Abstract
Natural populations consist of phenotypically diverse individuals that exhibit variation in their demographic parameters and intra- and interspecific interactions. Recent experimental work suggests that such variation can have significant ecological effects. However, ecological models typically disregard this variation and focus instead on trait means and total population density. Under what situations is this simplification appropriate? Why might intraspecific variation alter ecological dynamics? In this review, we synthesize recent theory, identifying six general mechanisms by which trait variation changes the outcome of ecological interactions. These include several direct effects of trait variation per se, and indirect effects arising from genetic variation’s role in trait evolution.
Keywords: Community dynamics, demographic stochasticity, eco-evolutionary dynamics, heterogeneity, individual specialization, Jensen’s inequality, niche variation, population dynamics, trait variation
Trait variation within natural populations
One of Charles Darwin’s greatest insights was recognizing that conspecific individuals differ in many traits, including obvious features such as gender, age, or size, and also more subtle traits such as shape, behavior, or physiology. This variation provides the raw material for natural selection and thus is a key focus of evolutionary theory. In contrast, ecological theory typically focuses on predicting the dynamics of species’ abundances over time without regard to particular phenotypes. Consequently, many models of species interactions implicitly assume that all conspecific individuals are effectively interchangeable. In this paper, we argue that this assumption is misleading, and that intraspecific trait variation can substantially alter ecological dynamics.
Ecologists have long recognized that sexes [1] and age classes [2] differ in ecologically significant ways. However, even within a sex and age class, phenotypic variation among individuals can generate variation in ecological attributes [3]. Within populations, individuals can differ in anti-predator defenses [4], parasite resistance [5, 6] or exposure [7], abiotic tolerances [8], resource use [3], or competitive ability [4, 9]. Such differences can, in turn, generate variance in demographic parameters [10, 11]. For example, Southern sea otter (Enhydra lutris nereis) individuals differ substantially in diet, controlling for age, sex, and morphology [12]. Some individuals specialize on snails, others on crabs and abalone, while others are generalists. Owing to this diet variation, individual otters differ in energetic income and pathogen exposure [7].
The proximate mechanisms underlying such diet variation can be complex. In sea otters, foraging is a learned behavior [12] that can respond flexibly to resource density [13]. In other organisms, diet variation reflects differences in morphology [14, 15], digestive physiology [16], individual social status [17], or search images from past experience [18]. Each of these causes of variation can have genetic and environmental sources, as most traits exhibit intermediate heritability [19, 20].
Ecological consequences of trait variation
Trait variation among conspecific individuals has long been recognized, forming the basis of classic work in ecological genetics [21] and niche evolution [22]. However, ecologists’ interest in such variation waned after the mid-1970s [23], and intraspecific trait variation has since been largely ignored by both empirical and theoretical ecologists. Recently, ecologists have gained renewed appreciation for the extent and community consequences of intraspecific variation [3, 24]. Several experiments have manipulated intraspecific genetic diversity and found corresponding changes in population productivity and stability [25], coexistence [9, 26, 27], and ecosystem processes [28]. Intraspecific variation can thus have large ecological effects [24–30]. These results motivate the central question of this review: when and why do such effects emerge?
Despite a fast-growing literature on the ecology of trait variation, we lack a general framework for understanding the mechanisms by which trait variation influences ecological dynamics. Developing such a framework is key to determining when, and to what extent, intraspecific trait variation will alter population densities, transient dynamics, and persistence. As there are currently too few studies for a meta-analysis, these general questions are best answered with theory. Here, we synthesize the relevant theoretical literature to build a framework for understanding the ecological effects of individual variation. Specifically, we consider the following thought experiment: take any ecological model, and contrast the community dynamics with and without intraspecific trait variation. When and why does variation change the dynamics?
A modest number of theoretical studies have investigated how intraspecific trait variation affects population dynamics [31–34], interspecific competition [35–37], and predator-prey or host-parasitoid systems [38–40]. These studies consider variation in diverse traits, including (1) traditional phenotypes like size or morphology; (2) emergent traits like competitive ability [41], prey attack rate [39], or vulnerability to enemies [42]; and (3) fitness-related traits like fecundity or survival [33]. Variation in these traits might be stochastic [32], environmentally induced [43], or genetic (Mendelian or quantitative; sexual or asexual [31, 44]). These studies have found very different (and sometimes conflicting) results. For instance, different models suggest that trait variation can increase or decrease extinction risk, depending on the specified trait and its heritability [32, 34]. Variation in competitive ability can reduce or increase equilibrium densities [45, 46].
Despite these models’ diverse assumptions and predictions, a few generalities emerge. All relevant models invoke at least one of six general mechanisms by which trait variation alters ecological dynamics (Table 1). By clearly delineating these mechanisms, we can better understand when it is necessary to consider trait variation. Below, we describe each mechanism and highlight representative theoretical models. The mechanisms fall into two general categories: (1) direct ecological effects of trait variation per se, regardless of its heritability (mechanisms 1–3), and (2) indirect effects arising because genetic variation permits eco-evolutionary dynamics (mechanisms 4–6).
Table 1.
Summary of the six mechanisms by which intraspecific trait variation could alter community structure or dynamics.
| Mechanism | Name | Summary: Occurs when: |
Variation alters: | Genetic process | Examples: e |
|---|---|---|---|---|---|
| 1 | Jensen’s inequality | Ecological interactions depend non-linearly on a variable trait. | Mean interaction strength. | NA | [31, 38, 40, 41] |
| 2 | Increased degree | Trait variation changes the strength or number of species interactions. | Niche breadth. | NA | [36, 42, 46, 94] |
| 3 | Portfolio effect | Phenotypes exhibit negative covariation in density over time. | Fluctuation in total population density. | NA a | [31, 44, 65] |
| 4 | Phenotypic subsidy | Genetic or developmental exchange between phenotypes decouples each phenotype’s growth rates from its fitness. | Feasibility of ecological equilibria. | Recombination, assortment, or mutation b | [31, 44, 65, 67] |
| 5 | Eco- evolutionary dynamics | Heritable trait variation permits rapid evolution. | Mean interaction strength. | Selection c | [39, 69, 75, 77, 95] |
| 6 | Sampling effects | Small population size permits stochastic sampling from trait distributions | Mean fitness or interaction strength. | Drift d | [32, 33, 37, 78, 86, 96] |
Notes:
Portfolio effect may be facilitated by a heritable basis distinguishing phenotypes.
Phenotypic subsidy may also arise via phenotypic plasticity (developmental or behavioral).
Adaptive phenotypic plasticity may drive similar outcomes.
Sampling effects may arise via demographic stochasticity or genetic drift.
This list is intended to be illustrative, not exhaustive, due to space constraints. We focus on relevant theory, because empirical studies of the ecological effects of intraspecific variation rarely distinguish among the mechanisms listed here.
Why does intraspecific variation affect ecological dynamics?
Mechanism 1: Jensen’s Inequality
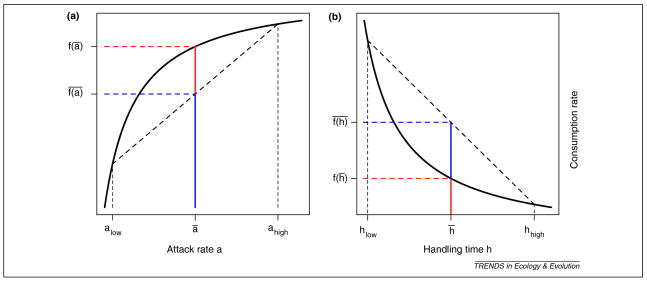
When an ecological interaction depends nonlinearly on a species’ trait, variation around the trait mean can alter the average interaction strength. Jensen’s Inequality [47] states that when x is a variable trait with mean x̄, and f(x) is a concave (convex) function of x, the average value of the function, , will be smaller (larger) than the function evaluated at the trait mean, f(x̄) (Fig. 1). Therefore, if f(x) is nonlinear and x is variable, then will typically differ from f(x̄). Hence, populations with identical trait means but different variances will have different mean interaction strengths, thereby altering demographic rates and community dynamics.
Figure 1.
The effects of individual predator variation in attack rates (a) and handling times (b) on rates of prey consumption. In both figures, the feeding rate f -- a function of an individual’s attack rate and handling time -- is plotted as a solid black line. For a dimorphic population consisting of individuals with low and high trait attack rates (or handling times), the average feeding rate f̄(a) is highlighted in blue and the feeding rate of the average individual f(ā) is highlighted in red. For any composition of individuals with low and high trait values, the average feeding rate lies on the thick dashed line. When the feeding rate is a concave function of the attack rate, the average feeding rate is lower than the feeding rate of the average individual. Conversely, feeding rate is a convex function of handling time and so variance in handling means the average feeding rate is higher than the feeding rate of the average individual.
Consider the type II functional response often used in predator-prey models to relate an individual predator’s feeding rate to its prey density. The type II functional response saturates with increasing prey density R,
| (eq. 1) |
where a is a predator’s attack rate, and h is its handling time. If predators vary in attack or handling rates, the population’s functional response is not simply f(R,ā,h̄), as is typically assumed in ecology [48], but instead is the average of individuals’ functional responses . Since (eq. 1) is concave with respect to a, variation in attack rates will lower overall predation pressure (Fig. 1a), potentially preventing predator-prey oscillations [38]. The same equation is convex with respect to h, so handling time variation increases predation pressure and destabilizes predator-prey interactions (Fig. 1b). This example illustrates how variation in two traits can yield contrasting ecological effects within a single model framework.
Non-linear (convex or concave) functions such as the Type II functional response are widespread in ecological models [47, 49]. Hence, Jensen’s Inequality might be of nearly ubiquitous importance in ecology (Box 1). Importantly, ecological effects of Jensen’s Inequality emerge irrespective of the genetic or environmental basis of trait variance, representing a direct consequence of trait variance rather than an effect of evolution (except where evolution alters the ecological function’s curvature).
Box 1. Examples of non-linear functions of variable traits.
When fitness depends non-linearly on any trait. When a population is subject to stabilizing selection on a quantitative trait, any variance around the optimal trait value imposes a fitness load because most individuals deviate from the average (e.g. optimum) and hence have suboptimal fitness. Thus, the average fitness of the population w̄(x) is less than the fitness of the average phenotype w(x̄). The opposite is true for populations under disruptive selection, w(x̄)< w̄ (x). Phenotypic variance can thus increase or decrease population growth rates depending on the mode and strength of selection acting on any given trait.
When population growth depends non-linearly on fluctuating demographic rates. Many models of population growth are non-linear functions of individual fitness [86]. Consequently, among-individual variation in fecundity or survival alters mean population growth and abundance. Genetic variation in demographic rates is typically assumed to be small because selection eliminates low-fitness genotypes. However, additional fitness variation arises from stochastic events such as random dispersal and settlement in a heterogeneous environment, random variation in gene expression, as well as stochastic sampling of realized trait values (e.g., clutch size, longevity) from a shared probability distribution.
When food web links depend on body size. Recent food web theory makes wide use of size-based niche models, which describe the probability that a species with mean body size x̄i consumes prey with mean size x̄j [54, 87]. However, if attack rate is a non-linear function of two species’ body sizes f(xi, xj), the appropriate attack rate is not f(x̄i, x̄j) but rather the weighted average attack rate across all predator/prey phenotype combinations. Consequently size variation alters food web structure (including the addition of size-dependent cannibalism [88]).
When any parameter is subject to metabolic scaling laws. Size-structured food web models often use metabolic scaling laws to obtain species-specific demographic parameters such as birth or death rates and total consumption rates [54]. These rates are assumed to scale with body mass (M) according to a power law y ~ aMb. Such power laws are considered a fundamental feature of biological systems [89]. However, their nonlinearity makes the use of mean body size misleading, because a population’s mean metabolic rate is not predicted by its mean body size (ȳ ≠ aM̄b).
Other examples include non-linear relationships between (1) light availability and photosynthesis; (2) nutrient concentration and chemostat population growth, (3) phenotypic similarity and intraspecific competition (Box 3).
Mechanism 2: Increased degree
Intraspecific trait variation can alter the number and strength of a species’ interspecific interactions, thereby changing the topology and dynamics of species interaction networks. Roughgarden [22] pointed out that a species’ diet diversity, D, could be decomposed into (1) the average within-individual diet diversity, and (2) between-individual variation (Dtotal = Dwithin + Dbetween). Consequently, increased between-individual variation (Dbetween) inflates the population’s total diet diversity (Dtotal), making the population more generalized even if individuals remain specialized (i.e., Dwithin is small and constant) [50].
Consider a food web in which each predator attacks all potential prey within a specified relative size range [51, 52]. Intraspecific variation in consumer body size increases the range of acceptable prey sizes and thus the number of species the consumer population attacks. Equivalently, heterogeneous resources are vulnerable to more diverse predators. We thus expect more variable species to interact with more species; this greater connectivity is known as ‘increased degree’ in network theory (degree is the number of links from one node to other nodes in a network). Conversely, each pairwise link might be weaker because only a subset of conspecific individuals participates in a given interaction.
Changes in population diet diversity due to trait variation can have profound effects on density-dependence, interspecific competition, and food web structure. Greater population niche width alleviates intraspecific competition, particularly among divergent phenotypes (a phenomenon known as niche complementarity; Box 2). Increased niche width also affects the outcome of interspecific competition [36] by simultaneously increasing diet overlap with other species, and reducing the impact of this overlap because only a subset of individuals in each species are affected. Existing food web models provide some insight into how network structure might affect multitrophic dynamics. Increased average node degree of a network (more generalist species) enhances its structural robustness to secondary extinctions [53] and can stabilize network dynamics [54, 55]. Even a few species with increased degree can alter network dynamics because generalists often serve as keystone species or link subnetworks [56]. Decreased interaction strength also promotes network persistence and stability by dampening consumer-resource oscillations [57, 58]. Therefore among-individual trait variation should promote persistence and reduce oscillations by changing network connectance and interaction strengths. As with Jensen’s inequality, this stabilizing effect is independent of trait heritability.
Box 2. Niche Complementarity.
Niche complementarity is the tendency for phenotypically divergent individuals (or species [90]) to compete less strongly. Models of intraspecific competition typically assume that population growth declines as a function of the total population density, N:
| (eq. 1) |
The function f (N) represents density-dependent population growth, such as:
| (eq. 2) |
where λ is the population’s intrinsic growth rate, a measures the impact of density on individual fitness, and b reflects the type of competition (e.g., scramble versus contest [41]). The important point is that as total population size N increases, the denominator overwhelms the numerator and population growth approaches zero.
This approach presumes that individuals compete equally with all members of the population. However, in variable populations competition may be strongest between phenotypically similar individuals [15, 31], and weak or absent between phenotypically divergent individuals. For instance, imagine two predator phenotypes that each specialize on a different prey species. Competition between the predator phenotypes that eat species A will compete with each other and will not compete with the predator phenotype that exclusively eats species B. Thus, the fitness of an individual with phenotype z depends not on the total population size, but only on the abundance of competitively relevant phenotypes. This frequency-dependence can be modeled by replacing total population density N in (eq. 2) with an ‘effective population density’ experienced by each phenotype z:
| (eq. 3) |
where ρ(z′) denotes the density of an alternative phenotype z′. This equation considers the density of every other phenotype (ρ(z′)), weighted by its competitive effect on the focal phenotype z. This competitive effect, α(z, z′), is the per capita decrease in population growth of phenotype z per individual of phenotype z′, which is a declining function of the phenotypic difference between individuals:
| (eq. 4) |
where determines how quickly competition declines with phenotypic divergence. All else being equal, a population with greater phenotypic variance will experience weaker competition (diet specialization [low ] has a similar effect). Consequently, a variable population reaches a higher carrying capacity and might be more stable than a homogenous one [31].
Niche complementarity is tightly tied to the changes in population niche width (degree) described in Mechanism 2. The model outlined above implicitly assumes that a more phenotypically variable population uses a wider range of resources, thereby reducing niche overlap among individuals. We therefore do not consider niche complementarity to be a distinct mechanism in its own right.
Some models of niche complementarity also have elements that invoke other mechanisms. Niche complementarity has aspects that can be described as a portfolio effect. Competition generates positive covariances among phenotype densities over time, because all phenotypes grow slower as density increases. By mitigating competition among phenotypes, niche complementarity reduces this density-dependence covariance in abundance, thus reducing fluctuations in overall population density (see eq. 2 in the main text). Finally, niche complementarity sets the stage for frequency-dependent competition that can drive eco-evolutionary feedbacks, particularly in the form of disruptive selection that can maintain or inflate trait variance through time.
Mechanism 3: Portfolio effect
Intraspecific trait variation can protect populations from extreme temporal fluctuations in population density. A population’s density (N) is the sum of the densities of its constituent phenotypes (Ni). Fluctuations in population density (across time or space) depends on fluctuations in phenotype densities [59]:
| (eq. 2) |
where pi is the frequency of phenotype i, is the temporal or spatial variance in its density, and σNi,Nj is the covariance between phenotype densities.When phenotype densities exhibit negative covariances, the second term of (eq. 2) is negative and the whole population exhibits less density variation than individual phenotypes. This portfolio effect has been implicated in the long-term sustainability of the Alaskan Sockeye salmon fishery which depends on hundreds of discrete salmon runs [60]. Such negative covariances can have many sources, including intraspecific competition among individuals with diverse diets. When individuals compete more strongly with phenotypically similar conspecifics (Box 2), rare phenotypes are more fit than common ones. Doebeli [31] showed that this frequency dependence generates negative covariance between densities of asexual phenotypes, stabilizing total population size even when each phenotype exhibited oscillatory dynamics with its respective prey. Note that negative covariance among genotype abundances implies fluctuating genotype frequencies (evolution). However, the ecological effect can arise primarily via stabilized total population size rather than via trait evolution (see Mechanism 5).
Mechanism 4: Phenotypic subsidy
Various population genetic processes allow reproduction by each phenotype to increase the abundance of (‘subsidize’) other phenotypes within the same population, thereby altering interactions with other species. Offspring are often genetically different from their parents, owing to mutation, Mendelian segregation of alleles, independent assortment, or recombination among loci. Even clonal offspring can differ from their parents due to phenotypic plasticity [61–63]. This parent-offspring difference leads to a ‘phenotypic subsidy’, which we define as the increased abundance of one phenotype owing to reproduction (or plasticity) by a different phenotype, which therefore produces fewer offspring of its own type. This subsidy is best illustrated by contrasting the dynamics of phenotypes produced via asexual versus sexual reproduction. In a strictly asexual system, the change in density of phenotype i depends only on its own abundance Ni, fecundity bi and death rate di:
| (eq. 3) |
However in a sexual organism, the change in each phenotype’s density is partially decoupled from its own fitness because of subsidies (see Box 3 for a detailed example). Consequently, the growth rate of each phenotype i depends on the fecundity, density, and the subsidy rates of all j phenotypes in the population to phenotype i:
| (eq. 4) |
Box 3. A simple example of phenotypic subsidy.
In a panmictic population with three Mendelian genotypes (AA, Aa, aa), reproduction by heterozygotes can produce homozygote offspring, and vice versa. This subsidy means that changes in each phenotype’s abundance depends on the fitness (and relative abundance) of every phenotype. To illustrate the potential ecological impact of these phenotypic subsidies, we can contrast asexual versus sexual reproduction in a polymorphic consumer population. Let P1, P2, and P3 denote the density of three consumer phenotypes which, respectively, consume resource species R1, R2, and R3. Assuming Lotka-Volterra predator-prey dynamics, the abundance of each clonal consumer phenotype (Pi) and its resource (Ri) can be modeled as:
| (eq. 1a) |
| (eq. 1b) |
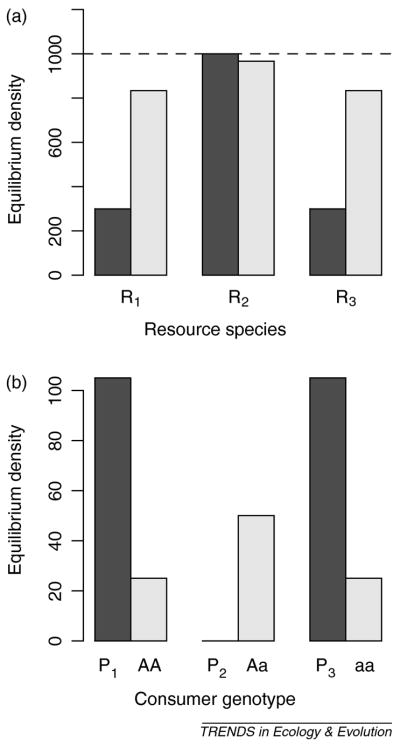
where ai is the attack rate of predator i on resource i; bi is the fecundity of the predator for each prey eaten; di is the predator per-capita death rate; ri is the intrinsic growth rate of resource i; and Ki is the carrying capacity of the resource. Each consumer-resource pair converges on its own independent equilibrium. The dark bars in Fig. I represent the equilibrium densities of resource and consumer species with asexual reproduction in a numerical example in which P2 is a much less efficient forager than P1 or P3 (P2 goes extinct).
Sexual reproduction can be incorporated using some simple Mendelian rules, by assuming an equal sex ratio and random mating. For example, consider the birth rate of aa individuals. There are female parents with genotype aa but only a fraction Saa→aa of their offspring will be aa:
| (eq. 2) |
where paa and pAa are genotype frequencies. Equation 2 takes into account the probability an aa female mates with each possible male genotype. It also accounts for the fraction of aa offspring that result from each possible pair of parents (1 from aa mating with aa; 0.5 from aa mating with Aa; 0 from aa mating with AA). Similarly, and SAA→aa = 0. The total birth rate of genotype aa individuals (Baa) can then be calculated, summing across all possible female genotypes, accounting for their densities, fecundities (fi =2aibiRi), and the subsidy fractions Sj→i:
| (eq. 3) |
This birth rate is substituted in for the birth term aibiPiRi in (eq. 1a).
Incorporating sexual reproduction dramatically alters the ratio of phenotypes and total population sizes, as illustrated in Fig. I (light bars). Because the homozygotes are equally fit and abundant in this numerical example, Mendelian segregation dictates a 1:2:1 ratio of predator phenotypes: random mating between homozygotes subsidizes the abundance of unfit heterozygotes. This genetic constraint on phenotype ratios is a substantial deviation from the P1:0:P3 ratio seen in the asexual model (dark bars, Fig. I b). The higher abundance of unfit heterozygotes places a substantial fitness load on the consumer population, whose total density is less than half that observed for asexual consumers. Correspondingly, resources abundances are on average both higher and more even.
Many other population genetic processes can generate ecologically relevant phenotypic subsidies. (1) Quantitative phenotypic traits are typically the result of additive effects of many loci of small effect. Segregation and recombination of these genes determine the trait distribution (often normally distributed) which might not match the ecologically-determined optimal trait distribution [31]. (2) Sexual reproduction is a form of subsidy because the production of new males is determined by female fecundity. When males and females consume different prey [1], the males’ prey might contribute little to the species’ abundance because male abundance is determined by the availability of the females’ prey.
Figure 1.
Equilibrium abundance of three consumer genotypes (B: asexual, dark bars; sexual, light bars) and their respective three prey resources (A). In (A) a horizontal line indicates the carrying capacity of all three resources (K = 1000). All parameters are identical across consumer-resource pairs (r = 1.5, a = 0.01, b = 0.05, d = 0.15) except that P2 (sexual genotype Aa) has attack rate a = 0.001.
The subsidy Sji is the fraction of phenotype j’s offspring contributed to phenotype i. This fraction depends on the mating system and genetic mechanism of the subsidy.
By coupling population dynamics of different phenotypes, a phenotypic subsidy can alter the equilibria and population dynamics of a species, as well as that of its resources and predators (Box 3). Phenotypic subsidies can sustain a given phenotype above the ecological equilibrium density it would experience in isolation, potentially preventing extinction of ecologically unfit forms. Conversely, a phenotype that would otherwise persist might go extinct by subsidizing unfit (‘sink’) phenotypes. Aside from influencing phenotypic variance, these subsidies can prevent populations from reaching ecologically determined equilibrium densities or trait distributions [22]. The resulting mismatch between ecologically determined equilibria and genetically permissible states can have cascading effects across other species (Box 3). For instance, if subsidies prevent extinction of an unfit phenotype, that phenotype’s resources (and predators) would reach a lower (higher) abundance than would be the case without the subsidy. The coupling of genetically differentiated forms via sexual reproduction or mutation introduces a negative feedback akin to self-limitation, stabilizing total population size by reducing the incidence and amplitude of fluctuations [64]. All else being equal, these population and community effects will likely be stronger with greater phenotypic diversity and stronger reproductive coupling (e.g., in sexual populations).
Few models specifically consider how reproductive subsidies affect community dynamics [31, 65]. However, we can extract some useful principles from spatial ecology models. The tendency for segregation and recombination to recreate otherwise moribund phenotypes resembles the ‘rescue effect’, in which immigration reduces the extinction risk of inherently unstable populations [66]. Models of patch dynamics can thus be reinterpreted to determine effects of subsidies: consumers in different patches can be redefined as different phenotypes, and dispersal between patches redefined as mutation. This reinterpretation suggests that phenotypic subsidies are not always stabilizing: dispersal (mutation) between predators in distinct patches (genotypes) often generates oscillatory or chaotic dynamics [67].
Mechanism 5: Adaptive eco-evolutionary dynamics
Heritable intraspecific variation enables adaptive evolution in response to ecological interactions. There is growing realization that evolution can occur on timescales commensurate with population dynamics [68]. Selection mediated by ecological interactions can change trait means, which in turn alters ecological interactions [69, 70]. Feedbacks between ecological processes and trait evolution (‘eco-evolutionary dynamics’) are of intense current interest [71, 72], though they have been recognized for decades (e.g., ecological character displacement [73]). Because most traits exhibit at least some heritable variation [19, 20], the most biologically relevant question is not whether eco-evolutionary feedbacks occur, but (1) how strongly does trait evolution affect ecology, and (2) how does the magnitude of additive genetic variation alter eco-evolutionary effects? Heritability determines evolutionary rates, and thus the relative speed of evolutionary and ecological dynamics. For instance, in a model of interspecific competition for multiple essential resources, high heritability allows rapid evolutionary character convergence thereby facilitating coexistence. However, low heritability slows evolution sufficiently that ecological dynamics dominate and exclusion ensues [74]. Trait variation also allows adaptation to changing environments. Greater additive genetic variance decreases extinction risk because more variable populations adapt faster and thus more closely track changing trait optima [75]. This faster adaptation to environmental change also alters the outcome of interspecfic competition [76]. A relatively underexplored question is whether similar feedbacks between ecology and trait distributions can arise from adaptive phenotypic plasticity instead of evolution.
Eco-evolutionary dynamics have been convincingly demonstrated in numerous controlled experiments [69, 72] and in theoretical models [77]. Fewer studies have specifically evaluated how genetic variance affects these dynamics [30, 39], and most fail to distinguish between eco-evolutionary versus non-evolutionary effects of variation (Mechanisms 1–3). Doing so requires factorial manipulation of both phenotypic variance and heritability, which is difficult. Theory can thus play a crucial role in partitioning effects arising from different components of phenotypic variation. For example, Schreiber et al. (unpublished) studied an apparent competition model (one predator, two prey) with genetic or environmental trait variation among predators that influenced attack rate on the two prey. Total phenotypic variance determined equilibrium population densities, whereas heritability affected the rate of transient dynamics and stability around equilibria. Hence, both genetic and environmental trait variance have appreciable and non-equivalent effects.
Mechanism 6: Trait sampling
In small populations, ecological and evolutionary sampling of individual variation can change trait means and variances thereby altering population dynamics. Demographic variance [32, 33, 78, 79] is the variance in a population’s vital rates arising from two sources: (1) stochastic sampling of individuals with different demographic rates, and (2) conventional demographic stochasticity arising from the discrete probabilistic nature of births and deaths. For instance, mean clutch size can fluctuate through time because the individuals who (randomly) succeed in reproducing in different years happen to have different expected fecundity, or because clutch size varies randomly among individuals with equal expected fecundity. Demographic variance (and trait sampling in general) will be most pronounced in small populations, and can affect long-term population dynamics [32, 33, 78], population persistence [79], and species coexistence [37].
An analogous process could drive maladaptive eco-evolutionary feedbacks. Genetic drift is the random change in allele frequencies resulting from stochastic sampling of alleles from the preceding generation (independent of demographic stochasticity). By altering trait means and variances, genetic drift might alter demographic rates or interspecific interactions. In small populations, drift can cause substantial departures from phenotypic optima thereby reducing mean fitness. Consequently, population density declines still further and drift becomes even more pronounced. This feedback might generate an ‘extinction vortex’ [80, 81] with cascading effects on an entire community.
Combining mechanisms
The six mechanisms outlined above imply that trait variation alters ecological dynamics when: (1) ecological parameters are non-linear functions of the trait(s); (2) trait variance affects niche width or network topology; (3) phenotypes exhibit asynchronous fluctuations in density over time; (4) phenotypic subsidies decouple genetic and ecological equilibria; (5) ecology both drives and responds to trait evolution, and (6) trait variation is stochastically sampled in small populations. Additional mechanisms might still be uncovered.
For trait variation to be ecologically unimportant, all of these mechanisms must be jointly ruled out or have weak effects. As each highlighted mechanism is quite general, intraspecific trait variation can have nearly ubiquitous ecological effects. These effects can cascade throughout a community of interacting species and substantially alter equilibrium densities [46] and change conditions for population stability or multispecies coexistence [31, 37].
The six mechanisms are not mutually exclusive. Heritable trait variation in both a predator and its prey can influence population dynamics simultaneously via Jensen’s inequality, a portfolio effect, phenotypic subsidy, and eco-evolutionary feedbacks [31]. However, little is known about the relative frequency or strengths of these different pathways. To complicate matters, the mechanisms listed above might interact. Fluctuations in one species’ population size due to demographic stochasticity can alter another species’ growth via Jensen’s Inequality. Interactions can also arise from eco-evolutionary change in trait variances, which alters the potential for other variation-based effects. Finally, intra- and inter-specific diversity could have parallel and perhaps interacting effects on population dynamics (Box 4).
Box 4. Intra- versus interspecific diversity.
The influence of intraspecific variation on population dynamics has a well studied analogue: multiple species within a community are sometimes aggregated into guilds to define a particular ecosystem service or function [91]. The mean total function (e.g. total biomass) or stability (temporal variability in function) of the system can change as species are added to the community: typically both measures improve but saturate as richness increases [91]. This community effect of species richness is commonly attributed to increased functional diversity as species are added, increasing resource availability or resistance to consumers or environmental conditions [91]. Similar effects can arise from functional diversity among individuals within populations, increasing equilibrium density in response to increasing variation [45, 46, 92].
Furthermore, when environmental conditions fluctuate over time, communities (or populations) possessing a diverse set of functional traits have a higher probability of including species (genotypes) that persist through particular environmental conditions, thereby stabilizing function [91]. For instance, negative covariance among edible and less-edible phytoplankton morphotypes dampens temporal fluctuations in total biomass during summer when competition is most intense [93]. Similar results have been found for competing phenotypes, where variation in a quantitative trait generated strong frequency-dependent competition and negative covariance among phenotypes due to niche complementarity [31]. This is an example of the portfolio effect, which can arise both for species diversity and intraspecific variation.
Our limited understanding of how genetic variation is maintained under stabilizing selection in real populations is fundamentally similar to the classic question in community ecology - Why do many species persist under what appears to be strong selection for a single or few species? Potential answers to these two questions have many similarities including temporal and/or spatial variation in selection, mutation-drift balance (colonization-extinction balance in community ecology), and negative frequency-dependence. Conversely, the two can differ in important ways. For example phenotypic subsidies due to interbreeding are possible within species but not between them.
Future directions
Given the general, multifaceted, and potentially large ecological effects of trait variation, we need to revisit classic ecological models and ask how the magnitude and heritability of trait variation alters previous conclusions. In doing so, we should evaluate (1) the magnitude of ecological effects of trait variance; (2) the relative contributions of the various mechanisms described above; and the consequences of (3) heritable, environmental, or stochastic variation, (4) the genetic architecture (one-locus, quantitative genetic, or explicit multi-locus models); (5) asexual versus sexual reproduction; and (6) variation in multiple traits. Addressing these questions will require new approaches for fusing ecological models with population and quantitative genetics. For discrete phenotypic variation, one could separately model the density of each phenotype within a species (coupled by reproduction), or track overall density and allele frequencies in each species. For continuous traits, one could describe each species with three equations that track changes in population density, trait mean, and trait (co)variance. However, modeling the evolution of trait (co)variances is a continuing challenge in quantitative genetics. Computationally intensive individual-based models with explicit multilocus genetics offer one solution.
Intraspecific variation has been particularly lacking in models of species-rich communities. Some food web models incorporate changing topologies via trait evolution or behavioral prey switching [61, 82], but even these typically assume homogenous conspecifics at each time step. Incorporating trait variation into food webs might change their topology (see Mechanism 2) and dynamics [83]. Such effects presumably depend on where in a food web one adds trait variance. Unfortunately, empirical studies of diet variation typically focus on single species, giving little guidance as to how variance is distributed across multiple interacting species [84].
One limitation for theoreticians is the scarcity of empirical data regarding the causes, patterns, and consequences of within-population ecological variance [3]. The majority of empirical studies of ecological variation among individuals simply test the null hypothesis that individuals are identical, but do not quantify the magnitude of variation or determine its mechanistic or genetic basis [3]. To predict the consequences of trait variation, ecologists need to (1) identify the genetic and phenotypic causes of ecological variation, and (2) quantify ecological variation across multiple species, trophic levels, or entire communities. Ideally, such information could guide theoreticians in developing biologically realistic models that can be parameterized with empirical data to yield testable quantitative predictions.
Most importantly, we need more experiments that test the ecological effects of trait variation within species. The typical approach is to manipulate trait (or genetic) variance within populations, and measure the resulting difference in population or community dynamics. A major challenge for such studies is to clearly distinguish between the direct and indirect (evolutionary) effects of trait variance outlined in this paper. This will require careful experimental designs that either manipulate phenotypic variance among genetically identical individuals (thereby preventing evolution), or that compare experimental replicates in which evolution is allowed versus inhibited (e.g., [85]). Clearly, many of the mechanisms outlined above will be best tested by studies that fuse quantitative models with empirical data. For instance, the ecological effects of Jensen’s Inequality are best studied by empirically estimating the non-linear relationships between variable traits and their functional consequences. Such studies will be a challenging and possibly unwelcome complication for ecologists. However, theory suggests that trait variation could have sufficiently large ecological effects that we cannot safely ignore intraspecific variation if we wish to understand the dynamics and conservation of populations, communities, and ecosystems.
Acknowledgments
This review was written as a part of the Ecology of Niche Variation Working Group at the National Institute for Mathematical and Biological Synthesis, sponsored by the National Science Foundation, the U.S. Department of Homeland Security, and the U.S. Department of Agriculture through NSF Award #EF-0832858, with additional support from The University of Tennessee, Knoxville. Work on the paper was also supported by the David and Lucile Packard Foundation (DIB). We thank all members of the working group for feedback. We thank Mark Vellend and Yuexin Jiang for comments on manuscript drafts.
GLOSSARY
- Eco-evolutionary dynamics
effects of ecological changes on evolutionary dynamics or the effects of evolutionary changes on ecological dynamics; feedbacks arise when a loop links both directions of effect
- Node degree
number of predator and prey links for a (trophic) species represented within a food web. Degree distribution describes the mean and variance of node degree across all nodes in a network
- Genetic drift
change in allele frequencies owing to random sampling during reproduction
- especially strong in small
isolated populations
- Jensen’s inequality
mathematical rule stating that the mean of a concave (convex) function of a variable trait is smaller (larger) than the value obtained by applying the function to the mean of the variable
- Narrow-sense heritability
the proportion of total phenotypic variation attributed to additive genetic variation; proportional to the rate of short-term responses to selection
- Niche complementarity
the tendency for phenotypically divergent individuals (or species) to compete less strongly
- Phenotype
the outward expression of an individual’s genotype as affected by the environment
- may include size
morphology, behavior, physiology
- Portfolio effect
negative covariances between the dynamics of subunits leads to a smaller total variance through averaging; applied here to phenotypic frequencies within a population
- Quantitative genetics
evolutionary approach to predicting the evolution of continuous traits assuming the action of many genes of small phenotypic effect
- Phenotypic subsidy
a process by which the density of one phenotype is augmented by reproduction, behavioral switching, phenotypic plasticity, or migration by another phenotype
- Trait
any measurable feature of an individual organism, including phenotype as well as demographic parameters such as clutch size, growth, or longevity
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shine R. Ecological causes for the evolution of sexual dimorphism: a review of the evidence. Quart Rev Biol. 1989;64:419–461. doi: 10.1086/416458. [DOI] [PubMed] [Google Scholar]
- 2.Polis GA. Age structure component of niche width and intraspecific resource partitioning - can age-groups function as ecological species. Am Nat. 1984;123:541–564. [Google Scholar]
- 3.Bolnick DI, et al. The ecology of individuals: incidence and implications of individual specialization. Am Nat. 2003;161:1–28. doi: 10.1086/343878. [DOI] [PubMed] [Google Scholar]
- 4.Duffy MA. Ecological consequences of intraspecific variation in lake Daphnia. Freshw Biol. 2010;55:995–1004. [Google Scholar]
- 5.Ganz HH, Ebert D. Benefits of host genetic diversity for resistance to infection depend on parasite diversity. Ecology. 2010;91:1263–1268. doi: 10.1890/09-1243.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearl S, et al. Nongenetic individuality in the host-phage interaction. PLOS Biology. 2008;6:957–964. doi: 10.1371/journal.pbio.0060120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson CK, et al. Prey choice and habitat use drive sea otter pathogen exposure in a resource-limited coastal system. Proc Nat Acad Sci. 2009;106:242–2247. doi: 10.1073/pnas.0806449106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer E, et al. Genetic variation in responses to a settlement cue and elevated temperature in the reef-building coral Acropora millepora. Mar Eco Prog Ser. 2009;392:81–92. [Google Scholar]
- 9.Lankau RA, Strauss SY. Mutual feedbacks maintain both genetic and species diversity in a plant community. Science. 2007;317:1561–1563. doi: 10.1126/science.1147455. [DOI] [PubMed] [Google Scholar]
- 10.Ortego J, et al. Egg production and individual genetic diversity in lesser kestrels. Molec Ecol. 2007;16:2383–2392. doi: 10.1111/j.1365-294X.2007.03322.x. [DOI] [PubMed] [Google Scholar]
- 11.Peacor SD, et al. Mechanisms of nonlethal predator effect on cohort size variation: ecological and evolutionary implications. Ecology. 2007;88:1536–1547. doi: 10.1890/06-1066. [DOI] [PubMed] [Google Scholar]
- 12.Estes JA, et al. Individual variation in prey selection by sea otters: patterns, causes, and implications. J Anim Ecol. 2003;72:144–155. [Google Scholar]
- 13.Tinker MT, et al. Food limitation leads to behavioral diversification and dietary specialization in sea otters. Proc Nat Acad Sci. 2008;105:560–565. doi: 10.1073/pnas.0709263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price T. Diet variation in a population of Darwin’s finches. Ecology. 1987;68:1015–1028. [Google Scholar]
- 15.Bolnick DI, Paull J. Diet similarity declines with morphological distance between conspecific individuals. Evol Ecol Res. 2009;11:1217–1233. [Google Scholar]
- 16.Afik D, Karasov WH. The trade-offs between digestion rate and efficiency in warblers and their ecological implications. Ecology. 1995;76:2247–2257. [Google Scholar]
- 17.Holbrook SJ, Schmitt RJ. Causes and consequences of dietary specialization in surfperches: patch choice and intraspecific competition. Ecology. 1992;73:402–412. [Google Scholar]
- 18.Lewis AC. Memory constraints and flower choice in Pieris rapae. Science. 1986;232:863–864. doi: 10.1126/science.232.4752.863. [DOI] [PubMed] [Google Scholar]
- 19.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Inc; 1998. [Google Scholar]
- 20.Roff DA. Evolutionary Quantitative Genetics. Chapman and Hall; 1997. [Google Scholar]
- 21.Ford EB. Ecological Genetics. Chapman and Hall; 1964. [Google Scholar]
- 22.Roughgarden J. Evolution of niche width. Am Nat. 1972;106:683–718. [Google Scholar]
- 23.Grant PR, Price TD. Population variation in continuously varying traits as an ecological genetics problem. Am Zool. 1981;21:795–811. [Google Scholar]
- 24.Hughes AR, et al. Ecological consequences of genetic diversity. Ecol Lett. 2008;11:609–623. doi: 10.1111/j.1461-0248.2008.01179.x. [DOI] [PubMed] [Google Scholar]
- 25.Agashe D. The stabilizing effect of intraspecific genetic variation on population dynamics in novel and ancestral habitats. Am Nat. 2009;174:255–267. doi: 10.1086/600085. [DOI] [PubMed] [Google Scholar]
- 26.Fridley JD, Grime JP. Community and ecosystem effects of intraspecific genetic diversity in grassland microcosms of varying species diversity. Ecology. 2010;8:2272–2283. doi: 10.1890/09-1240.1. [DOI] [PubMed] [Google Scholar]
- 27.Imura D, et al. Genetic variation can promote system persistence in an experimental host-parasitoid system. Pop Ecol. 2003;45:205–212. [Google Scholar]
- 28.Crutzinger GM, et al. Plant genotypic diversity predicts community structure and governs an ecosystem process. Science. 2006;313:966–968. doi: 10.1126/science.1128326. [DOI] [PubMed] [Google Scholar]
- 29.Johnson MTJ. Bottom-up effects of plant genotype on aphids, ants, and predators. Ecology. 2008;89:145–154. doi: 10.1890/07-0395.1. [DOI] [PubMed] [Google Scholar]
- 30.Becks L, et al. Reduction of adaptive genetic diversity radically alters eco-evolutionary community dynamics. Ecol Lett. 2010;13:989–997. doi: 10.1111/j.1461-0248.2010.01490.x. [DOI] [PubMed] [Google Scholar]
- 31.Doebeli M. Quantitative genetics and population dynamics. Evolution. 1996;50:532–546. doi: 10.1111/j.1558-5646.1996.tb03866.x. [DOI] [PubMed] [Google Scholar]
- 32.Fox GA, Kendall BE. Demographic stochasticity and the variance reduction effect. Ecology. 2002;83:1928–1934. [Google Scholar]
- 33.Vindenes Y, et al. Individual heterogeneity in vital parameters and demographic stochasticity. Amm Nat. 2008;171:455–467. doi: 10.1086/528965. [DOI] [PubMed] [Google Scholar]
- 34.Lloyd-Smith J, et al. Superseeding and the impact of individual variation on disease emergence. Nature. 2005;438:355–359. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lankau RA. Genetic variation promotes long-term coexistence of Brassica nigra and its competitors. Am Nat. 2009;174:E40–E53. doi: 10.1086/600083. [DOI] [PubMed] [Google Scholar]
- 36.Vellend M. The consequences of genetic diversity in competitive communities. Ecology. 2006;87:304–311. doi: 10.1890/05-0173. [DOI] [PubMed] [Google Scholar]
- 37.Lichstein JW, et al. Intraspecific variation and species coexistence. Am Nat. 2007;170:807–818. doi: 10.1086/522937. [DOI] [PubMed] [Google Scholar]
- 38.Okuyama T. Individual behavioral variation in predator-prey models. Ecol Res. 2008;23:665–671. [Google Scholar]
- 39.Saloniemi I. A coevolutionary predator-prey model with quantitative characters. Am Nat. 1993;141:880–896. doi: 10.1086/285514. [DOI] [PubMed] [Google Scholar]
- 40.Doebeli M. Genetic variation and the persistence of predator-prey interactions in the Nicholson-Bailey model. J Theor Biol. 1997;188:109–120. [Google Scholar]
- 41.Doebeli M, de Jong G. Genetic variability in sensitivity to population density affects the dynamics of simple ecological models. Theor Pop Biol. 1999;55:37–52. doi: 10.1006/tpbi.1998.1385. [DOI] [PubMed] [Google Scholar]
- 42.Lively CM. Effect of host genetic diversity on disease spread. Am Nat. 2010;175:E149–E152. doi: 10.1086/652430. [DOI] [PubMed] [Google Scholar]
- 43.West-Eberhard MJ. Developmental Plasticity and Evolution. Oxford University Press; 2003. [Google Scholar]
- 44.Doebeli M, Koella JC. Sex and population dynamics. Proc Roy Soc Lond Ser B. 1994;257:17–23. [Google Scholar]
- 45.Bürger R. A multilocus analysis of intraspecific competition and stabilizing selection on a quantitative trait. J Mat Biol. 2005;50:355–396. doi: 10.1007/s00285-004-0294-2. [DOI] [PubMed] [Google Scholar]
- 46.Kopp M, Hermisson J. The evolution of genetic architecture under frequency-dependent disruptive selection. Evolution. 2006;60:1537–1550. [PubMed] [Google Scholar]
- 47.Ruel JJ, Ayres MP. Jensen’s inequality predicts effects of environmental variation. TREE. 1999;14:361–366. doi: 10.1016/s0169-5347(99)01664-x. [DOI] [PubMed] [Google Scholar]
- 48.Sherratt TN, Macdougall AD. Some population consequences of variation in preference among individual predators. Biol J Linn Soc. 1995;55:93–107. [Google Scholar]
- 49.Kolokotrones T, et al. Curvature in metabolic scaling. Nature. 2010;464:753–756. doi: 10.1038/nature08920. [DOI] [PubMed] [Google Scholar]
- 50.Bolnick DI, et al. More generalized populations are also more heterogeneous: comparative support for the niche variation hypothesis. Proc Nat Acad Sci. 2007;104:10075–10079. doi: 10.1073/pnas.0703743104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams RJ, Martinez ND. Simple rules yield complex food webs. Nature. 2000;404:180–183. doi: 10.1038/35004572. [DOI] [PubMed] [Google Scholar]
- 52.Allesina S, et al. A general model for food web selection. Science. 2008;320:658–661. doi: 10.1126/science.1156269. [DOI] [PubMed] [Google Scholar]
- 53.Dunne JA, et al. Network structure and biodiversity loss in food webs: robustness increases with connectance. Ecol Lett. 2002;5:558–567. [Google Scholar]
- 54.Otto SB, et al. Allometric degree distributions facilitate food-web stability. Nature. 2007;450:1226–1229. doi: 10.1038/nature06359. [DOI] [PubMed] [Google Scholar]
- 55.Gross T, et al. Generalized models reveal stabilizing factors in food webs. Science. 2009;325:747–750. doi: 10.1126/science.1173536. [DOI] [PubMed] [Google Scholar]
- 56.Gonzalez AMM, et al. Centrality measures and the importance of generalist species in pollination networks. Ecol Complex. 2010;7:36–43. [Google Scholar]
- 57.McCann K, et al. Weak trophic interactions and the balance of nature. Nature. 1998;395:794–798. [Google Scholar]
- 58.Emerson MC, Raffaelli D. Predator-prey body size, interaction strength and the stability of a real food web. J Anim Ecol. 2004;73:399–409. [Google Scholar]
- 59.Tilman D, et al. Diversity-stability relationships: statistical inevitability or ecological consequence? American Naturalist. 1998;151:277–282. doi: 10.1086/286118. [DOI] [PubMed] [Google Scholar]
- 60.Schindler DE, et al. Population diversity and the portfolio effect in an exploited species. Nature. 2010;465:609–612. doi: 10.1038/nature09060. [DOI] [PubMed] [Google Scholar]
- 61.Abrams PA. Implications of flexible foraging for interspecific interactions: lessons from simple models. Func Ecol. 2010;24:7–17. [Google Scholar]
- 62.Beckerman AP, et al. Adaptive foragers and community ecology: linking individuals to communities to ecosystems. Func Ecol. 2010;24:1–6. [Google Scholar]
- 63.Schreiber SJ, et al. Dancing between the devil and the deep blue sea: the stabilizing effect of enemy-free sinks and victimless sinks. Oikos. 2006;113:67–81. [Google Scholar]
- 64.Holt RD. Ecology at the mesoscale: the influence of regional processes on local communities. In: Ricklefs RE, Schluter D, editors. Species Diversity in Ecological Communities: Historical and Geographical Perspectives. University of Chicago Press; 1993. pp. 7–88. [Google Scholar]
- 65.Ruxton GD. Population models with sexual reproduction show a reduced propensity to exhibit chaos. J Theor Biol. 1995;175:595–601. [Google Scholar]
- 66.Brown JH, Kodric-Brown A. Turnover rates in insular biogeography: effect of immigration on extinction. Ecology. 1977;58:445–449. [Google Scholar]
- 67.Jansen VAA. The dynamics of two diffusively coupled predator-prey populations. Theor Pop Biol. 2001;59:119–131. doi: 10.1006/tpbi.2000.1506. [DOI] [PubMed] [Google Scholar]
- 68.Hendry AP, Kinnison MT. The pace of modern life: measuring rates of contemporary microevolution. Evolution. 1999;53:1637–1653. doi: 10.1111/j.1558-5646.1999.tb04550.x. [DOI] [PubMed] [Google Scholar]
- 69.Yoshida T, et al. Rapid evolution drives ecological dynamics in a predator-prey system. Nature. 2003;424:303–306. doi: 10.1038/nature01767. [DOI] [PubMed] [Google Scholar]
- 70.terHorst CP, et al. Evolution of prey in ecological time reduces the effect size of predators in experimental microcosms. Ecology. 2010;9:629–636. doi: 10.1890/09-1481.1. [DOI] [PubMed] [Google Scholar]
- 71.Pelletier F, et al. Eco-evolutionary dynamics. Phil Trans Roy Soc Lond Ser B. 2009;364:1483–1489. doi: 10.1098/rstb.2009.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Post DM, Palkovacs EP. Eco-evolutionary feedbacks in community and ecosystem ecology: interactions between the ecological theater and the evolutionary play. Phil Trans Roy Soc Lond Ser B. 2009;364:1629–1640. doi: 10.1098/rstb.2009.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grant PR. Convergent and divergent character displacement. Biol J Linn Soc. 1972;4:39–69. [Google Scholar]
- 74.Fox J, Vasseur DA. Character convergence under competition for essential resources. Am Nat. 2008;172:667–680. doi: 10.1086/591689. [DOI] [PubMed] [Google Scholar]
- 75.Bürger R. Evolution of genetic variability and the advantage of sex and recombination in changing environments. Genetics. 1999;153:1055–1069. doi: 10.1093/genetics/153.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Urban MC, et al. The evolutionary ecology of metacommunities. TREE. 2008;23:311–317. doi: 10.1016/j.tree.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 77.Jones LE, et al. Rapid contemporary evolution and clonal food web dynamics. Phil Trans Roy Soc Lond Ser B. 2009;364:1579–1591. doi: 10.1098/rstb.2009.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kendall BE, Fox GA. Unstructured individual variation and demographic stochasticity. Conserv Biol. 2003;17:1170–1172. doi: 10.1046/j.1523-1739.2002.00036.x. [DOI] [PubMed] [Google Scholar]
- 79.Lande R, et al. Stochastic Population Dynamics in Ecology and Conservation. Oxford University Press; 2003. [Google Scholar]
- 80.Gilpin ME, Soulé ME. Minimum Viable Populations: Processes of Species Extinction. In: Soulé ME, editor. Conservation Biology: The Science of Scarcity and Diversity. Sinauer; 1986. pp. 19–34. [Google Scholar]
- 81.Gabriel W, et al. Muller’s ratchet and mutational meltdowns. Evolution. 1993;47:1744–1757. doi: 10.1111/j.1558-5646.1993.tb01266.x. [DOI] [PubMed] [Google Scholar]
- 82.Ingram T, et al. Niche evolution, trophic structure, and species turnover in model food webs. Am Nat. 2009;174:56–67. doi: 10.1086/599301. [DOI] [PubMed] [Google Scholar]
- 83.Rudolf VHW, Lafferty KD. Stage structure alters how complexity afects stability of ecological networks. Ecol Lett. doi: 10.1111/j.1461-0248.2010.01558.x. (In press) [DOI] [PubMed] [Google Scholar]
- 84.Svanbäck R, et al. Intra-specific competition drives multiple species trophic polymorphism in fish communities. Oikos. 2008;117:114–124. [Google Scholar]
- 85.Rice WR. Sexually antagonistic male adaptation triggered by experimental arrest of female evolution. Nature. 1996;381:232–234. doi: 10.1038/381232a0. [DOI] [PubMed] [Google Scholar]
- 86.Kendall BE, Wittmann ME. A stochastic model for annual reproductive success. Am Nat. 2010;175:461–468. doi: 10.1086/650724. [DOI] [PubMed] [Google Scholar]
- 87.Petchey OL, et al. Size, foraging and food web structure. Proc Nat Acad Sci. 2008;105:4191–4196. doi: 10.1073/pnas.0710672105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rudolf VHW. Consequences of stage-structured predators: Cannibalism, behavioral effects and trophic cascades. Ecology. 2007;87:2991–3003. doi: 10.1890/07-0179.1. [DOI] [PubMed] [Google Scholar]
- 89.Duncan RP, et al. Testing the metabolic theory of ecology: allometric scaling exponents in mammals. Ecology. 2007;88:324–333. doi: 10.1890/0012-9658(2007)88[324:ttmtoe]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 90.Hughes AR, et al. Morphological and physiological variation among seagrass (Zostera marina) genotypes. Oecologia. 2009;159:725–733. doi: 10.1007/s00442-008-1251-3. [DOI] [PubMed] [Google Scholar]
- 91.Hooper DU, et al. Effects of biodiversity on ecosystem function: a consensus of current knowledge. Ecol Monogr. 2005;75:3–35. [Google Scholar]
- 92.Vellend M. Conceptual synthesis in community ecology. Quart Rev Biol. 2010;85:183–206. doi: 10.1086/652373. [DOI] [PubMed] [Google Scholar]
- 93.Vasseur DA, et al. A seasonal alternation of coherent and compensatory dynamics occurs in phytoplankton. Oikos. 2005;110:507–514. [Google Scholar]
- 94.Pachepsky E, et al. Consequences of intraspecific variation for the structure and function of ecological communities Part 2: linking diversity and function. Ecol Model. 2007;207:277–285. [Google Scholar]
- 95.Jones EI. Eco-evolutionary dynamics of mutualists and exploiters. Am Nat. 2009;174:780–794. doi: 10.1086/647971. [DOI] [PubMed] [Google Scholar]
- 96.Fox GA. Extinction risk of heterogeneous populations. Ecology. 2005;86:1191–1198. [Google Scholar]