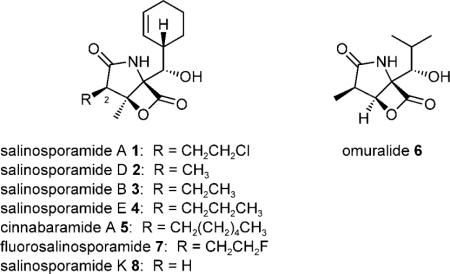
The γ-lactam-β-lactone natural product salinosporamide A (1) is a potent proteasome inhibitor produced by the marine bacterium Salinispora tropica.[1,2] This chlorinated anticancer agent dominates a family of natural structural analogues that primarily differ at the C-2 substituent.[3] In the case of 1, the C-2 chloroethyl group is a key functional group that enables the molecule to irreversibly bind to the 20S proteasome.[4] All γ-lactam-β-lactone natural products, including the salinosporamides (1–4), the cinnabaramides (5), and omuralide (6), share the initial reaction with the proteasome in which the N-terminal threo-nine residue of the catalytic β-subunit attacks the β-lactone group of the inhibitor to form an ester linkage.[3] While this covalent proteasome–inhibitor complex is susceptible to water hydrolysis, a subsequent reaction of the β-lactone-derived C-3 hydroxyl group with a C-2 side-chain leaving group such as in 1 yields a tetrahydrofuran adduct that is stable to hydrolysis.[4] Due to the mechanistic importance of the salinosporamide C-2 substituent, biosynthetic studies in S. tropica explored the origins of this small compound library to reveal that the salinosporamides are atypical products of a hybrid polyketide synthase–nonribosomal peptide synthetase (PKS–NRPS).[5,6] By accommodating different PKS building blocks such as chloroethyl-, methyl-, ethyl-, and propyl-malonyl-CoA, salinosporamides A(1), D (2), B (3) and E (4), respectively, are biosynthesized.[6,7] This understanding provided the logic to engineer the unnatural derivative fluorosalinosporamide[8,9] (7) as well as the molecular basis to explore new genome sequences for the discovery of novel salinosporamide derivatives. Herein we report the genome-inspired discovery and characterization of salinosporamide K (8) from a new source, “Salinispora pacifica” strain CNT-133, that provides insight into the evolution of the salinosporamide biosynthetic pathway. From the three proposed Salinispora species, S. tropica and “S. pacifica” are more closely related to each other than each is to S. arenicola.[10]
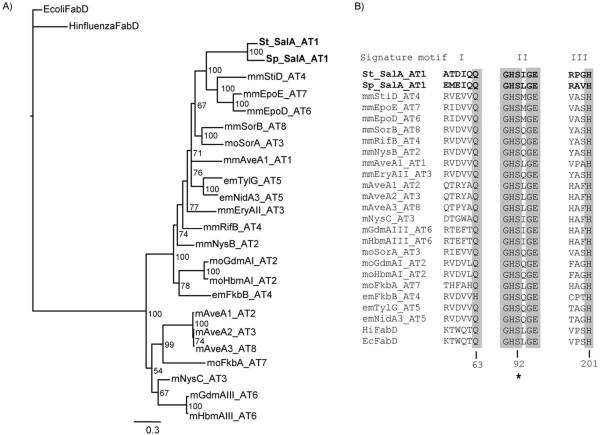
A draft genome of the marine-sediment-derived “S. pacifica” strain CNT-133 from Fiji was generated by a hybrid approach involving 454 pyrosequencing and Sanger sequencing to give 5.3 Mb of sequence assembled into 330 scaffolds. Bioinformatic analysis of the secondary metabolic pathways revealed a truncated biosynthetic gene cluster related to the 41 kb S. tropica strain CNB-440 sal (St_sal) locus responsible for salinosporamide A biosynthesis,[6] albeit with one significant change. While genes expected to be involved in the formation of the core γ-lactam-β-lactone nucleus and those involved in the biosynthesis of the nonproteinogenic amino acid residue cyclohexenylalanine are conserved in the two species, all genes coding for the enzymes in the chloroethylmalonyl-CoA pathway are absent from the 26 kb “S. pacifica” cluster (Sp_sal)—a difference of 15 kb DNA sequence. The order of the orthologous genes is also conserved except for the rearranged regulator salR3 and for the presence of transposases in the two locations where chloroethylmalonyl-CoA biosynthetic genes reside in S. tropica (Figure 1, Table S1 in the Supporting Information). Based on these observations, we anticipated that if “S. pacifica” indeed produces a salinosporamide molecule, it would be one in which the C-2 substituent is not halogenated.
Figure 1.
Organization of the salinosporamide biosynthetic clusters (sal)in “S. pacifica” strain CNT-133 (top) and in S. tropica strain CNB-440 (bottom) with the respective main products shown on the right. Genes putatively involved in the construction of the core γ-lactam-β-lactone ring system (gray), assembly of the nonproteinogenic amino acid cyclohexenylalanine (blue), chloroethylmalonyl-CoA pathway (red), regulation and resistance (yellow), unknown (white) and transposases (black) are color-coded. Blue and red genes correlate with the respective building blocks in the salinosporamide skeleton. Intact incorporation of [1,2-13C]acetate into salinosporamide K was detected in the green shaded regions and contrasts that previously measured for salinosporamide A.[11]
To test whether Sp_sal is expressed under laboratory growth conditions, we conducted transcript analyses of representative genes by semiquantitative RT-PCR. Transcription of the PKS gene salA, the prephenate decarboxylase salX, and the putative regulatory gene salR3 was confirmed in a seawater-based production medium in which transcription of the regulator salR3 precedes that of the structural genes salA and salX in agreement with its proposed role as a transcriptional regulator (Figure 2).[6] We next analyzed a seven-day “S. pacifica” culture for salinosporamide molecules. While no known salinosporamides were detected, we did identify a novel compound (8) with a salinosporamide-like UV profile. Chemical workup of a 20 L culture yielded 23 mg of salinosporamide K (8) with the molecular composition C13H17NO4 as established by high-resolution ESI-MS (m/z [M+H]+ 252.1227 obs., 0.3 mmu error). Analysis of the proton and carbon NMR spectra, with the aid of gradient-enhanced HMBC data, clearly indicated that 8 was a novel salinosporamide without substitution at C-2 (Table S2). The connectivity and stereochemistry were further confirmed by single-crystal X-ray analysis (Figure 3), which showed a framework identical to those of the S. tropica salinosporamides 1–4. Salinosporamide K (8) exhibited 20S proteasome (chymotrypsin-like) inhibitory activity ((4.6±0.4) nM) comparable to that of 3 ((5.2±0.3) nM) and, as expected, was markedly less potent than 1 ((0.7±0.05) nM).[8] Moreover, the cytotoxicity of 8 was evaluated ex vivo against the human-colon-carcinoma cell line HCT-116 and shown, at (988±155) nM, to be 100 times less potent than 1 ((9.5±1.6) nM) yet slightly more potent than 3 ((6100±300) nM).[8] These data confirm the mechanistic importance of the C-2 leaving group in 1 and suggest that 8 could be a convenient synthon for the rapid generation of chemical diversity at C-2 in order to further explore the chemistry of this side chain as it interacts with the protea-some S2 binding site.
Figure 2.
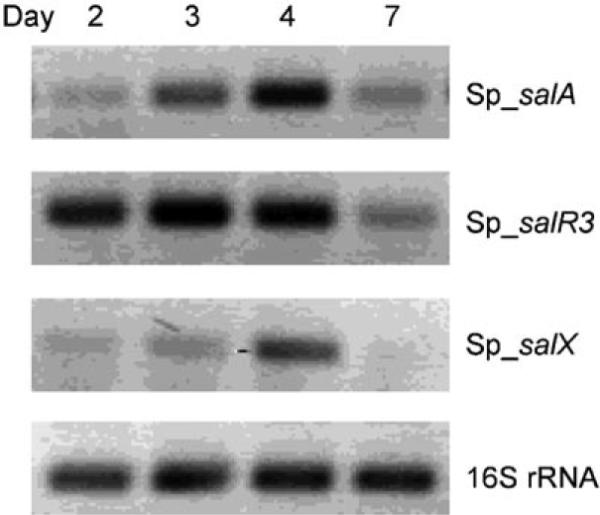
Transcript analysis of representative genes from the S. pacifica salinosporamide gene cluster. The absence of genomic DNA contamination in the RNA samples and cDNA quality (shown) were assessed by using 16S rRNA primers.
Figure 3.
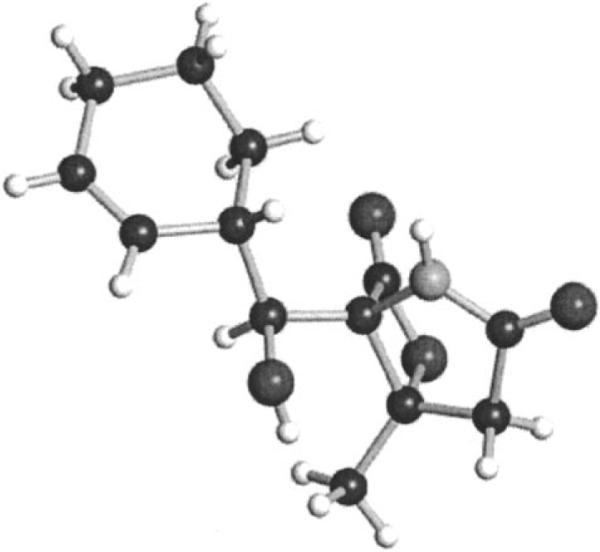
ORTEP plot at the 99.6% level of the final X-ray structure of salinosporamide K depicting its relative stereochemistry.
The structure of 8 is suggestive of a biosynthetic pathway in which the non-branched PKS extender unit malonyl-CoA is alternatively employed by the Sp_salA didomain PKS rather than the branched extenders preferred by St_salA.[7] To explore this scenario, we fed [1,2-13C]acetate to a culture of “S. pacifica” and isolated the resultant 8, which upon 13C NMR analysis showed the intact incorporation of two acetate molecules at C-1/C-2 (J1,2 = 47 Hz) and C-3/C-12 (J3,12 = 42 Hz) with relative enrichments of ~ 40 % (Figure 1). This result nicely complemented the biosynthesis of 1 in which C-3/C-14 is also acetate-derived,[11] while the C-2 fragment originates from the chlorination of S-adenosylmethionine.[12] To further explore whether S. tropica-produced salinosporamides such as 2 and 3 can be produced in “S. pacifica” strain CNT-133, we administered propionate and crotonate, respectively, to the culture, as these precursors selectively specify the methyl- and ethyl-branched salinosporamides 2 and 3.[7] Propionate was the only precursor tested that yielded a product, which was verified as 2 when compared with an authentic standard derived from S. tropica. Thus, the extender unit specifying acyltransferase (AT) domain salA-AT1 likely governs the natural product outcome in the two systems where the “S. pacifica” AT1 prefers smaller substrates than the analogous AT1 domain of S. tropica.
Despite this difference in specificity, the St and Sp_salA_AT1 domains are 76 % identical at the amino acid level, with the next closest homologues at 41–43 % identity (see Table S3). The more common malonyl- and methylmalonyl-specific ATs are thought to share a common ancestor that diverged to form two distinct groups, whereas the relatively rare ethylmalonyl- and methoxymalonyl-specific ATs apparently evolved more than once and fall in either clade.[13] We previously showed that St_salA_AT1 belongs to the methylmalonyl-CoA group,[6] and a similar phylogenetic analysis reveals that Sp_ salA_AT1 shares a close common ancestor in spite of its distinct substrate discrimination (Figure 4); this suggests a case of convergent evolution for malonyl-CoA specificity.
Figure 4.
Phylogenetic analysis of SalA_AT1. A) Tree generated by Bayesian inference of codon-aligned DNA sequences from Sp and St SalA_AT1 and representative AT extender domains specific for malonyl-CoA (m), methylmalonyl-CoA (mm), ethylmalonyl-CoA (em), and methoxymalonyl-ACP (mo). The malonyl-CoA:ACP transacylase component of fatty acid synthase, FabD, from E. coli and Haemophilus influenzae was used as an outgroup to root the tree. The scale bar indicates changes per nucleotide. Values on nodes are representative of the reliability of a particular grouping (i.e. posterior probabilities in %). B) Sequence alignments showing three signature motifs known to be involved in substrate discrimination.[16] Both Salinispora SalA_AT1 domains have rather distinct signature motifs, and a prediction of substrate specificity based solely on these motifs is not possible. Numbers below the alignment correspond to E. coli FadD, the catalytic serine is indicated by an asterisk. See ref. [6] for details on sequences used to construct the phylogeny.
With the genome-aided discovery of 8 from a new Salinispora species not previously known to produce salinosporamides, we have the rare opportunity to compare their biosyntheses at the molecular level; this, in turn, gives insight into pathway evolution of these natural products. The striking difference between the syntenic St_sal and Sp_sal loci is the exact replacement of the chloroethylmalonyl-CoA biosynthetic genes that reside on either side of the salinosporamide synthetase encoding genes salAB with putative transposase genes belonging to the IS3, IS4, and IS2606 families[14] (Figure 1). Thus, without the ability of Sp_sal to generate a dedicated PKS extender unit such as chloroethyl-[6] or propylmalonyl-CoA[7] that gives rise to 1 and 4 in S. tropica, the Sp_sal system likely evolved to take advantage of readily available cellular pools of malonyl-CoA and, to a lesser extent, methylmalonyl-CoA in the biosynthesis of its products 8 and 2, respectively. A similar scenario might also operate in Streptomyces sp. JS360, which produces cinnabaramides, such as 5, that contain a C-2 hexyl side chain,[15] in which genes conferring hexylmalonyl-CoA biosynthesis might be encoded in a biosynthetic gene cluster resembling that of Sp_sal. This study provides the first glimpse of pathway evolution in the salinosporamides by dictating precursor supply and gives insight into γ-lactam-β-lactone pathway engineering for designer proteasome inhibitors.
Supplementary Material
Acknowledgements
We thank Jonathan H. Badger, JCVI, for the draft genome sequence data of “S. pacifica” strain CNT-133, Arnold L. Rheingold, UCSD, for crystallography data, Lauren A. Paul, UCSD, for cytotoxicity data, and William Aalbersberg, University of the South Pacific, for providing laboratory space and facilitating field collections. We also thank the Government of Fiji for permission to work in their territorial waters. Financial support was provided by the National Institutes of Health (CA127622 to B.S.M., GM085770 to B.S.M. and P.R.J., CA044848 to W.F., and U01-TW007401 from the Fogerty Center's International Cooperative Biodiversity Groups program to P.R.J. and W.F.), the Life Sciences Research Foundation via a Tularik postdoctoral fellowship to A.S.E., and the Albert and Anneliese Konanz Foundation, Mann-heim, for a graduate fellowship to A.L.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cbic.201000564: full experimental details, NMR tables and spectra, and bioactivity data.
References
- [1].Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W. Angew. Chem. 2003;115:369. doi: 10.1002/anie.200390115. Angew. Chem. Int. Ed. 2003, 42, 355. [DOI] [PubMed] [Google Scholar]
- [2].Fenical W, Jensen PR, Palladino MA, Lam KS, Lloyd KG, Potts BC. Bioorg. Med. Chem. 2009;17:2175. doi: 10.1016/j.bmc.2008.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gulder TAM, Moore BS. Angew. Chem. 2010;122:9534–9556. Angew. Chem. Int. Ed. 2010, 49, 9346-9367. [Google Scholar]
- [4].Groll M, Huber R, Potts BCM. J. Am. Chem. Soc. 2006;128:5136. doi: 10.1021/ja058320b. [DOI] [PubMed] [Google Scholar]
- [5].Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. Proc. Natl. Acad. Sci. USA. 2007;104:10376. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Eustáaquio AS, McGlinchey RP, Liu Y, Hazzard C, Beer LL, Florova G, Alhamadsheh MM, Lechner A, Kale AJ, Kobayashi Y, Reynolds KA, Moore BS. Proc. Natl. Acad. Sci. USA. 2009;106:12295. doi: 10.1073/pnas.0901237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Liu Y, Hazzard C, Eustáquio AS, Reynolds KA, Moore BS. J. Am. Chem. Soc. 2009;131:10376. doi: 10.1021/ja9042824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Eustáquio AS, Moore BS. Angew. Chem. 2008;120:4000. Angew. Chem. Int. Ed. 2008, 47, 3936. [Google Scholar]
- [9].Eustáquio AS, O'Hagan D, Moore BS. J. Nat. Prod. 2010;73:378. doi: 10.1021/np900719u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jensen PR, Williams PG, Oh D-C, Zeigler L, Fenical W. Appl. Environ. Microbiol. 2007;73:1146. doi: 10.1128/AEM.01891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Beer LL, Moore BS. Org. Lett. 2007;9:845. doi: 10.1021/ol063102o. [DOI] [PubMed] [Google Scholar]
- [12].Eustáquio AS, Pojer F, Noel JP, Moore BS. Nat. Chem. Biol. 2008;4:69. doi: 10.1038/nchembio.2007.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ridley CP, Khosla C. Proc. Natl. Acad. Sci. USA. 2008;105:4595. doi: 10.1073/pnas.0710107105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mahillon J, Chandler M. Microbiol. Mol. Biol. Rev. 1998;62:725. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Stadler M, Bitzer J, Mayer-Bartschmid A, üMller H, Benet-Buchholz J, Gantner F, Tichy H-V, Reinemer P, Baco KB. J. Nat. Prod. 2007;70:246. doi: 10.1021/np060162u. [DOI] [PubMed] [Google Scholar]
- [16].Reeves CD, Murli S, Ashley GW, Piagentini M, Hutchinson CR, McDaniel R. Biochemistry. 2001;40:15464. doi: 10.1021/bi015864r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.