Abstract
The University of California Santa Cruz (UCSC) Table Browser (http://genome.ucsc.edu/cgi-bin/hgText) provides text-based access to a large collection of genome assemblies and annotation data stored in the Genome Browser Database. A flexible alternative to the graphical-based Genome Browser, this tool offers an enhanced level of query support that includes restrictions based on field values, free-form SQL queries and combined queries on multiple tables. Output can be filtered to restrict the fields and lines returned, and may be organized into one of several formats, including a simple tab- delimited file that can be loaded into a spreadsheet or database as well as advanced formats that may be uploaded into the Genome Browser as custom annotation tracks. The Table Browser User’s Guide located on the UCSC website provides instructions and detailed examples for constructing queries and configuring output.
INTRODUCTION
The UCSC Table Browser data retrieval tool is built on top of the Genome Browser Database, a set of MySQL relational databases that each store sequence and annotation data for one genome assembly (1). Tables within the databases may be differentiated by whether the data are based on genomic start–stop coordinates or are independent of position.
Positional tables contain data associated with specific locations in the genome, such as mRNA alignments, gene predictions, cross-species alignments and various other annotations. Each of the annotation ‘tracks’ displayed in the graphical Genome Browser is based on one or more positional tables. Data associated with custom annotation tracks active within the user’s Table Browser session are also available as positional tables.
Non-positional tables contain data not tied to genomic location, for example a table that correlates a Genethon marker name with a Marshfield marker name. Some non-positional tables relate internal numeric mRNA IDs to extended information such as author, tissue or keyword. Other ‘meta’ tables contain information about the structure of the database itself or describe external files containing sequence data.
Because of the large size of the data set stored in each database, particular attention has been paid to maintaining adequate interactive performance. The databases contain optimizations to support range-based queries from the Table Browser and Genome Browser. Smaller tables are indexed on a few critical fields and the data are presorted prior to loading into the database. With larger tables, the data are separated by chromosome into smaller tables, and a binning scheme is implemented on the larger chromosome tables.
The document http://genome.ucsc.edu/goldenPath/gbdDescriptions.html contains a detailed description of the database tables and fields, which are dumped weekly into downloadable tab-delimited files.
In addition to the inclusion of the latest human and mouse assemblies, the Genome Browser Database has expanded in the past year to include rat, worm and a collection of species targeted by the NISC Comparative Sequencing Program (2), with plans to add support for several additional genomes in the coming year.
Recently, the UCSC Genome Bioinformatics group has placed considerable emphasis on comparative genome analysis. The group has been active in the analysis of evolutionary conservation and divergence among species (3,4), phylogenetic analysis of rates of substitution (5) and multiple species alignments. This research has resulted in the addition of several new types of annotation data to the Genome Browser Database.
The axtChain program written by Jim Kent produces chained BLASTZ alignments between two species (6). This alignment tool uses a gap scoring system that allows longer gaps than traditional affine gap scoring systems and can also tolerate gaps in both species simultaneously. Further processing of the chained alignments with the chainNet program outputs an alignment net that shows the best chain for every part of the genome.
UCSC has also been collaborating closely with the Penn State University Bioinformatics Group to produce three-way multiple species alignments using Webb Miller’s multiz program, which takes BLASTZ and axtBest alignments as input (7,8).
Many research scientists are familiar with the UCSC Genome Browser (9), the graphical interface to the Genome Browser Database that displays requested portions of genome assemblies together with a series of aligned annotation ‘tracks’. Despite its ease of use, situations exist in which a graphical browser may not be the optimal tool for working with genomic data. A user might wish to view the raw data or examine the relationships between the tables underlying the browser. It is often desirable to filter the display output with greater restrictions than are offered by the Genome Browser, or to output the data in a text-based format that can be imported into other programs.
The UCSC Table Browser—which may be accessed directly at http://genome.ucsc.edu/cgi-bin/hgText or through the Tables link on the UCSC Genome Bioinformatics home page (http://genome.ucsc.edu)—provides a powerful and flexible alternative for querying and manipulating the annotation tables within the Genome Browser Database. Using Table Browser form-based or free-form queries, one may quickly and easily extract subsets of the database, in many cases eliminating the need to set up a local copy of the MySQL database. By configuring the tool’s output options, the user can generate a custom annotation track that may be automatically added to the graphical browser session, or create a file in one of several output formats that can be used as input into other programs. The Table Browser can also display basic statistics calculated over a selected subset of data.
BASIC DATA QUERIES
In its most basic form, the Table Browser can be used to retrieve a specific subset of records from a table in a selected genome assembly. The user specifies a position of interest within the assembly (or the keyword ‘genome’ to access data from the entire assembly), selects a table, and then chooses the ‘Get all fields’ option. The Table Browser displays the query results in a tab-delimited text format that can be easily downloaded and imported into text editors, spreadsheets and other databases, or may be further processed by the user’s own scripts.
For example, a user who is examining alternative splicing in the human genome might be interested in downloading the indices of all mRNA sequences that align to a chromosomal region containing a particular gene. One would set the Table Browser to the gene position, select the chrN_mrna positional table, and then click the Get all fields button. This query produces a tab-delimited list of names and positions of mRNAs that align to the specified location.
ADVANCED QUERIES
Although basic data retrieval is useful, the real power of the Table Browser lies in the ability to filter and refine queries, intersect query results from different tables and configure the resulting output. These options may be accessed through the Table Browser’s set of advanced query features.
The available query formats and output options vary by table. Many apply only to tables in which the data is position-oriented, thus preserving the database distinction between positional and non-positional tables. Position-based tables may be further differentiated by the types of data they characterize. For example, alignment tables describe a block structure for each element, but other tables may describe only a starting and ending position. Still others may specify translation start and end positions as well as transcription start and end points.
Output format configuration
The Table Browser offers a variety of data configuration formats. In addition to the tab-separated output provided by basic queries, a user can choose from several file formats that may be uploaded as aligned custom annotation tracks in the Genome Browser: Gene Transfer Format (GTF), Browser Extensible Data (BED) and Custom Track format.
The custom annotation tracks generated by the Table Browser are a valuable research tool, offering the ability to view the results of a complex customized query in alignment with the standard annotation tracks in the Genome Browser. Because custom annotations are temporary, they persist for only 8 h after they were last accessed. The tracks never become part of the Genome Browser Database, and therefore are accessible only on the machine from which they were uploaded.
The Table Browser FASTA output option allows the user to format and retrieve DNA sequence for a selected region of the genome, similar to the Get DNA utility in the Genome Browser. Other output options enable the user to generate a list of hyperlinks to the Genome Browser corresponding to the locations of features identified by a query on a positional table, or display statistical information about the query, including the number and size of matches and type information about table fields.
Filtering
The most flexible feature in the Table Browser is its filtering mechanism. The form-based filter provides a straightforward interface for configuring simple SQL-based queries of the data. By default, a Table Browser search retrieves all records for a specified coordinate range or position. Using the filter, the user may set constraints on the values of some or all of the fields within a table to restrict the set of records retrieved from the query range.
The text fields within the filter support wildcard pattern matching and multiple entries. If any word or pattern within the text field matches the value, then the record meets the constraint on that field. Numeric field comparisons support the operators <, >, and != (not equal) and allow comparisons with ranges of numbers.
To satisfy the needs of advanced users who find the form-based filtering options to be insufficient, the Table Browser also supports free-form queries allowing more complex constraints, typically to relate two or more fields within the selected table. These queries—which use SQL ‘where’ clause syntax—can combine simple constraints with AND, OR and NOT, using parentheses as needed for clarity. A basic free-form constraint consists of a field name (or an arithmetic expression of numeric field names), a comparison operator and a value.
For example, when searching for gene models in which a promoter region may be present, the simple free-form query (txStart != cdsStart) on the refGene table will produce a list of genes that have the expected 5′ untranslated region (UTR) upstream sequence. Note that if the strand is negative, this will search for cases of 3′ UTR downstream sequence.
In a more complex version of the previous query, (txStart != cdsStart) AND (txEnd != cdsEnd) AND (exonCount = 1) will return a list of single exon genes with both 5′ and 3′ flanking UTRs.
Multiple table comparisons
At times one may wish to compare the data between two tables to determine whether any features have positions in common within the genome. The Table Browser provides a simple interface offering the choice of several types of table comparisons based on feature positions.
One class of comparisons preserves the gene or alignment structure of the primary table, resulting in output that describes the same type of feature as is shown in that table. Primary table features are kept or discarded based on the amount of positional overlap with features contained in the secondary table. The user controls the query output by specifying the threshold of overlap: any, none or a percentage.
For example, one might want to identify all the spliced ESTs that align to a particular region in the Known Genes annotation track. The user would select the location of interest in the Table Browser, choose the chrN_intronEst table, and then proceed to the advanced query options. Intersecting the EST table with the knownGene table results in the desired list.
A second class of intersections and unions compares the positions of table features one base position at a time. These queries return only position ranges and do not preserve the structure of the primary table. A base-by-base intersection of two tables will include the base in the output if the nucleotide position is covered by at least one feature of both tables. In a union, the base position need only be covered by the feature of one table.
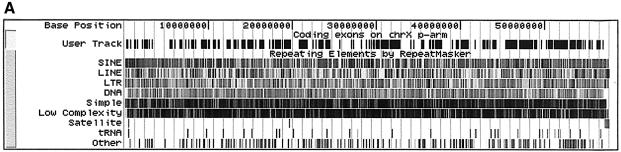
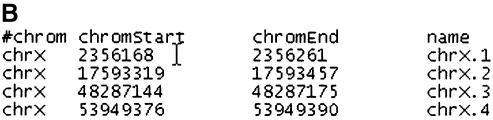
A case in which this kind of comparison is appropriate is a density estimation of a certain feature, e.g. the number of bases within a genomic given region that are repeats or the number of genes within a chromosome that overlap with a repeat. Figure 1 shows an example in which a user wishes to obtain a list of positions in the human chromosome X p-arm in which a SINE repeat overlaps the coding sequence of a gene. This query also illustrates the use of the Table Browser’s custom annotation output format.
Figure 1.
Example of an advanced Table Browser query illustrating the use of a base-by-base table intersection between a standard table and a user- created custom annotation table. The goal is to obtain a list of positions in the p-arm of human chromosome X (Build 34 assembly) in which a SINE repeat overlaps the coding sequence of a gene. (a) The user first creates a custom annotation table called tb_refGene that contains all coding exon entries from the refGene table in the genomic region chrX:1–58500000 (p-arm). To generate the table, one selects the refGene positional table, chooses the Custom Track output option and then selects the Coding Exons BED record option. The custom annotation table may be loaded into the UCSC Genome Browser for further inspection (labeled ‘User Track’ in this figure). (b) The tb_refGene table is then intersected with the chrN_rmsk table using the base-pair-wise (AND) intersection option. To limit the output to only SINE repeats, the chrN_rmsk table is filtered on the repClass field, with the value set to ‘SINE’. The BED format output file is configured to create one record per whole gene. The resulting tab-delimited output indicates four positions that meet the query criteria. This output can be loaded into another program or displayed in the Genome Browser as a custom annotation track.
The set of positions covered by one of the above tables can be complemented (inverted) prior to making the comparison to give the user more flexibility. The user also has the option to set constraints on the field values of the secondary table.
Retrieving subregions of features
In addition to the SQL constraints on queries, the Table Browser allows the user to specify which subregions of features should be present in the output. For example, someone interested in promoters may want to view the region covered by a gene as well as 5000 additional bases upstream from the 5′ end (or downstream from the 3′ end on the negative strand).
The set of available subregion constraints varies among table types. For instance, gene prediction tables specify both exon structure and translated region. The user may constrain the output to show upstream and downstream regions, exons, introns, or 5′, 3′, or coding exons. Alternatively, alignment tables, which specify block structure but not translated region, offer only upstream, downstream, blocks or inter-block regions.
FUTURE DIRECTIONS
The leading feature request from Table Browser users is a batch query-processing interface. Such a tool accepts a file or list of keywords as input and outputs the matching table records. This extension is in development and should be available in late 2003 or early 2004.
Other enhancements under consideration include retrieving data from multiple positions simultaneously and joining together relational tables in a unified interface.
CONTACTING US
The mailing list genome@soe.ucsc.edu provides a forum for announcements of new releases and features, questions and discussion about the UCSC Table Browser, Genome Browser and databases. Users may subscribe to this list at http://www.cse.ucsc.edu/mailman/listinfo/genome. To report problems accessing the website, servers or mirror sites, or for correspondence inappropriate for the public forum, send email to genome-www@soe.ucsc.edu.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank the many collaborators who have contributed sequence and annotation data to our project, as well as our users for their feedback and support. The UCSC Table Browser project is funded by the National Human Genome Research Institute (NHGRI) Grant 1P41HG02371 and the Howard Hughes Medical Institute (HHMI). C.S. is a Howard Hughes Medical Institute Predoctoral Fellow.
REFERENCES
- 1.Karolchik D., Baertsch,R., Diekhans,M., Furey,T.S., Hinrichs,A., Lu,Y.T., Roskin,K.M., Schwartz,M., Sugnet,C.W., Thomas,D.J. et al. (2003) The UCSC Genome Browser Database. Nucleic Acids Res., 31, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas J.W., Touchman,J.W., Blakesley,R.W., Bouffard,G.G., Beckstrom-Sternberg,S.M., Margulies,E.H., Blanchette,M., Siepel,A.C., Thomas,P.J., McDowell,J.C. et al. (2003) Comparative analyses of multi-species sequences from targeted genomic regions. Nature, 424, 788–793. [DOI] [PubMed] [Google Scholar]
- 3.Waterston R.H., Lindblad-Toh,K., Birney,E., Rogers,J., Abril,J.F., Agarwal,P., Agarwala,R., Ainscough,R., Alexandersson,M., An,P. et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature, 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 4.Hardison R., Roskin,K.M., Yang,S., Diekhans,M., Kent,W.J., Weber,R., Elnitski,L., Li,J., O’Connor,M., Kolbe,D. et al. (2003) Covariation in frequencies of substitution, deletion, transposition and recombination during eutherian evolution. Genome Res., 13, 13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siepel A. and Haussler,D. (2003) Combining phylogenetic and Hidden Markov Models in biosequence analysis. Proceedings of the Seventh Annual International Conference on Research in Computational Molecular Biology (RECOMB 2003), pp. 277–286. [DOI] [PubMed]
- 6.Kent W.J., Baertsch,R., Hinrichs,A., Miller,W. and Haussler,D. (2003) Evolution’s cauldron: duplication, deletion and rearrangement in the mouse and human genomes. Proc. Natl Acad. Sci. USA, 100, 11484–11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz S., Kent,W.J., Smit,A., Zhang,Z., Baertsch,R., Hardison,R., Haussler,D. and Miller,W. (2003) Human–mouse alignments with BLASTZ. Genome Res., 13, 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiaromonte F., Yap,V.B. and Miller,W. (2002) Scoring pairwise genomic sequence alignments. Pac. Symp. Biocomput., 2002, 115–126. [DOI] [PubMed] [Google Scholar]
- 9.Kent W.J., Sugnet,C.W., Furey,T.S., Roskin,K.M., Pringle,T.H., Zahler,A.M. and Haussler,D. (2002) The Human Genome Browser at UCSC. Genome Res., 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]