Abstract
The CyberCell Database (CCDB: http://redpoll.pharmacy.ualberta.ca/CCDB) is a comprehensive, web-accessible database designed to support and coordinate international efforts in modeling an Escherichia coli cell on a computer. The CCDB brings together both observed and derived quantitative data from numerous independent sources covering many aspects of the genomic, proteomic and metabolomic character of E.coli (strain K12). The database is self-updating but also supports ‘community’ annotation, and provides an extensive array of viewing, querying and search options including a powerful, easy-to-use relational data extraction system.
BACKGROUND
Escherichia coli is perhaps the most completely characterized microorganism in existence. The quantity of information known about this Gram-negative bacterium, in combination with its amenability to wet-lab studies and relatively simplistic cellular structure has made it the organism of choice for several international efforts in cellular simulation (1). Project CyberCell (www.projectcybercell.com), which is part of the International E.coli Alliance, is one of these efforts. This large-scale multidisciplinary project involves both the acquisition of new quantiative data about E.coli (strain K12) and the collation or back-filling of nearly 50 years of pre-existing E.coli information covering all aspects of the genomic, proteomic and metabolomic character of this organism. In an effort to coordinate both the back-filling and ongoing experimental studies being conducted on E.coli for these simulation efforts, we have built a web-accessible data repository called the CyberCell Database (CCDB). The intent of the CCDB is not to duplicate the many excellent E.coli resources that already exist [such as EcoCyc (2), SwissProt (3), EcoGene (4) and MultiFun (5)], but to facilitate the collection, correction, coordination and storage of the key information needed to simulate E.coli on a computer.
Cellular simulation is an intrinsically data-intensive endeavour, requiring a very broad range of data and data types. This requirement has made it essential to integrate and compile as much available molecular data describing all aspects of E.coli (strain K12) into a single easily accessible resource, including: (i) DNA, RNA and protein sequence data; (ii) gene and protein names, alternative names or abbreviations; (iii) extensive functional or ontological information; (iv) gene position and protein location; (v) macromolecular secondary, tertiary and quaternary structure data; (vi) protein, metabolite and RNA expression levels, copy numbers and concentrations; (vii) protein interaction and protein stoichiometry information; (viii) enzyme rate constants; (ix) metabolite structures, reactions and pathways; (x) lists of cofactors and ligands as well as dozens of other pieces of quantitative molecular data. To compile, confirm and validate this comprehensive collection of data, several hundred journal articles, more than two dozen different electronic databases and a dozen in-house or web-based programs were searched, accessed, compared, written or run over the course of 2 years. On average, each gene, protein or metabolite entry in the CCDB contains more than 70 separate biomolecular data fields, filled to varying levels of completeness. As the scope of the CyberCell project expands, the number and completeness of the data fields are also expected to expand, with some information being updated continuously as new experimental data becomes available. A complete listing of the current data fields as well as the web resources and programs used to assemble the CyberCell database is provided at the CCDB home page.
DATABASE DESCRIPTION
The CCDB is actually a composite of four browsable databases; (i) the main CyberCell database (CCDB, containing gene and protein information); (ii) the 3D structure database (CC3D, containing information for structural proteomics); (iii) the RNA database (CCRD, containing tRNA and rRNA information) and (iv) the metabolite database (CCMD, containing metabolite information). Each of these is accessible through hyperlinked buttons located at the top of the CCDB home page. All the CCDB sub-databases are fully web enabled, permitting a wide variety of interactive browsing, search and display operations. To facilitate browsing, each CyberCell database is divided into synoptic summary tables which, in turn, are linked to more detailed ‘ColiCards’—in analogy to the very successful GeneCards concept (6). The CCDB summary tables can be rapidly browsed, sorted or reformatted (using up to 10 different criteria) in a manner similar to the way PubMed abstracts may be viewed. Clicking on the ColiCard (or RNACard, MetaboCard, etc.) button found in the left-most column of any given summary table opens a web page describing the gene/protein/molecule of interest in much greater detail. In addition to providing comprehensive numeric and textual data, each ColiCard also contains hyperlinks to other databases, abstracts, digital images and interactive applets for viewing molecular structures (2D and 3D) or chromosomal maps (Fig. 1). Users may also electronically edit or update a ColiCard by filling out a simple (‘Edit ColiCard’) web form. All submissions, suggestions and corrections are screened by the CCDB archivist before being permanently added to the database. In addition to this community-based updating process, the CCDB also employs web-bot technology to automatically self-update on a weekly basis. This process, which keeps the CCDB current, is restricted to sequential, structural and functional corrections or additions to major public databases. The CCDB website also provides general information on E.coli (see ‘E. coli’), numerous flat files for downloading (see ‘Download’), as well as extensive statistics (see ‘Stats’) on E.coli dimensions, properties, copy numbers, features, structures and other data which are critical to in silico modeling efforts.
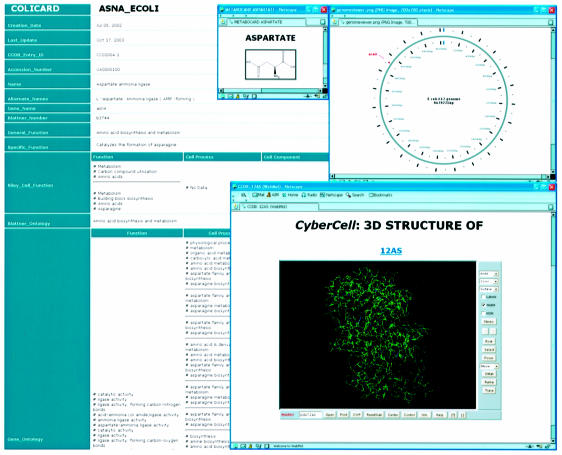
Figure 1.
A screenshot montage of the CCDB showing several possible views of information describing the protein AsnA (aspartate ammonia ligase). Not all fields are shown.
A key distinguishing feature of the CCDB is its extensive support for database searching and sorting functions. In addition to the data viewing and sorting features already described, the CCDB also offers a local BLAST search (protein and DNA against E.coli plus four other model organisms), a Boolean text search (using GLIMPSE) (7), a chemical structure search utility (see‘ChemQuery’) and a relational data extraction tool. The data extraction utility employs a simple relational database system that allows users to select one or more data fields and to search for ranges, occurrences or partial occurrences of words, strings or numbers. The data extractor uses clickable web forms so that users may intuitively construct SQL-like queries. Using a few mouse clicks it is relatively simple to construct very complex queries (Fig. 2). The output from these queries is provided in multiple formats including an HTML table format, a circular chromosome applet view and a tab-delimited Excel format for subsequent downloading, analysis or graphical display.
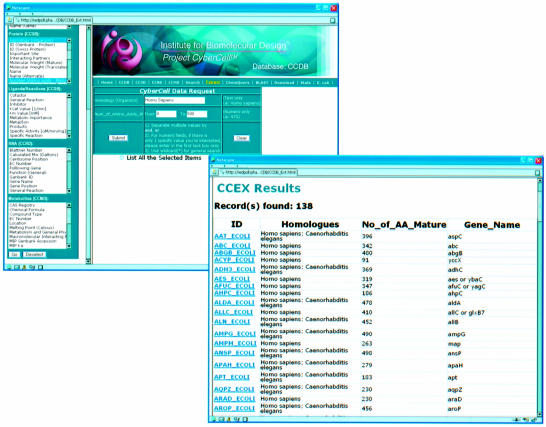
Figure 2.
An example of a complex query conducted using the CCDB’s data extractor. Results are shown for a query asking for all proteins in E.coli that are both <500 residues and homologous (>40% identity) to human proteins.
In summary, the CCDB is a comprehensive, web-accessible database that brings together both observed and derived quantitative data covering nearly all aspects of the genomic, proteomic and metabolomic character of E.coli. The database is self-updating and supports an extensive array of visualizing, querying and search options including a powerful, easy-to-use relational data extraction system. It is hoped that the CCDB will serve as a useful resource not only to cellular simulation aficionados but also to many other members of the E.coli community.
REFERENCES
- 1.Holden C. (2002) Cell biology alliance launched to model E.coli. Science, 297, 1459–1460. [DOI] [PubMed] [Google Scholar]
- 2.Karp P.D., Riley,M., Saier,M., Paulsen,I.T., Collado-Vides,J., Paley,S.M., Pellegrini-Toole,A., Bonavides,C. and Gama-Castro S. (2002) The EcoCyc Database. Nucleic Acids Res., 30, 56–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bairoch A. and Apweiler R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudd K.E. (2000) EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res., 28, 60–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serres M.H. and Riley,M. (2000) MultiFun, a multifunctional classification scheme for Escherichia coli K-12 gene products. Microb. Comp. Genomics, 5, 205–222. [DOI] [PubMed] [Google Scholar]
- 6.Rebhan M., Chalifa-Caspi,V., Prilusky,J. and Lancet,D. (1998) GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics, 14, 656–664. [DOI] [PubMed] [Google Scholar]
- 7.Manber U. and Wu,S. (1994) GLIMPSE: A tool to search through entire file systems. Proceedings from the Usenix Winter 1994 Technical Conference, San Francisco, CA, pp. 23–32.