Summary
In eukaryotes, cytokinesis generally involves an acto-myosin ring, the contraction of which promotes daughter cells segregation. Assembly of the contractile ring is tightly controlled in space and time (see[1–4] for reviews). In the fission yeast, contractile ring components are first organized by the Anillin-like protein Mid1 [5, 6] and Cdr2 kinase [7–9] into medial cortical nodes. These nodes then coalesce laterally into a functional compact contractile ring [10–13]. Although Mid1 is present at the medial cortex throughout G2 phase [14], recruitment of contractile ring components to nodes starts only at mitotic onset [12] indicating that this event is cell cycle regulated. Polo kinases are key temporal coordinators of mitosis and cytokinesis [1] and the Polo-like kinase Plo1 [15] has long been implicated in Mid1 regulation [16]: Plo1 activates Mid1 nuclear export at mitotic onset [16], coupling division plane specification to nuclear position [7]. Here, we provide evidence that Plo1 also triggers the recruitment of contractile ring components into medial cortical nodes. Plo1 binds at least two independent sites on Mid1, including a consensus site phosphorylated by Cdc2. Plo1 phosphorylates several residues within the first 100 amino acids of Mid1, which directly interact with the IQGAP Rng2 [17], and influences the timing of Myosin II recruitment. Plo1 thereby facilitates contractile ring assembly at mitotic onset.
Results and discussion
Medial cortical nodes organized by Cdr2 kinase pre-establish the division plane during interphase by recruiting Mid1 [5–7, 9, 14]. Yet, the further recruitment of contractile ring components to medial cortical nodes by Mid1 does not take place until mitotic onset [10, 12, 13]. Several lines of evidence have connected the polo-like kinase Plo1 to Mid1: certain plo1 mutations phenocopy mid1 mutants [18, 19]; Mid1 is hyper-phosphorylated in mitosis [6] or upon Plo1 overexpression [16]; Mid1 export from the nucleus is driven by Plo1 [16]; Plo1 localization to the contractile ring depends on Mid1 [16]. Therefore, we wondered if Plo1 could regulate Mid1-dependent recruitment of contractile ring components to medial cortical nodes at mitotic onset. Consistent with this hypothesis, a Mid1 mutant deficient for nuclear localization does not rescue the division plane position defects of the plo1-1 mutant and does not compact into the contractile ring [14].
Mid1 contains at least two plo1-binding sites
We first analyzed how Plo1 binds to Mid1. Similar to other substrates of Polo kinases, Mid1 interaction with Plo1 depends on Plo1 polo box domains (PBD) [22] (see [20, 21]). The Plo1 PBD bound to the Mid1 N-terminus (GST-Nter, aa 1–422; Fig 1A–B) as well as the C-terminus (GST-Cter, aa 443–920; Fig 1A–B) in vitro, indicating that Mid1 contains at least two independent Plo1 interaction sites. Similar results were obtained by immunoprecipitation (Fig S1A).
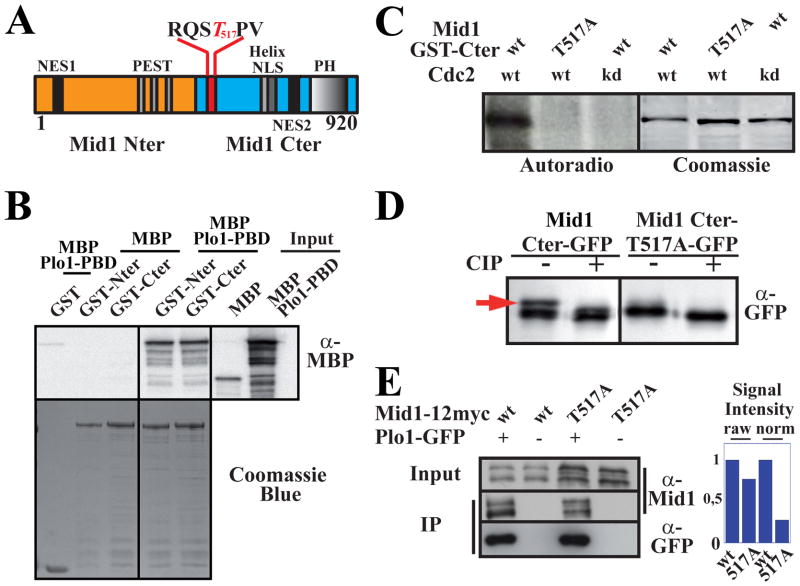
Figure 1. Plo1 binding to Mid1 involves two sites and is favored by Cdc2 phosphorylation of threonine 517 within the RQST517PV consensus.
A: Mid1 molecule (Nter, orange; Cter, blue). The consensus Plo1 binding site RQSTPV is detailed with phospho-T517 in red.
B: In vitro binding of Plo1 PBD (MBP-Plo1-PBD) on Mid1 Nter and Cter (GST-Nter, aa 1–422; GST-Cter, aa 443–920). Top: MBP-Plo1-PBD and MBP (negative control) were revealed using α-MBP Abs. Bottom: loading controls (coomassie blue staining).
C: Cdc2 kinase assay on Mid1 Cter. Left: Mid1 GST-Cter (aa 443–920) or GST-T517A-Cter were incubated with Cdc2 and Cdc2 kinase dead (kd). Phosphorylation is detected by 32P autoradiography. Right: loading controls (coomassie blue staining).
D: Migration pattern of Mid1 Cter-GFP (aa 501–920) and Cter-T517A-GFP immunoprecipitated with an anti-GFP mAb and treated or not with CIP. WB: anti-GFP mAb. Red arrow: phospho-Cter-GFP.
E: Coimmunoprecipitation of Mid1-12myc or Mid1-T517A-12myc with Plo1-GFP. IPs and WB for Plo1-GFP were performed with an anti-GFP mAb. Mid1 was detected using anti-Mid1 affinity purified Ab (α-Mid1). Negative controls: anti-GFP IPs on extracts from cells expressing untagged Plo1. Right: signal quantification. Raw signals of Mid1-12myc and Mid1-T517A-12myc in Plo1-GFP IP samples (left). Normalized signals relative to Mid1 and Mid1-T517A concentration in input (2.8 ratio; right).
See also Figure S1.
PBDs generally bind substrates through a consensus sequence, MQSTPL, wherein the threonine is phosphorylated [23]. Accordingly, deletion of a related motif within the Mid1 Cter fragment (RQSTPV, aa 514–519) or the surrounding region (Mid1 Δ507–625), but not the C-terminal part of Mid1 (Mid1 Δ626–920), reduced Mid1 co-immunoprecipitation with Plo1-GFP (Fig S1B). However, it did not fully abolish Plo1 binding, confirming the presence of a second interaction site.
T517 phosphorylation by Cdc2 favors Plo1 interaction with Mid1
Cyclin-dependent kinases can prime substrates for Polo kinases [23] and Cdc2 phosphorylated the Mid1 Cter (GST-Cter, aa 443–920, Fig 1C). Phosphorylation by Cdc2 was abolished when threonine 517 was replaced by alanine (Fig 1C). Moreover, SDS-PAGE of Mid1 Cter (Cter-GFP, aa 506–920; [24]) expressed in vivo revealed a slow migrating band eliminated by phosphatase treatment and absent in the Cter-T517A mutant (Fig 1D), indicating that Mid1 Cter is phosphorylated on T517 in vivo. PhosphoT517 was also detected by mass spectrometric analysis of GFP-Mid1 purified from mitotic cells (see Table S2).
Furthermore, Plo1-GFP coimmunoprecipitated less Mid1-T517A-12myc than Mid1-12myc (~30 % of Mid1-12myc IP levels after normalization according to Mid1 and Mid1-T517A expression levels; Fig 1E) and Plo1 localization to the contractile ring was abolished in the Mid1-T517A mutant (Fig S1C–D, 0/66 mitotic T517A cells with Plo1-GFP at the contractile ring compared to 26/58 in control mitotic cells) as in mid1 deleted cells [16]. In contrast, Mid1-T517A localization was similar to wild type Mid1 in interphase or mitosis (Fig S1E) indicating that the residual binding of Mid1-T517A to Plo1 is sufficient to promote Mid1 nuclear export and Mid1 compaction into the contractile ring during mitosis. Nevertheless, using the separation of SPBs stained for Sfi1 [25] as a marker for mitosis onset and the Myosin II light chain Rlc1 [26, 27], we observed a small but significant delay of ~3 minutes in Myosin II recruitment to medial cortical nodes in this mutant (Fig S1F).
In conclusion, Plo1 binds to Mid1 through at least two independent sites. Phosphorylation of T517 by Cdc2 within the C-terminal site favors Plo1 interaction with Mid1 during mitosis, stable Plo1 localization at the contractile ring and Mid1 activity at the cortex.
Plo1 phosphorylates several residues within Mid1 1–100 fragment
Plo1 phosphorylated Mid1 Nter in vitro (GST-Nter, aa 1–422) but not Mid1 Cter (GST-Cter, aa 443–920; Fig 2A). Two-dimensional phosphopeptide mapping identified four major and several minor tryptic phosphopeptides in Mid1 Nter (Fig S2C). Consistent with Plo1 activity affecting the Mid1 Nter, Plo1 overexpression drove its nuclear export and assembly into transient filamentous clusters before septation (Fig 2D, S2A), whereas the Mid1 Cter remained at the medial cortex [24] (Fig S2A).
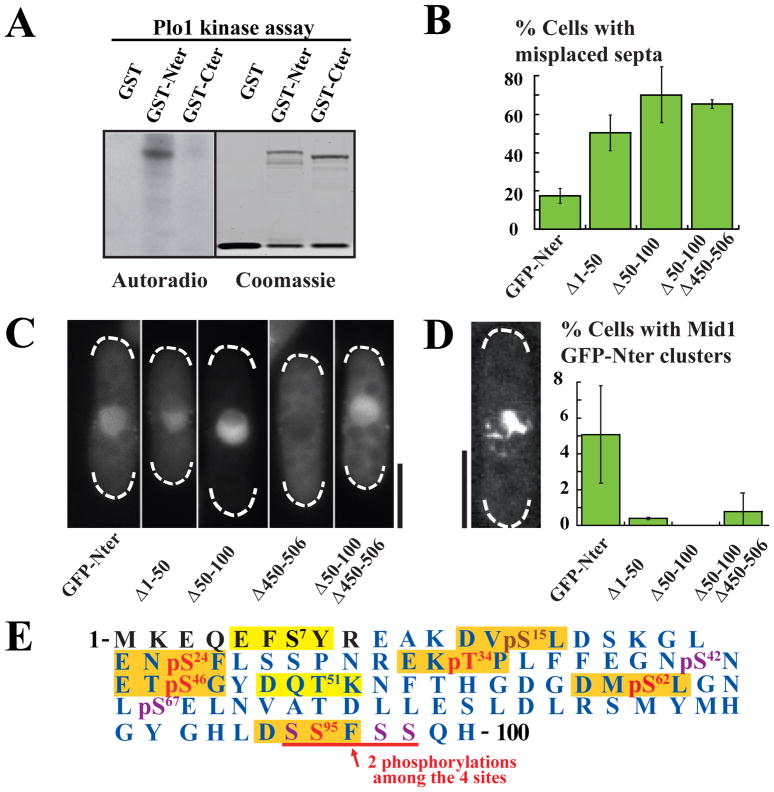
Figure 2. Plo1 phosphorylates Mid1 Nter residues 1–100.
A: Plo1 in vitro kinase assay on Mid1 GST-Nter (aa 1–422) or Mid1 GST-Cter (aa 443–920). Phosphorylation is detected by 32P autoradiography (left). Right: loading controls (coomassie blue staining).
B: Percentage of misplaced septa in Mid1 GFP-Nter, GFP-Nter Δ1–50, Δ50–100 and Δ50–100Δ450–506 mutants. Cells were grown at 30°C. Error bars: SD, 9 independent counts of 100 cells.
C: Localization of Mid1 GFP-Nter, GFP-Nter Δ1–50, Δ50–100, Δ450–506 and Δ50–100Δ450–506 deletion mutants expressed in mid1Δ cells during interphase. Bar: 5 μm.
D: Percentage of GFP-Nter, GFP-NterΔ1–50, Δ50–100 and Δ50–100Δ450–506 cells assembling clusters of filaments upon Plo1 overexpression for 20 hours at 25°C. Error bars: SD, 3 independent experiments, n > 400. Left: GFP-Nter fluorescence in a cell showing a cluster. Bar: 5 μm.
E: Phospho-map of Mid1 residues 1–100. Consensus Plo1 phosphorylation sites are boxed in orange when mutated in Mid1-6Ala mutant, or in yellow. Phosphosites detected by mass spectrometric analysis of mitotic GFP-Mid1 are shown in red in Plo1 consensus sites or purple. Sequence coverage is shown in blue. pS15 (in brown) was detected on Mid1 Nter phosphorylated in vitro by Plo1.
See also Figure S2.
If Plo1 activates Mid1 by phosphorylating its Nter, mutants of Mid1 Nter lacking key Plo1 phosphorylation sites should exhibit strong functional defects. We thus analyzed our collection of 50 amino acid deletion mutants within the Mid1 Nter [7]. Besides the two well characterized cortex anchoring mutants (Δ300–350 and Δ400–450, Fig S2B) [7], Δ1–50 and Δ50–100 exhibited strong division plane position defects (50 and 70% of misplaced septa respectively; 17% for full length Mid1 Nter, Fig 2B, S2B). As Mid1 residues 50–100 contain a nuclear export sequence [14], Mid1 NterΔ50–100 was strongly concentrated in the nucleus (nucleo/cytoplasmic ratio of ~5 instead of ~2 for Mid1 Nter; Fig 2C). When we additionally deleted domain 450–506 which contains a nuclear import sequence and is not necessary for Mid1 Nter function, to force the mutant protein to remain cytosolic (Mid1 NterΔ50–100 Δ450–506; Fig 2C, S2B) [7], a normal nucleo-cytoplasmic ratio was restored (~1.5; Fig 2C), but this did not rescue division plane position defects (65% misplaced septa, Fig 2B, S2B). We conclude that residues 50–100 have important functions in addition to driving Mid1 nuclear export.
When Plo1 was overexpressed in the Mid1 Nter mutants Δ1–50 and Δ50–100 or Δ50–100 Δ450–506, they did not assemble filamentous clusters, or very few as compared to cells expressing the complete Mid1 Nter (Fig 2D), consistent with the hypothesis that Mid1 1–100 fragment contains key Plo1 phosphosites.
Within this region are 8 sites with strong similarity to Plo1 consensus phosphorylation motifs ((D/E)−2-(X)−1-(S/T)0-Φ+1-(X)+2-(D/E)+3 where Φ is an hydrophobic amino acid; Fig 2E) [28]. Mutation of these sites to alanine (aa 1–422; GST-Nter-8ala) eliminated two major and one minor tryptic phosphopeptides (Fig S2C) corresponding to S15, S24 and T34 phosphosites (data not shown). Mass spectrometric analysis analysis of GST-Mid1 Nter phosphorylated by Plo1 in vitro confirmed S24 and T34 and identified also S46 as a Plo1 phosphosite (Table S3). Finally, mass spectrometry revealed that five consensus sites were phosphorylated in vivo during mitosis (S24, T34, S46, S62 and 2 phosphosites among S94-S95-S97-S98, Fig 2E and Table S2). Altogether, we confirmed six of the eight consensus Plo1 phosphosites (S15, S24, T34, S46, S62 and S95; Fig 2E).
Plo1-dependent phosphorylation of Mid1 1–100 triggers Myosin II recruitment
Mutation of the six Plo1 phosphosites to alanine (Mid1-6Ala) successfully reduced Mid1 hyperphosphorylation upon Plo1 overexpression in vivo (Fig S2D) [16]. In addition, the Mid1-6Ala phosphodefective mutant was not fully exported from the nucleus in ~60% mitotic cells and always produced weak rings which did not compact properly in ~70% of the cases (Fig S2E–F), reminiscent of Mid1 behaviour in plo1-1 [14, 16].
Myosin II is not recruited to medial cortical nodes at mitotic entry in plo1-1 cells (Fig S3A) [16]. We thus analyzed Myosin II recruitment in the Mid1 6Ala mutant, which also contained mutations that force the protein to remain cytosolic (nsm) [7]. Rlc1 was recruited ~4 minutes after SPB separation in the Mid1nsm_6Ala mutant, with a delay of ~9 minutes as compared to control Mid1nsm cells (Fig 3A–B). Rlc1 recruitment pattern was irregular (Fig 3A) and Mid1nsm-6Ala could not compact properly into a ring structure as observed for the Mid1-6Ala mutant (Fig 3A, see also kymographs on Fig S3B).
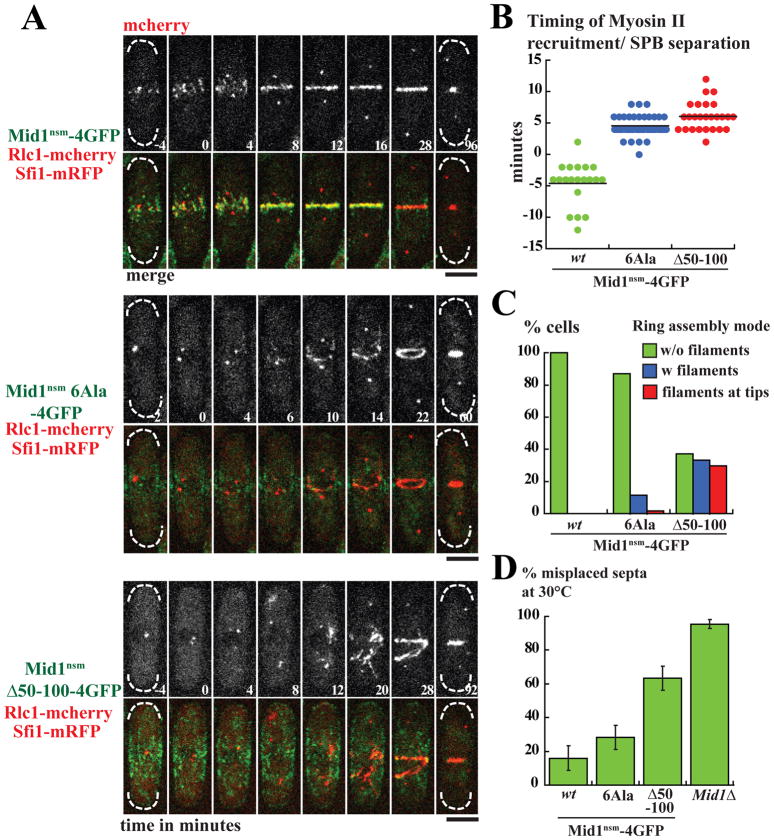
Figure 3. Mid1 activation by Plo1 controls the timing of Myosin II recruitment to medial cortical nodes.
A: Time-lapse movies of cells deleted for endogenous mid1 expressing Mid1nsm-4GFP (top panel), Mid1nsm6Ala-4GFP (medium) or Mid1nsmΔ50–100-4GFP (bottom), Rlc1-mcherry and SPB component Sfi1-mRFP as a marker for mitotic entry. Time is in minutes and time 0 corresponds to SPB separation. Maximum projections of z stacks. Bars: 4 μm.
B: Timing of Rlc1-mcherry cortical recruitment in same experiments as in A. Mid1nsm-4GFP: n=20. Mid1nsm6Ala-4GFP: n=39. Mid1nsmΔ50–100-4GFP: n=26. P value in Student t-test for equality between Mid1nsm6Ala or Mid1nsmΔ50–100 and Mid1nsm <10−10 and 10−12 respectively.
C: Contractile ring assembly modes in same experiments as in A. Green: without filaments, blue: with filaments, red: with filaments starting at tips. Mid1nsm-4GFP: n=28. Mid1nsm6Ala-4GFP: n=62. Mid1nsmΔ50–100-4GFP: n=27.
D: Percentage of misplaced septa in same strains as in A and in mid1Δ cells. Cells were grown at 30°C. Error bars: SD; 6 independent counts of 100 cells.
See also Figure S3.
In Mid1nsmΔ50–100 mutant (Fig 3A bottom panel), which lacks half the region containing Plo1 phosphosites, Rlc1 recruitment occurred with a mean delay of ~11 minutes compared to control cells (Fig 3A–B), similar to mid1Δ cells (Fig S3F). As in plo1-1 cells, recruitment frequently started away from the medial cortex, sometimes at the very cell tip, leading to the formation of disorganized Myosin II filaments [14]. Moreover, the Mid1nsmΔ50–100 mutant showed strong ring compaction defects (Fig 3A,C) and was excluded from contractile rings during all steps of ring formation (Fig 3A, S3B). Division plane positioning was also strongly defective (~60% misplaced septa, Fig 3D, S3C). However, these defects were less severe than in mid1Δ cells (~60% and ~95% misplaced septa, Fig 3D, S3C) suggesting that Mid1 can promote contractile ring assembly independently of Myosin II recruitment.
The relatively mild phenotype of the Mid1nsm-6ala mutant suggested that Plo1 may phosphorylate additional residues. Indeed, Mid1-1–100-6Ala could still be phosphorylated by Plo1 in vitro (Fig S3D). Introducing the T517A mutation, which reduced Mid1 interaction with Plo1, aggravated the Mid1nsm6Ala phenotype (Fig S3E–H). However, mutating other Plo1 consensus sites located within the Mid1 1–100 fragment (S7 and T51; Fig 2E), or Plo1 consensus sites outside of Mid1 1–100 fragment phosphorylated in vivo during mitosis (S293, S347, S388) or in vitro by Plo1 (S167, S371) (data not shown) did not aggravate the Mid1nsm-6ala phenotype (data not shown).
In contrast, mutation to alanine of S42 and S67, which were detected as phosphorylated in vivo during mitosis (see Fig 2E, Table S2), produced defects very similar to the Mid1nsmΔ50–100 mutant when added to the Mid1nsm6Ala mutant (Fig S3E–H) indicating that phosphorylation of S42 and S67 participates in Mid1 activation. Although S42 and S67 do not match typical Plo1 consensus sites, this does not exclude them as potential Plo1 targets as Polo kinases can phosphorylate sites only loosely related to the consensus (see [21]).
Mid1 residues 1–100 associate with the contractile ring and interact with a C-terminal fragment of Rng2
We next tested if Mid1 residues 1–100 are sufficient for Myosin II recruitment by fusing residues 1–100 to Mid1 Cter (Mid1 1–100-Cter). Mid1 Cter provides a cortical anchor for residues 1–100 [24] and contains a Plo1 binding site (RQSTPV site, aa 514–519). As shown previously, it did not associate with the contractile ring (Fig 4A left panel and S4A, top panel) [24]. In contrast, Mid1 1–100-Cter colocalized with Rlc1 at the contractile ring (Fig 4A, right panel and S4A–C) and rescued mid1 deletion to a great extent (Fig 4B). Myosin II recruitment was still delayed and its recruitment pattern was disorganized (Fig S4A) suggesting that phosphorylation of Mid1 1–100 might be less efficient in this context. Of note, Mid1 1–100-Cter cannot associate with medial cortical nodes and lacks a nuclear localization domain (aa 450–506), impairing Mid1 spatial regulation [7]. It cannot interact either with Clp1, altering the dynamics of contractile ring components [29].
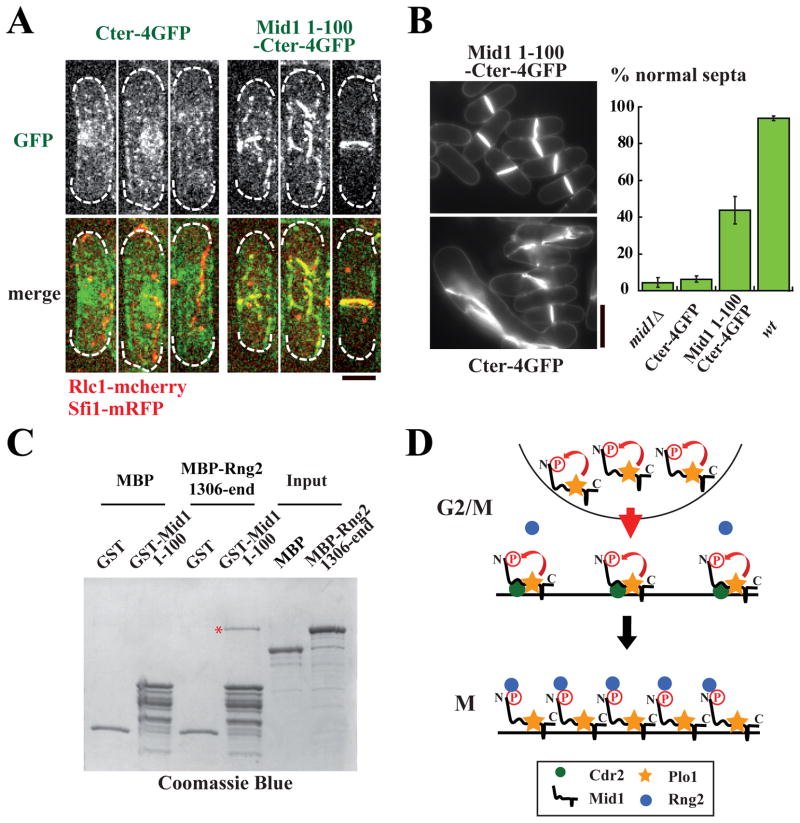
Figure 4. Mid1 residues 1–100 drive Mid1 localization at the contractile ring and interact with a Rng2 fragment.
A: Localization of Cter-4GFP (left panel) and Mid1 1–100-Cter-4GFP (right panel) during mitosis in cells deleted for mid1 expressing Rlc1-mcherry and Sfi1-mRFP. Maximum projections of z stacks. Bar: 4 μm.
B: Cell wall and septum staining (left) and percentage of normally placed septa (right) in same cells as in A, wild type (wt) or mid1Δ cells. Cells were grown at 30°C. Bar: 10 μm. Error bars: SD, 6 independent counts of 100 cells.
C: In vitro binding of Rng2 C-termibrnal fragment (MBP-Rng2-1306-end) on Mid1 1–100 fragment (GST-Mid1 1–100). Proteins are stained with coomassie blue. GST and MBP are used as negative controls. Red star: MBP-Rng2-1306-end bound to GST-Mid1 1–100.
D: Model for Mid1 activation by Plo1 at the onset of mitosis. In G2/M, Cdc2-dependent phosphorylation of Plo1 binding site RQST517PV favors Plo1 interaction with Mid1. Phosphorylation of Mid1 by Plo1 triggers Mid1 export from the nucleus, reinforces its localization at the medial cortex and couples the position of the division plane to nuclear position. Plo1 phosphorylation of domain 1–100, which interacts with a C-terminal fragment of Rng2, also triggers myosin II recruitment to medial cortical nodes and initiates the process of contractile ring assembly.
See also Figure S4.
When we introduced the 6Ala mutations in Mid1 1–100-Cter, in combination or not with S42A and S67A, the ability of Mid1 1–100-Cter to colocalize with Rlc1 at the contractile ring was reduced (Fig S4B–C). Of note, these constructs retained the ability to partially complement mid1 deletion (Fig S4D), raising the possibility that hypo-phosphorylated Mid1 1–100 can interact with ring components, at least transiently.
Finally, we asked which contractile ring component directly interacted with Mid1 1–100. We found that a C-terminal fragment of Rng2 (aa 1306 to end), an IQGAP recruited to medial cortical nodes at the same time as Myosin II in a Mid1-dependent manner [12, 13, 17], could interact with Mid1 1–100 in vitro (Fig 4C). This interaction was observed in the absence of Mid1 1–100 phosphorylation by Plo1 at high protein concentrations. Nevertheless, our in vivo analysis of phospho-defective Mid1nsm and Mid1 1–100-Cter suggests that this interaction is finely tuned by Plo1 in vivo.
In conclusion, our work provides evidence that Mid1 amino acids 1–100 are targeted by Plo1, which phosphorylates multiple serine and threonine residues to indirectly trigger Myosin II recruitment a few minutes before SPBs separate. We propose that this recruitment may be mediated by Rng2, a C-terminal fragment of which directly interacts with this Mid1 fragment (see Fig 4D). Whether Rng2 directly interacts with Myosin II remains an open question that will be important to address in the future.
As Plo1 gets quickly activated at mitotic entry [30], Plo1-dependent regulation of Mid1 export and Myosin II recruitment to medial cortical nodes provides a controlled mechanism to couple early stages of contractile ring assembly with mitotic onset. In addition, Plo1 activates the SIN pathway which triggers septation [30, 31] and functions in parallel to Mid1 to promote contractile ring assembly [32, 33] (see also [4]). These data altogether reveal mechanisms by which Plo1 acts as a key temporal coordinator of contractile ring assembly events in fission yeast.
Polo kinase Plk1, the metazoan counterpart to Plo1, is also a key regulator of cytokinesis in animal cells, where it regulates Cyk4/MgcRacGAP interaction with Ect2 RhoGef, leading to RhoA activation and contractile ring assembly at the cell equator (see [1, 2]). Anillin, the functional homolog of Mid1, physically interacts with RhoA and Cyk4/MgcRacGAP and serves as a scaffold for RhoA signalling and contractile ring assembly (see [34] for a review). It will be important to determine if Plk1 regulates Anillin scaffolding activities in animal cells, similar to Plo1-dependent regulation of Mid1 activity.
Supplementary Material
Acknowledgments
We thank P. Russell and D. McCollum for strains and plasmids, Mercè Guzmán-Vendrell and Anna Feoktistova for experimental assistance, the Curie imaging facility for spinning-disc microscope maintenance and technical help and Phong Tran and Sergio Rincón for critical reading of the manuscript. This work was supported by ANR, ARC, LNCC “programme labellisation” and Mairie de Paris “programme emergence” as well as the HHMI, of which KLG is an Investigator. Maria Almonacid received doctoral fellowships from MESR and ARC. Support was provided to JLJ through a GVSU Presidential Grant and DMC by the National Institutes of Health grant F32-GM076897 and GVSU CSCE Research and Development grant.
Footnotes
Supplemental data contain experimental procedures, 4 supplementary figures and 3 supplementary tables. Figure S1 to S4 relate to Figure 1 to 4 respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- 2.Oliferenko S, Chew TG, Balasubramanian MK. Positioning cytokinesis. Genes Dev. 2009;23:660–674. doi: 10.1101/gad.1772009. [DOI] [PubMed] [Google Scholar]
- 3.Pollard TD, Wu JQ. Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts-Galbraith RH, Gould KL. Stepping into the ring: the SIN takes on contractile ring assembly. Genes Dev. 2008;22:3082–3088. doi: 10.1101/gad.1748908. [DOI] [PubMed] [Google Scholar]
- 5.Chang F, Woollard A, Nurse P. Identification and characterization of fission yeast mutants defective in actin ring assembly and placement. J Cell Sci. 1996;109:131–142. doi: 10.1242/jcs.109.1.131. [DOI] [PubMed] [Google Scholar]
- 6.Sohrmann M, Fankhauser C, Brodbeck C, Simanis V. The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev. 1996;10:2707–2719. doi: 10.1101/gad.10.21.2707. [DOI] [PubMed] [Google Scholar]
- 7.Almonacid M, Moseley JB, Janvore J, Mayeux A, Fraisier V, Nurse P, Paoletti A. Spatial control of cytokinesis by Cdr2 kinase and Mid1/anillin nuclear export. Curr Biol. 2009;19:961–966. doi: 10.1016/j.cub.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 8.Morrell JL, Nichols CB, Gould KL. The GIN4 family kinase, Cdr2p, acts independently of septins in fission yeast. J Cell Sci. 2004;117:5293–5302. doi: 10.1242/jcs.01409. [DOI] [PubMed] [Google Scholar]
- 9.Moseley JB, Mayeux A, Paoletti A, Nurse P. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature. 2009;459:857–860. doi: 10.1038/nature08074. [DOI] [PubMed] [Google Scholar]
- 10.Motegi F, Mishra M, Balasubramanian MK, Mabuchi I. Myosin-II reorganization during mitosis is controlled temporally by its dephosphorylation and spatially by Mid1 in fission yeast. J Cell Biol. 2004;165:685–695. doi: 10.1083/jcb.200402097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vavylonis D, Wu JQ, Hao S, O’Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 12.Wu JQ, Kuhn JR, Kovar DR, Pollard TD. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell. 2003;5:723–734. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- 13.Wu J-Q, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol. 2006:175. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paoletti A, Chang F. Analysis of mid1p, a protein required for placement of the cell division site, reveals a link between the nucleus and the cell surface in fission yeast. Mol Biol Cell. 2000;11:2757–2773. doi: 10.1091/mbc.11.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohkura H, Hagan IM, Glover DM. The conserved Schizosaccharomyces pombe kinase plo1, required to form a bipolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2 cells. Genes Dev. 1995;9:1059–1073. doi: 10.1101/gad.9.9.1059. [DOI] [PubMed] [Google Scholar]
- 16.Bahler J, Steever AB, Wheatley S, Wang Y, Pringle JR, Gould KL, McCollum D. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J Cell Biol. 1998;143:1603–1616. doi: 10.1083/jcb.143.6.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eng K, Naqvi NI, Wong KC, Balasubramanian MK. Rng2p, a protein required for cytokinesis in fission yeast, is a component of the actomyosin ring and the spindle pole body. Curr Biol. 1998;8:611–621. doi: 10.1016/s0960-9822(98)70248-9. [DOI] [PubMed] [Google Scholar]
- 18.Bahler J, Nurse P. Fission yeast Pom1p kinase activity is cell cycle regulated and essential for cellular symmetry during growth and division. EMBO J. 2001;20:1064–1073. doi: 10.1093/emboj/20.5.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacIver FH, Glover DM, Hagan IM. A ‘marker switch’ approach for targeted mutagenesis of genes in Schizosaccharomyces pombe. Yeast. 2003;20:587–594. doi: 10.1002/yea.983. [DOI] [PubMed] [Google Scholar]
- 20.Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10:265–275. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- 21.Park JE, Soung NK, Johmura Y, Kang YH, Liao C, Lee KH, Park CH, Nicklaus MC, Lee KS. Polo-box domain: a versatile mediator of polo-like kinase function. Cell Mol Life Sci. 2010;67:1957–1970. doi: 10.1007/s00018-010-0279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reynolds N, Ohkura H. Polo boxes form a single functional domain that mediates interactions with multiple proteins in fission yeast polo kinase. J Cell Sci. 2003;116:1377–1387. doi: 10.1242/jcs.00314. [DOI] [PubMed] [Google Scholar]
- 23.Elia AE, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 24.Celton-Morizur S, Bordes N, Fraisier V, Tran PT, Paoletti A. C-terminal anchoring of mid1p to membranes stabilizes cytokinetic ring position in early mitosis in fission yeast. Mol Cell Biol. 2004;24:10621–10635. doi: 10.1128/MCB.24.24.10621-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kilmartin JV. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J Cell Biol. 2003;162:1211–1221. doi: 10.1083/jcb.200307064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Goff X, Motegi F, Salimova E, Mabuchi I, Simanis V. The S. pombe rlc1 gene encodes a putative myosin regulatory light chain that binds the type II myosins myo3p and myo2p. J Cell Sci. 2000;113(Pt 23):4157–4163. doi: 10.1242/jcs.113.23.4157. [DOI] [PubMed] [Google Scholar]
- 27.Naqvi NI, Wong KC, Tang X, Balasubramanian MK. Type II myosin regulatory light chain relieves auto-inhibition of myosin-heavy-chain function. Nat Cell Biol. 2000;2:855–858. doi: 10.1038/35041107. [DOI] [PubMed] [Google Scholar]
- 28.Nakajima H, Toyoshima-Morimoto F, Taniguchi E, Nishida E. Identification of a consensus motif for Plk (Polo-like kinase) phosphorylation reveals Myt1 as a Plk1 substrate. J Biol Chem. 2003;278:25277–25280. doi: 10.1074/jbc.C300126200. [DOI] [PubMed] [Google Scholar]
- 29.Clifford DM, Wolfe BA, Roberts-Galbraith RH, McDonald WH, Yates JR, 3rd, Gould KL. The Clp1/Cdc14 phosphatase contributes to the robustness of cytokinesis by association with anillin-related Mid1. J Cell Biol. 2008;181:79–88. doi: 10.1083/jcb.200709060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka K, Petersen J, MacIver F, Mulvihill DP, Glover DM, Hagan IM. The role of Plo1 kinase in mitotic commitment and septation in Schizosaccharomyces pombe. EMBO J. 2001;20:1259–1270. doi: 10.1093/emboj/20.6.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulvihill DP, Petersen J, Ohkura H, Glover DM, Hagan IM. Plo1 kinase recruitment to the spindle pole body and its role in cell division in Schizosaccharomyces pombe. Mol Biol Cell. 1999;10:2771–2785. doi: 10.1091/mbc.10.8.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hachet O, Simanis V. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev. 2008;22:3205–3216. doi: 10.1101/gad.1697208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Y, Yan H, Balasubramanian MK. Assembly of normal actomyosin rings in the absence of Mid1p and cortical nodes in fission yeast. J Cell Biol. 2008;183:979–988. doi: 10.1083/jcb.200806151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piekny AJ, Maddox AS. The myriad roles of Anillin during cytokinesis. Seminars in Cell & Developmental Biology. 2010;21:881–891. doi: 10.1016/j.semcdb.2010.08.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.