down into classes and sub-classes of repeats (Supplementary table 1). The collected repetitive sequences from genera within the Brassicaceae (Arabidopsis, Brassica), Fabaceae (Glycine, Lotus, Medicago), Gramineae (Hordeum, Oryza, Sorghum, Triticum, Zea) and Solanaceae (Lycopersicon, Solanum) families were combined into a repeat database for the plant family. As shown in Table 1, we were able to collect and code 3993 repetitive sequences representing 5.33 Mb of sequence by querying public databases. The majority of these repetitive sequences were obtained from the Gramineae family [2658 entries (66.6%), representing 4.4 Mb], consistent with previous reports of the high degree of repetitive sequences within Gramineae species such as maize (Zea mays) (1,3) and rice (Oryza sativa) (9,10). The second highest number of repetitive sequences was obtained from the Brassicaceae family (775 entries, 19.4%), which is reflective of the availability of the entire Arabidopsis genome (11). A smaller number of repetitive sequences were obtained from the Fabaceae (308 entries, 7.7%) and Solanaceae (252 entries, 6.3%) families. Throughout the entire set of family databases, the highest number of entries was in the transposable element super-class with a total of 2068 sequences (Fig. 1). Within the transposable element super-class, retrotransposons were the most abundant (1550 entries, ∼75% of the total transposable element entries). The second most abundant repeat super-class was the rDNA super-class with 1610 entries in total. Detailed tables showing the composition of each family repeat database can be obtained on the project web page at www.tigr.org/tdb/e2k1/plant.repeats/rp.stat.shtml.
Table 1. Statistics of the repetitive sequences within the family repeat databases.
| Family | No. entries | Length (kb) |
|---|---|---|
| Brassicaceae | 775 | 449.5 |
| Fabaceae | 308 | 292.6 |
| Gramineae | 2658 | 4434.8 |
| Solanaceae | 252 | 154.1 |
| Total | 3993 | 5331.0 |
Figure 1.
Composition of the plant family repeat databases. The numbers for each of the five repeat super-classes and major classes within the four plant family repeat databases are shown.
To capture repetitive sequences in the public databases that were not annotated as repetitive sequences and thus expand the database for a single genus (e.g. Oryza), we searched available genomic sequences (the HTGS and PLANT divisions of GenBank) with the nascent family repeat database (e.g. TIGR Gramineae Repeat Database) using FLAST (12). Sequences that matched a repetitive DNA sequence within the family repeat database with ∼75% identity and ∼95% in overall length were selected and coded accordingly. The sequences were combined with repetitive sequences obtained previously from public databases to create the TIGR Plant Repeat Database for that genus. There are a total of 29 561 entries in the 12 genera-specific repeat databases, representing 15 Mb of sequence, a 3-fold expansion of repetitive sequences compared with our family repeat databases (Table 2; detailed statistics available at www.tigr.org/tdb/e2k1/plant.repeats/rp.stat.shtml). The Orzya database contains the largest amount of repetitive sequence, measured either by the number of entries or by the length of repetitive sequence, reflecting the abundance of genomic DNA for Oryza sativa subsp. japonica as part of the International Rice Genome Sequencing Project (13–16).
Table 2. Statistics of the repetitive sequences within the 12 genera-specific repeat databases created from searching the HTGS and PLANT divisions of GenBank.
| Genus | No. entries | Total length (kb) | Total length of sequences searched (Mb)a | No. sequences extracted | Total length of sequences extracted (kb) |
|---|---|---|---|---|---|
| Arabidopsis | 1744 | 957.2 | 119.9 | 1198 | 588.2 |
| Brassica | 249 | 89.6 | 2.2 | 20 | 9.2 |
| Glycine | 130 | 165.2 | 3.7 | 14 | 9.0 |
| Hordeum | 708 | 670.7 | 3.0 | 186 | 103.0 |
| Lotus | 71 | 42.9 | 26.4 | 7 | 4.9 |
| Lycopersicon | 95 | 71.0 | 3.2 | 9 | 2.3 |
| Medicago | 452 | 222.1 | 68.7 | 324 | 123.7 |
| Oryza | 23637 | 8928.3 | 502.2 | 23102 | 8316.6 |
| Solanum | 167 | 85.9 | 1.7 | 1 | 0.4 |
| Sorghum | 233 | 192.9 | 1.9 | 28 | 6.9 |
| Triticum | 783 | 903.4 | 2.9 | 124 | 58.2 |
| Zea | 1292 | 2746.6 | 13.5 | 555 | 522.3 |
| Total | 29561 | 15075.7 | 749.3 | 25568 | 9744.7 |
aRepetitive sequences were identified by searching the HTGS and PLANT divisions of GenBank with the respective family repeat database, parsing the genomic sequences and collapsing overlapping sequences into one record.
While the HTGS and PLANT divisions of GenBank are repositories for sequences typically representative of whole genes or large stretches of genomic DNA [e.g. bacterial artificial chromosome (BAC) clones], the genome survey sequences (GSS) division of GenBank is a repository for single-pass sequences such as BAC end sequences and whole-genome shotgun sequences. Although the single pass and short length of GSS sequences have limitations due to sequence errors and lack of large contiguous stretches of sequence, GSS sequences can provide a broader sampling of an unfinished genome. To further identify repetitive sequences in the 12 plant genera represented in our databases, we searched the GSS division for repetitive sequences using the nascent family repeat sequence databases as the query. As shown in Table 3, 174 441 sequences representing 93.8 Mb were identified in a search of 1162 Mb of GSS sequence. The highest number (124 724) and length of repetitive sequences (69 Mb) identified in the GSS division were from Zea, consistent with the high degree of repetitive content in maize (1,3) and the deposition of a large number of sequences from two maize genomic sequencing projects (17). Brassica had the second most abundant number and length of repetitive sequences identified from the GSS sequences (34 314 sequences representing 19.2 Mb) attributable to the availability of a large number of whole shotgun sequences for Brassica oleraceae (http://www.tigr.org/tdb/e2k1/bog1/). Due to the limited amount of sequence in the GSS division for Hordeum and Solanum, no repetitive sequences were identified. The large number and total length of the GSS-derived sequences presents a logistical problem with the incorporation of these repetitive sequences into our database. Thus, we elected to keep the GSS- and HTGS/PLANT-division-derived repetitive sequences separate and not merge these into a single database.
Table 3. Summary of repetitive sequences extracted by searching the GSS division of GenBank.
| Genus | Total length of sequences searched (Mb) | No. of sequences extracteda | Total length (kb) of extracted sequences |
|---|---|---|---|
| Arabidopsis | 87.9 | 8018 | 2809.1 |
| Brassica | 386.2 | 34314 | 19255.2 |
| Glycine | 6.3 | 149 | 64.0 |
| Hordeum | 0.07 | 0 | 0 |
| Lotus | 19.8 | 479 | 201.1 |
| Lycopersicon | 5.8 | 343 | 174.9 |
| Medicago | 1.9 | 15 | 6.6 |
| Oryza | 61.5 | 4627 | 1989.8 |
| Solanum | 0.03 | 0 | 0 |
| Sorghum | 25.9 | 1755 | 464.6 |
| Triticum | 0.2 | 17 | 7.4 |
| Zea | 566.3 | 124724 | 68793.0 |
| Total | 1161.9 | 174441 | 93765.7 |
aRepetitive sequences were identified by searching the GSS division of GenBank with the respective family repeat database, parsing the genomic sequences and collapsing overlapping sequences into one record.
ACCESS TO THE DATABASES
Several modes of access are available for the TIGR Plant Repeat Databases. All databases, the four family databases, the 12 HTGS/PLANT-division-derived genera-specific databases, and the 10 GSS-division-derived genera-specific databases are available for BLAST searching at http://tigrblast.tigr.org/euk-blast/index.cgi?project=plant.repeats. All of these databases (26 in total) can be downloaded through anonymous FTP as a flat file (ftp://ftp.tigr.org/pub/data/TIGR_Plant_ Repeats/). For repetitive sequences obtained by querying GenBank using annotation, the header line contains the internal repeat code classification, the originating GenBank accession number and description. For sequences obtained through similarity searches, the header contains the internal repeat code classification, the GenBank GI number, the coordinate information of the parent sequence and the classification of the extracted repeat sequence. Additionally, the user can query the four family databases and 12 HTGS/PLANT-division-derived genera-specific databases for a subset of sequences (http://www.tigr.org/tdb/e2k1/plant.repeats/subset.shtml). Available selection criteria include the database, super-class or class of repetitive sequences, as well as sequences derived from querying GenBank by annotation or sequences derived by sequence similarity searches.
CONCLUSIONS
We have generated a set of repetitive sequence databases that represent four major plant families and a number of agriculturally significant plant genera including maize (Zea), rice (Oryza), wheat (Triticum), barley (Hordeum), sorghum (Sorghum), soybean (Glycine), tomato (Lycopersicon), potato (Solanum) and canola (Brassica). Model species such as Arabidopsis thaliana, Lotus japonicus and Medicago truncatula are represented in the Arabidopsis, Lotus and Medicago databases, respectively. The collective set of databases contains >100 Mb of repetitive sequence and with the ongoing genome sequencing efforts in plants, these databases can be expanded in the future. The broad nature of the databases with respect to taxon provides a starting point for the identification of repetitive sequences in additional species within these families.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Qiaoping Yuan, Jia Liu, Agnes Chan, Cathy Whitelaw and Chris Town for their assistance in querying public databases for repetitive sequences. We also wish to acknowledge the members of the TIGR Bioinformatics and IT departments for their contribution in supporting our efforts. In addition, we thank Phillip John SanMiguel, Jeff Bennetzen and Joachim Messing, for donating unpublished maize transposable element sequences to the TIGR Gramineae and Zea Repeat Databases. This work was supported by funds awarded to C.R.B. from the US Department of Agriculture (99-35317-8275), the National Science Foundation (DBI-998282) and the US Department of Energy (DE-FG02-99ER20357; DE-FG02-01ER15265).
REFERENCES
- 1.Flavell R.B., Bennett,M.D., Smith,J.B. and Smith,D.B. (1974) Genome size and the proportion of repeated nucleotide sequence DNA in plants. Biochem. Genet., 12, 257–269. [DOI] [PubMed] [Google Scholar]
- 2.Arumuganathan K. and Earle,E.D. (1991) Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep., 9, 208–218. [Google Scholar]
- 3.SanMiguel P., Tikhonov,A., Jin,Y.K., Motchoulskaia,N., Zakharov,D., Melake-Berhan,A., Springer,P.S., Edwards,K.J., Lee,M., Avramova,Z. et al. (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science, 274, 765–768. [DOI] [PubMed] [Google Scholar]
- 4.Feschotte C., Jiang,N. and Wessler,S.R. (2002) Plant transposable elements: where genetics meets genomics. Nature Rev. Genet., 3, 329–341. [DOI] [PubMed] [Google Scholar]
- 5.Nagaki K., Song,J., Stupar,R.M., Parokonny,A.S., Yuan,Q., Ouyang,S., Liu,J., Hsiao,J., Jones,K.M., Dawe,R.K. et al. (2003) Molecular and cytological analyses of large tracks of centromeric DNA reveal the structure and evolutionary dynamics of maize centromeres. Genetics, 163, 759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Copenhaver G.P., Nickel,K., Kuromori,T., Benito,M.I., Kaul,S., Lin,X., Bevan,M., Murphy,G., Harris,B., Parnell,L.D. et al. (1999) Genetic definition and sequence analysis of Arabidopsis centromeres. Science, 286, 2468–2474. [DOI] [PubMed] [Google Scholar]
- 7.McKnight T.D., Fitzgerald,M.S. and Shippen,D.E. (1997) Plant telomeres and telomerases. A review. Biochemistry, 62, 1432–1441. [PubMed] [Google Scholar]
- 8.Shishido R., Sano,Y. and Fukui,K. (2000) Ribosomal DNAs: an exception to the conservation of gene order in rice genomes. Mol. Gen Genet., 263, 586–591. [DOI] [PubMed] [Google Scholar]
- 9.Deshpande V.G., Ranjekar,P.K. (1988) Repetitive DNA in three Gramineae species with low DNA content. Hoppe Seylers Z. Physiol. Chem., 361, 1223–1233. [DOI] [PubMed] [Google Scholar]
- 10.McCouch S.R., Kochert,G., Yu,Z.H., Wang,Z.Y., Khush,G.S., Coffman,W.R. and Tanksley,S.D. (1988) Molecular mapping of rice chromosomes. Theor. Appl. Genet., 76, 815–829. [DOI] [PubMed] [Google Scholar]
- 11.The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 12.Yuan Q., Quackenbush,J., Sultana,R., Pertea,M., Salzberg,S.L. and Buell,C.R. (2001) Rice bioinformatics. Analysis of rice sequence data and leveraging the data to other plant species. Plant Physiol., 125, 1166–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng Q., Zhang,Y., Hao,P., Wang,S., Fu,G., Huang,Y., Li,Y., Zhu,J., Liu,Y., Hu,X. et al. (2002) Sequence and analysis of rice chromosome 4. Nature, 420, 316–320. [DOI] [PubMed] [Google Scholar]
- 14.The Rice Chromosome 10 Sequencing Consortium (2003) In-depth view of structure, activity and evolution of rice chromosome 10. Science, 300, 1566–1569. [DOI] [PubMed] [Google Scholar]
- 15.Sasaki T., Matsumoto,T., Yamamoto,K., Sakata,K., Baba,T., Katayose,Y., Wu,J., Niimura,Y., Cheng,Z., Nagamura,Y. et al. (2002) The genome sequence and structure of rice chromosome 1. Nature, 420, 312–316. [DOI] [PubMed] [Google Scholar]
- 16.Sasaki T. and Burr,B. (2000) International Rice Genome Sequencing Project: the effort to completely sequence the rice genome. Curr. Opin. Plant Biol., 3, 138–141. [DOI] [PubMed] [Google Scholar]
- 17.Chandler V.L. and Brendel,V. (2002) The maize genome sequencing project. Plant Physiol., 130, 1594–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.