Abstract
RNA interference (RNAi) is being used in large-scale genomic studies as a rapid way to obtain in vivo functional information associated with specific genes. How best to archive and mine the complex data derived from these studies provides a series of challenges associated with both the methods used to elicit the RNAi response and the functional data gathered. RNAiDB (RNAi Database; http://www.rnai.org) has been created for the archival, distribution and analysis of phenotypic data from large-scale RNAi analyses in Caenorhabditis elegans. The database contains a compendium of publicly available data and provides information on experimental methods and phenotypic results, including raw data in the form of images and streaming time-lapse movies. Phenotypic summaries together with graphical displays of RNAi to gene mappings allow quick intuitive comparison of results from different RNAi assays and visualization of the gene product(s) potentially inhibited by each RNAi experiment based on multiple sequence analysis methods. RNAiDB can be searched using combinatorial queries and using the novel tool PhenoBlast, which ranks genes according to their overall phenotypic similarity. RNAiDB could serve as a model database for distributing and navigating in vivo functional information from large-scale systematic phenotypic analyses in different organisms.
INTRODUCTION
One of the major goals of functional genomics is to fully characterize the function of every gene found in the genome. Because ‘function’ is multifaceted (1,2), a variety of experimental approaches characterizing different aspects of gene function—such as where and when a gene is expressed, the biochemical functions of its gene product(s) and to what other cellular components it binds—must be combined to achieve this goal (3,4). Since the completion of the Caenorhabditis elegans genomic sequence and identification of its ∼20 000 protein-coding genes in 1998 (5), numerous studies of gene expression profiles [(6–10), reviewed in (11)] and protein interactions (4,12,13) have been carried out with the ultimate goal of generating comprehensive functional genomic and proteomic ‘maps’ (14,15). Together with sequence analysis, these approaches may reveal that a certain protein is a kinase, interacts with a cytoskeletal component and is expressed in dividing cells. However, these data cannot by themselves tell us, for example, that this protein is required to properly initiate cytokinesis or that it is also required for cell motility in a different cell or at a different time.
To understand the cellular and developmental roles a gene product plays in the context of an organism, empirical in vivo functional evidence is necessary. Classically, such data have been gathered using forward genetic approaches which, at the time the C.elegans genome was published, had revealed some in vivo information for only ∼7% of the predicted genes (16). The ability to collect more information rapidly and on a large scale has been augmented through the use of RNAi, a process that depletes gene functions using double-stranded RNA (dsRNA) (17,18). Recently, several large-scale systematic analyses of in vivo function have been carried out in C.elegans using RNAi to induce gene knock-downs (19–26). Such in vivo studies are crucial to reveal in which processes proteins operate within living organisms. Currently the resolution of RNAi phenotypic data for most genes is still relatively low (e.g. ‘sterile’ or ‘embryonic lethal’), but innovations in high-throughput high-content assay development promise to increase the amount and level of detail of RNAi phenotypic data in the near future.
Hand in hand with these developments, the challenge of archiving, distributing and mining the results of functional genomic studies has become an important issue. To complement other functional mapping projects, tools analogous to the digital representation and database archival of sequence data, expression patterns or protein–protein interactions are needed for the complex types of information obtained through phenotypic analysis. However, the development of phenotype-based maps has been hindered by two factors: the non-systematic way in which these data are usually gathered and the complex phenomenological nature of the results. Pheno typic studies have traditionally been conducted as screens for a small range of specific defects. Although powerful, this approach leads to a highly filtered view of function by focusing on a limited range of phenomena while ignoring others. In addition, the resulting phenotypic phenomena are typically recorded using free-form natural language descriptions, limiting the potential for computational analysis.
To realize the goal of constructing phenotypic maps, phenotypic data need to be gathered and archived in a systematic and comprehensive way that is amenable to downstream analyses. An explicit system of phenotypic documentation is also necessary, in which observable defects arising from the depletion of a gene’s function are described and annotated using a controlled structured vocabulary. The Gene Ontology Consortium has undertaken an effort to standardize gene functional annotations across different biological systems (27,28), but the application of this idea to phenotypic data remains a challenge. By systematically encoding phenotypic data from the early C.elegans embryo, we have demonstrated previously that genes can be clustered on the basis of phenotypic data, resulting in groups of genes enriched for different functional classes (29). We have also used these data to begin integrating phenotypic, protein interaction and expression maps in the early embryo (4).
The accelerating accumulation of RNAi-based data from ongoing large-scale analyses calls for a database system focused on phenotypes. Such a system should ideally be freely available, web-accessible, user-friendly, adhere to community standards and provide flexible query options and tools for analysis of the data. Here we describe RNAiDB, an online database system, and PhenoBlast, an associated search/analysis tool, for genome-scale RNAi phenotypic studies in C.elegans.
DESCRIPTION OF THE DATABASE
One of the ideas behind public sharing of genome information is the distributed database model (30), in which interconnected databases can also act as portals displaying specific types of information from other databases that are curated and developed by the community of people involved in populating them. Guided by this vision, we designed a database system that can exchange information easily with WormBase [the central repository for genomic information from C.elegans (31)] by using the same object-oriented database engine, AceDB [R. Durbin and J. Thierry-Mieg: A C.elegans Database (1991); documentation and code available at http://www.acedb.org]. The RNAiDB data models are largely compatible with those in WormBase, although extensions to the RNAi and other object models have been made to enhance functionality for RNAi data in particular. RNAiDB is currently served through the Web using AcePerl (32) and Perl CGI (http://stein.cshl.org/WWW/software/CGI/) scripts running on a Linux system with an Apache web server.
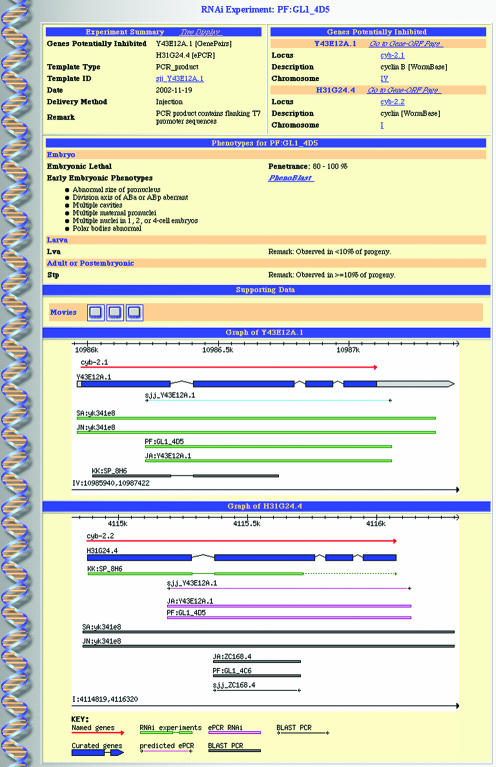
Data currently contained in RNAiDB were assembled from a combination of locally generated RNAi experimental data and publicly available data imported from WormBase, including RNAi results, gene annotations and physical mapping data. For each RNAi experiment in the database, phenotypic summaries organized by life stage are displayed along with associated experimental details, RNAi to gene mappings and supporting raw data such as still images or streaming Quicktime time-lapse movies of embryogenesis (Fig. 1). Reciprocal links to WormBase are provided as well as links to other external resources, such as WorfDB (33) and WormGenes at NCBI (http://www.wormgenes.org; D. Thierry-Mieg, J. Thierry-Mieg and Y. Thierry-Mieg, M. Potdevin, M. Sienkiewicz, V. Simonyan, unpublished).
Figure 1.
An example screenshot of an ‘RNAi Experiment Card’ page. The card contains several panels, each providing a different set of information. Top row, left panel: summary of information associated with the experimental protocol, including genes potentially inhibited and the sequence analysis method used to identify them (e.g. ‘GenePairs’, the canonical location of the PCR product used in this experiment; ‘ePCR’, additional location identified by ePCR). Top row, right panel: summary data on genes potentially inhibited by this RNAi experiment, with links to WormBase and other external resources. Second row: summary of phenotypes elicited by this RNAi experiment, organized by life stage. The link to PhenoBlast will automatically run PhenoBlast on the early embryonic phenotypic signature for this experiment (see text for details). Third row: links to supporting images and time-lapse movies available for this experiment, which can be viewed in a separate window by clicking on the icons provided. Fourth and subsequent rows: clickable graphical maps of genes potentially inhibited by this RNAi experiment showing chromosome coordinates, gene models and reagents mapped to this location as indicated in the key below each diagram. Select features are hyperlinked to related pages.
RNAi is a very powerful technique for gene function depletion but for best interpretation of the results it is important to consider some important technical issues. These are mostly related to the sequence used to trigger the RNAi response, the way in which the dsRNA is delivered to the animal and the way in which the affected animals are scored. Extensive cross-validation analyses for RNAi-derived phenotypes have suggested a generally low level of false positive results but a higher level of false negative results (26,29). Differences in experimental methods can lead to variability in results of phenotypic assays for the same gene, much as different genetic mutations can give rise to allelic series with defects of differing severity. In addition, because RNAi elicits its effects in a sequence-specific manner, in some cases more than one gene may potentially be inhibited by a single dsRNA reagent. This could occur for two reasons: (i) PCR primers using genomic DNA as the template may amplify more than one distinct genomic interval by annealing at multiple genomic locations, and (ii) heterologous mRNAs with high sequence similarity to the primary mRNA target may also be targeted for degradation. These points are important for drawing genome-wide conclusions on the types of genes that can elicit specific types of defects as well as for single-gene analyses used to explore the function of specific genes.
To address such issues in the display of the RNAi results we have designed two main views for the visualization of RNAi data: an ‘RNAi Experiment’ card (Fig. 1) and a ‘Gene Product/ORF’ card (not shown). These cards provide complementary views of the data that show, respectively, all gene mappings for a single RNAi experiment, or all RNAi experiments corresponding to a single gene, allowing the user both to critically review the possibility that a single RNAi assay could affect multiple genes and to quickly compare the results from different studies.
To determine when a single RNAi experiment might target more than one gene, we have supplemented the ‘canonical’ RNAi–gene mappings provided in WormBase with additional evidential support based on either electronic PCR (‘ePCR’) (34) of primer pairs used to generate PCR products and/or BLAST (35) analysis of PCR products and cDNAs from which dsRNA reagents were produced for the RNAi assays contained in RNAiDB. Together, the ePCR and BLAST analyses define the set of genes potentially inhibited by each RNAi experiment. Both analyses were performed using in-house pipelines designed to retrieve all genomic coordinates that could be targeted simultaneously in the same RNAi experiment, based on ePCR or strong sequence similarity identified by BLAST. The resulting mappings are stored in a genome coordinate-based general feature format (GFF) database implemented in MySQL (36). RNAi to gene mappings along with the type of evidential support are indicated in the text of the RNAi and Gene Product card pages and are also displayed graphically (Fig. 1) using the Bio::DB::GFF and Bio::Graphics Perl modules distributed with the BioPerl toolkit (37).
SEARCHING THE DATABASE
A variety of entry points to the database are available. The home page contains a simple ‘quick’ search form for finding RNAi experiments that provides drop-down menus for selecting phenotypes by life stage and an optional text box for specifying RNAi experiments, genes, phenotypes, laboratories and experimental reagents by name. RNAi experiments can also be searched by phenotype using either a simple menu-driven form or an advanced phenotype search form that provides a combinatorial query builder. Additional search options provide the ability to query any object represented in the database (genes, PCR products, etc.) using either a simple class browser with optional name or wildcard pattern, a text/keyword search, or an Ace Query Language (AQL) statement. Related objects are cross-referenced in the database, and these connections can be navigated via hyperlinked text.
Digital phenotypic screening
The ability to perform combinatorial searches of large-scale phenotypic mapping projects is one of the most powerful foreseeable outcomes of such projects. When phenotypic scoring is carried out systematically [e.g. see (25), in which embryos were scored using a comprehensive list of 47 phenotypes], they can then be mined to identify groups of genes that elicit specific combinations of phenotypes. There are over 1014 potential complete phenotypic signatures that could theoretically result from different combinations of these 47 phenotypes (if scored as either wild type or mutant, i.e. not taking into account penetrance or expressivity). The idea behind ‘digital phenotypic screening’ is to have the ability to search for any subset of desired phenotypes among these combinations.
The phenotype query builder was designed to allow phenotype-based searches to be conducted in a way that parallels the logic behind genetic screens, which typically begin with a primary survey to identify and retain mutants displaying a specific set of desired characteristics while weeding out others. Similarly, with the advanced search option users can construct complex queries on specific characteristics of interest and can explicitly exclude undesired phenotypes. In essence this enables users to perform ‘digital phenotypic screens’ for specific syndromes. For example, users can search for genes that display RNAi phenotypes indicative of defects in cytokinesis but not other aspects of mitosis. This search, which would take months on the bench, takes only minutes on the computer.
Mining phenotypic data: PhenoBlast
A basic tenet of many genetic studies is that genes whose mutant phenotypes are similar are often components of the same pathway. Consequently developmental geneticists often have a body of knowledge that permits them to perform associative phenotype studies in their head. This means that they may see a trend in a series of phenotypes elicited by a particular mutation that resembles those of other mutations they already know about. However, as more and more genome-wide phenotype-based studies are performed, it is becoming impossible to do this effectively in a non-systematic way. Inspired by the development of tools for large-scale sequence searching and alignment necessitated by the DNA sequencing revolution, to begin to address this issue we have developed a tool that can help perform phenotype-associative studies using RNAiDB.
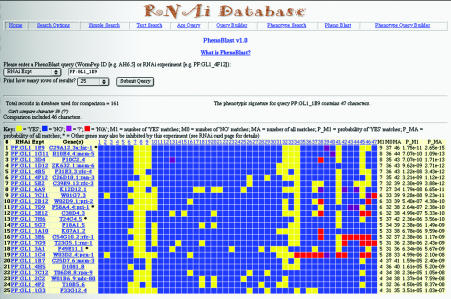
If RNAi experiments have been scored systematically for the presence or absence of a set of discrete phenotypes (or ‘phenotypic characters’) (25), then it is possible to examine this set as a composite and ask how similar the overall ‘phenotypic signatures’ are among different genes. We have implemented a novel tool in RNAiDB to permit searching for genes that elicit a similar range of phenotypes. Given a single query gene or RNAi assay, this tool dynamically generates a list of genes ranked according to how similar their phenotypic signature is to that of the query (Fig. 2). Because it performs a function analogous to that of BLAST for nucleotide or amino acid sequence searching, we have designated this tool ‘PhenoBlast’ to highlight this similarity (although the algorithms are unrelated). The genes are ranked in relation to the query based on a series of measures, the most significant of which is the number of phenotypic defects shared between the query and target genes. By scanning the PhenoBlast result page, users can identify which other genes in the database display the most similar composite phenotypic signature relative to a single particular gene of interest. PhenoBlast can be accessed either from RNAiDB’s main search menu or from Gene and RNAi card pages.
Figure 2.
An example of a ‘PhenoBlast’ result page. The phenotypic signature of the query is shown in the first row of the result list, followed by the 24 best matches in the database. In this example, the query (RNAi experiment ‘PF:GL1_1E9’, corresponding to gene ‘lig-1’) is thought to be involved in DNA synthesis, as are many top-ranking genes (based on sequence similarity to genes of known function in other organisms). Wild-type states are represented by a blue square while mutant phenotypes are represented by yellow squares. Red and purple squares represent cases where the phenotype state for that character is, respectively, missing or unknowable under these assay conditions. Results are ranked by similarity to the query signature and are sorted lexicographically based on the number of shared phenotypic defects (M1), the total number of shared phenotypic characters (MA) and the probability of observing this combination of shared defects (P_M1) or all shared characters (P_MA) in the population of experiments in the database. Results with the same number of shared characters are thus ranked in a weighted fashion that is inversely proportional to the frequency of character states in the population. Each result row is linked to the corresponding RNAi Experiment and Gene Product card pages.
FUTURE of RNAiDB
We are continuing to develop RNAiDB to enhance its functionality, focusing initially on three main areas. First, we are in the process of expanding RNAiDB to provide a web-based community ‘lab notebook’ system that will allow users from different laboratories to upload raw data and RNAi results, to perform computer-assisted phenotypic scoring using controlled data entry forms and to generate phenotypic signatures that can then immediately be compared with other entries in the database. Second, we are generalizing the data models to allow the definition and scoring of any arbitrary set of phenotypic characters in addition to the early embryo set we have previously defined (29). For example, we anticipate that the current collection of raw data, generated using differential interference contrast (DIC) microscopy, will be supplemented in the future with fluorescent images using tagged cellular markers for different cellular components, which will provide additional levels of detail in phenotypic descriptors. Finally we plan to develop and provide additional data analysis tools to facilitate mining of these data, such as online phenoclustering (29). This work will be done in conjunction with continued efforts to develop and refine methods for the systematization of phenotypic data.
CONCLUSION
The prospect of a comprehensive database of phenotypic descriptors in a simple system like C.elegans, including all genetic aberrations and reverse-genetic defects induced by RNAi, holds promise to benefit other areas of study in which phenotypes play an important role, such as chemical genetics and medical research. Model systems are especially suited to the discovery not only of individual key proteins that are required for basic cellular and developmental processes, but also to the identification of network components through genetic analysis and integration of functional genomic data from comprehensive data sets. Consequently, the development of effective phenotypic maps for model organisms will not only contribute a vital view of genome function in those particular systems but may also lead to a better way of identifying core molecular pathways underlying cellular pathologies associated with disease or the effects of drugs that elicit specific cellular responses. Such benefits may prove especially relevant for highly conserved genes whose in vivo function is not yet known in any animal. We believe RNAiDB could serve as a model for phenome projects in other organisms.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Fritz Roth for suggesting the name ‘PhenoBlast’, Jean and Danielle Thierry-Mieg for help with AceDB, Marc Vidal’s group and Philippe Vaglio in particular for sharing their experience with BioGraphics, and Lincoln Stein for his help with RNAiDB and ePCR and for authoring extremely useful open-source software for web and genome applications. This work was supported by NSF award DBI-0137617 to K.C.G.
REFERENCES
- 1.Vidal M. (2001) A biological atlas of functional maps. Cell, 104, 333–339. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B., Lane,D. and Levine,A.J. (2000) Surfing the p53 network. Nature, 408, 307–310. [DOI] [PubMed] [Google Scholar]
- 3.Tyers M. and Mann,M. (2003) From genomics to proteomics. Nature, 422, 193–197. [DOI] [PubMed] [Google Scholar]
- 4.Walhout A.J., Reboul,J., Shtanko,O., Bertin,N., Vaglio,P., Ge,H., Lee,H., Doucette-Stamm,L., Gunsalus,K.C., Schetter,A.J. et al. (2002) Integrating interactome, phenome and transcriptome mapping data for the C. elegans germline. Curr. Biol., 12, 1952–1958. [DOI] [PubMed] [Google Scholar]
- 5.The Caenorhabditis elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 6.Hill A.A., Hunter,C.P., Tsung,B.T., Tucker-Kellogg,G. and Brown,E.L. (2000) Genomic analysis of gene expression in C. elegans. Science, 290, 809–812. [DOI] [PubMed] [Google Scholar]
- 7.Reinke V., Smith,H.E., Nance,J., Wang,J., Van Doren,C., Begley,R., Jones,S.J., Davis,E.B., Scherer,S., Ward,S. et al. (2000) A global profile of germline gene expression in C. elegans. Mol. Cell, 6, 605–616. [DOI] [PubMed] [Google Scholar]
- 8.Jiang M., Ryu,J., Kiraly,M., Duke,K., Reinke,V. and Kim,S.K. (2001) Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA, 98, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roy P.J., Stuart,J.M., Lund,J. and Kim,S.K. (2002) Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature, 418, 975–979. [DOI] [PubMed] [Google Scholar]
- 10.Baugh L.R., Hill,A.A., Slonim,D.K., Brown,E.L. and Hunter,C.P. (2003) Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development, 130, 889–900. [DOI] [PubMed] [Google Scholar]
- 11.Reinke V. (2002) Functional exploration of the C. elegans genome using DNA microarrays. Nature Genet., 32 (Suppl.), 541–546. [DOI] [PubMed] [Google Scholar]
- 12.Boulton S.J., Gartner,A., Reboul,J., Vaglio,P., Dyson,N., Hill,D.E. and Vidal,M. (2002) Combined functional genomic maps of the C. elegans DNA damage response. Science, 295, 127–131. [DOI] [PubMed] [Google Scholar]
- 13.Walhout A.J., Sordella,R., Lu,X., Hartley,J.L., Temple,G.F., Brasch,M.A., Thierry-Mieg,N. and Vidal,M. (2000) Protein interaction mapping in C. elegans using proteins involved in vulval development. Science, 287, 116–122. [DOI] [PubMed] [Google Scholar]
- 14.Kim S.K., Lund,J., Kiraly,M., Duke,K., Jiang,M., Stuart,J.M., Eizinger,A., Wylie,B.N. and Davidson,G.S. (2001) A gene expression map for Caenorhabditis elegans. Science, 293, 2087–2092. [DOI] [PubMed] [Google Scholar]
- 15.Grant B.D. and Wilkinson,H.A. (2003) Functional genomic maps in Caenorhabditis elegans. Curr. Opin. Cell Biol., 15, 206–212. [DOI] [PubMed] [Google Scholar]
- 16.Costanzo M.C., Hogan,J.D., Cusick,M.E., Davis,B.P., Fancher,A.M., Hodges,P.E., Kondu,P., Lengieza,C., Lew-Smith,J.E., Lingner,C. et al. (2000) The yeast proteome database (YPD) and Caenorhabditis elegans proteome database (WormPD): comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res., 28, 73–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo S. and Kemphues,K.J. (1995) par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell, 81, 611–620. [DOI] [PubMed] [Google Scholar]
- 18.Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 19.Gönczy P., Echeverri,G., Oegema,K., Coulson,A., Jones,S.J., Copley,R.R., Duperon,J., Oegema,J., Brehm,M., Cassin,E. et al. (2000) Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature, 408, 331–336. [DOI] [PubMed] [Google Scholar]
- 20.Fraser A.G., Kamath,R.S., Zipperlen,P., Martinez-Campos,M., Sohrmann,M. and Ahringer,J. (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature, 408, 325–330. [DOI] [PubMed] [Google Scholar]
- 21.Piano F., Schetter,A.J., Mangone,M., Stein,L. and Kemphues,K.J. (2000) RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr. Biol., 10, 1619–1622. [DOI] [PubMed] [Google Scholar]
- 22.Hanazawa M., Mochii,M., Ueno,N., Kohara,Y. and Iino,Y. (2001) Use of cDNA subtraction and RNA interference screens in combination reveals genes required for germ-line development in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA, 98, 8686–8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeda I., Kohara,Y., Yamamoto,M. and Sugimoto,A. (2001) Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol., 11, 171–176. [DOI] [PubMed] [Google Scholar]
- 24.Zipperlen P., Fraser,A.G., Kamath,R.S., Martinez-Campos,M. and Ahringer,J. (2001) Roles for 147 embryonic lethal genes on C. elegans chromosome I identified by RNA interference and video microscopy. EMBO J., 20, 3984–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piano F., Schetter,A.J., Morton,D.G., Gunsalus,K.C., Reinke,V., Kim,S.K. and Kemphues,K.J. (2002) Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Curr. Biol., 12, 1959–1964. [DOI] [PubMed] [Google Scholar]
- 26.Kamath R.S., Fraser,A.G., Dong,Y., Poulin,G., Durbin,R., Gotta,M., Kanapin,A., Le Bot,N., Moreno,S., Sohrmann,M. et al. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature, 421, 231–237. [DOI] [PubMed] [Google Scholar]
- 27.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T. et al. The Gene Ontology Consortium (2000) Gene Ontology: tool for the unification of biology. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill D.P., Blake,J.A., Richardson,J.E. and Ringwald,M. (2002) Extension and integration of the gene ontology (GO): combining GO vocabularies with external vocabularies. Genome Res., 12, 1982–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piano F. and Gunsalus,K.C. (2002) RNAi-based functional genomics in C. elegans. Curr. Genomics, 3, 69–81. [Google Scholar]
- 30.Dowell R.D., Jokerst,R.M., Day,A., Eddy,S.R. and Stein,L. (2001) The Distributed Annotation System. BMC Bioinformatics, 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris T.W., Lee,R., Schwarz,E., Bradnam,K., Lawson,D., Chen,W., Blasier,D., Kenny,E., Cunningham,F., Kishore,R. et al. (2003) WormBase: a cross-species database for comparative genomics. Nucleic Acids Res., 31, 133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stein L.D. and Thierry-Mieg,J. (1998) Scriptable access to the Caenorhabditis elegans genome sequence and other ACEDB databases. Genome Res., 8, 1308–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaglio P., Lamesch,P., Reboul,J., Rual,J.F., Martinez,M., Hill,D. and Vidal,M. (2003) WorfDB: the Caenorhabditis elegans ORFeome Database. Nucleic Acids Res., 31, 237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuler G.D. (1997) Sequence mapping by electronic PCR. Genome Res., 7, 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stein L.D., Mungall,C., Shu,S., Caudy,M., Mangone,M., Day,A., Nickerson,E., Stajich,J.E., Harris,T.W., Arva,A. et al. (2002) The generic genome browser: a building block for a model organism system database. Genome Res., 12, 1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stajich J.E., Block,D., Boulez,K., Brenner,S.E., Chervitz,S.A., Dagdigian,C., Fuellen,G., Gilbert,J.G., Korf,I., Lapp,H. et al. (2002) The Bioperl toolkit: Perl modules for the life sciences. Genome Res., 12, 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]