Abstract
Full-malaria (http://fullmal.ims.u-tokyo.ac.jp), a database for full-length cDNAs from the human malaria parasite, Plasmodium falciparum has been updated in at least three points. (i) We added 8934 sequences generated from the addition of new libraries, so that our collection of 11 424 full-length cDNAs covers 1375 (25%) of the estimated number of the entire 5409 parasite genes. (ii) All of our full-length cDNAs and GenBank EST sequences were mapped to genomic sequences together with publicly available annotated genes and other predictions. This precisely determined the gene structures and positions of the transcriptional start sites, which are indispensable for the identification of the promoter regions. (iii) A total of 4257 cDNA sequences were newly generated from murine malaria parasites, Plasmodium yoelii yoelii. The genome/cDNA sequences were compared at both nucleotide and amino acid levels, with those of P.falciparum, and the sequence alignment for each gene is presented graphically. This part of the database serves as a versatile platform to elucidate the function(s) of malaria genes by a comparative genomic approach. It should also be noted that all of the cDNAs represented in this database are supported by physical cDNA clones, which are publicly and freely available, and should serve as indispensable resources to explore functional analyses of malaria genomes.
INTRODUCTION
Malaria is the most devastating parasitic disease in the world; it kills more than a million people every year. Plasmodium falciparum is the causative agent of the lethal form of malaria in humans. Thus, the recent completion of the genome sequencing for P.falciparum, ∼23 Mb on 14 chromosomes (seven finished and seven unfinished) has been a great milestone, which provides invaluable information about this organism (1–5). Mass spectrometry and oligonucleotide array techniques have been utilized to characterize ∼5000 candidate genes (6,7). However, these techniques depend upon the correct annotation of the gene structure. Furthermore, to understand the mechanism(s) by which the parasite controls expression of genes throughout its complicated life cycle, the elucidation of transcription factors and binding motifs are mandatory.
Full-malaria started as a database for full-length cDNA clones produced from the erythrocyte-stage parasite of P.falciparum using the oligo-capping method, while the genome sequencing efforts were concurrently underway (8,9). It consisted of 5′ one-pass information, supported by corresponding physical plasmid clones, which are deposited at MR4 (http://www.malaria.mr4.org/).
NEW FEATURES
In this update, we made two additional libraries from P.falciparum and determined 8934 sequences. Originally we used a full-length enriched library from erythrocyte-stage parasites of P.falciparum and reported 5′ end one-pass sequence of 2490 random clones (8). Since then, we have produced two additional libraries from parasites, which were grown under different condition(s), and determined a total of 11 424 clones. Determined sequences were compared with genome nucleotide sequences and displayed on the graphical map along with annotated and predicted genes with three different software packages (PlasmoDB). In total, 1375 genes were represented by full-length clones. Their physical plasmids are available for various experiments (Table 1).
Table 1. The numbers of predicted annotated genes and genes represented by full-length clones are shown for Plasmodium falciparum and Plasmodium yoelii.
| Chromosome | P.falciparum | P.yoelii orthologues | ||||||
|---|---|---|---|---|---|---|---|---|
| Annotated genes | Genes represented by full cDNAs | Genes represented by ESTs | Total represented genes | Annotated genes | Genes represented by full cDNAs | Genes represented by ESTs | Total represented genes | |
| Chr1 | 155 | 28 | 18 | 44 | 81 | 10 | 53 | 53 |
| Chr2 | 224 | 50 | 43 | 80 | 136 | 26 | 83 | 87 |
| Chr3 | 245 | 61 | 59 | 114 | 195 | 35 | 120 | 129 |
| Chr4 | 249 | 63 | 42 | 91 | 163 | 40 | 107 | 114 |
| Chr5 | 330 | 69 | 84 | 146 | 261 | 59 | 157 | 172 |
| Chr6 | 319 | 99 | 71 | 153 | 253 | 46 | 155 | 163 |
| Chr7 | 297 | 59 | 53 | 104 | 214 | 58 | 120 | 137 |
| Chr8 | 299 | 82 | 73 | 150 | 233 | 50 | 136 | 144 |
| Chr9 | 366 | 97 | 114 | 176 | 281 | 49 | 142 | 157 |
| Chr10 | 404 | 105 | 106 | 191 | 295 | 65 | 173 | 184 |
| Chr11 | 514 | 132 | 127 | 230 | 380 | 61 | 217 | 225 |
| Chr12 | 533 | 132 | 120 | 230 | 425 | 88 | 243 | 259 |
| Chr13_1 | 682 | 194 | 166 | 327 | 573 | 116 | 312 | 344 |
| Chr13_2 | 5 | 0 | 0 | 0 | 3 | 1 | 2 | 2 |
| Chr14 | 776 | 204 | 171 | 327 | 636 | 122 | 326 | 353 |
| Unmapped1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unmapped2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unmapped3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Unmapped4 | 7 | 0 | 0 | 0 | 7 | 1 | 1 | 1 |
| Total | 5409 | 1375 | 1247 | 2363 | 4136 | 827 | 2347 | 2524 |
As the genome sequences became publicly available, all the cDNA sequences were mapped on 14 chromosomes using BLAT and sim4 programs (10,11) and the exact alignment was graphically presented.
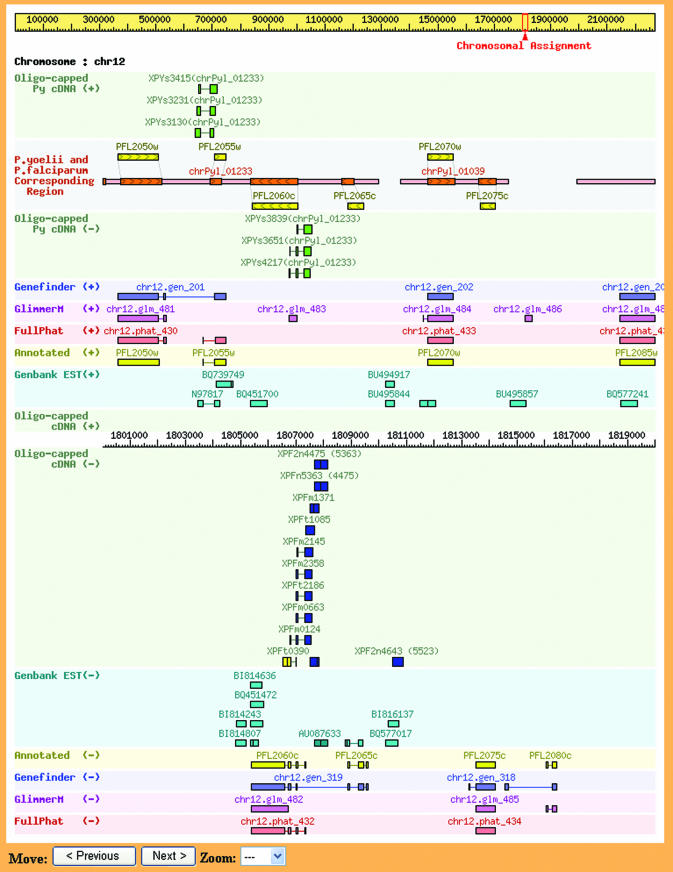
The chromosome map is viewed by choosing the chromosome number and the positions of both ends of the region of interest, or by searching for the Full-malaria clone name or the annotated gene name (Fig. 1). The magnification level can easily be changed. Alternatively, BLASTN will search for similar sequences within the database, enabling the location of the gene to be determined. Regarding each of the genes, hydropathy plot analysis and motif searches (Pfam: http://www.ebi.ac.uk/interpro/) were performed based on the deduced amino acid sequences and the results are represented graphically. Predictions of protein subcellular localization is also possible, using PSORT, PSORTII (http://psort.ims.u-tokyo.ac.jp) and SubLoc (http://www.bioinfo.tsinghua.edu.cn/SubLoc/eu_batchpredict.htm) (Fig. 1).
Figure 1.
(Next page) A view of the map showing a region of chromosome 12 (1800001–182000). The scale in the center shows the position within the P.falciparum genome sequence. Structures of the annotated genes and genes predicted by Genefinder, GlimmerM and FullPhat are shown as colored boxes. Boxes above the scale indicate that the genes are in the positive direction and those below are in the negative direction. Full-malaria clones are shown in the boxes nearest to the scale. Blue box, full-length clone; dark blue, probably full-length clone; light blue, possibly full-length clone; yellow, non-full clone. GenBank ESTs are shown in turquoise. In the upper part of the map, P.yoelii contigs are aligned with the P.falciparum genome, as described in the text. Red line, unique alignment; blue line, alignment with multiple sites; purple line, chimeric contig. Brown boxes represent the aligned P.yoelii predicted genes. Yellow boxes next to the contig line are the P.falciparum annotated genes. Boxes above the line are plus direction and those below the line are minus direction. Arrows in boxes also show the forward direction of the genes. A click on the contig line will open the alignment table.
We incorporated EST sequence data downloaded from GenBank and mapped on the chromosomes. Interestingly, some Full-malaria clones and ESTs represent different sets of genes. Using both Full-malaria cDNAs and ESTs, numerous modifications in gene structures were identified, including the existence of non-coding exon(s), alternative splicing events, correction of splicing and even the identification of hitherto unknown genes. A summary of the statistics from the current Full-malaria database is shown in Table 1.
Furthermore, in order to provide a useful platform for the comparative genomics of Plasmodium species, we constructed a full-length cDNA library from murine malaria parasite Plasmodium yoelii, which was propagated in vivo. As a result of random sequencing analysis, we determined 4257 5′end one-pass sequences. We also mapped those cDNA sequences along with 5×-coverage draft genome sequences of this organism (12) (Fig. 1 upper part). Comparisons of contig nucleotide sequences of P.yoelii with the amino acid sequences of annotated genes of P.falciparum using TBLASTN, successfully aligned 1740 contigs with 4136 genes (Figs 1 and 2). Synteny is conserved in all P.yoelii genes at the genomic level, except for one contig in which the gene order is reversed.
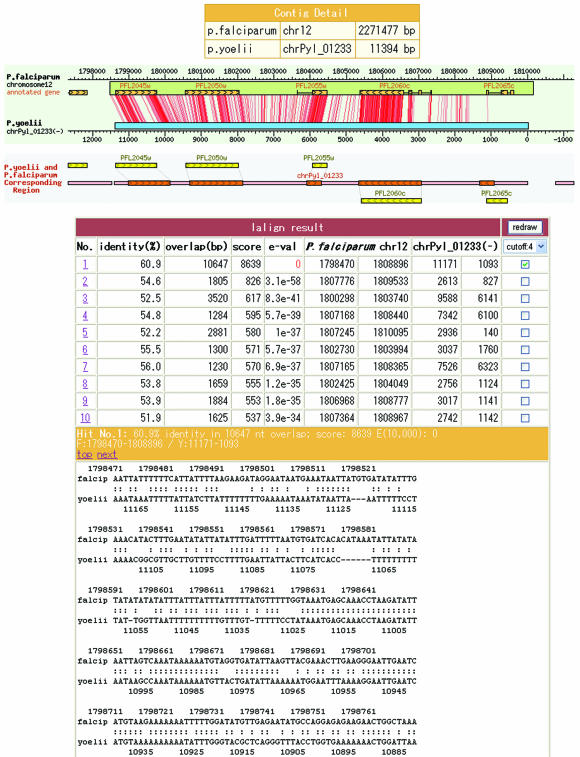
Figure 2.
The results of TBLASTN are shown in table and graphic view. A click of the Lalign button will show the results of Lalign (as in Fig. 3).
The sequence alignments were further analyzed at the nucleotide level using Lalign (13). These results are shown in the P.falciparum chromosome map and a click on the P.yoelii contig box will display the details of these comparisons (Fig. 3). Furthermore, at the nucleotide level synteny is quite well preserved between these two species. The locations of full-length clones are mostly in accordance with the predicted gene structures. Comparison of the promoter regions of both species is of great interest.
Figure 3.
Similarity of the local nucleotide sequences is shown as red lines. A click on the Redraw button will show a new picture of the alignment at a different level.
Comparative analysis of full-length cDNA of P.falciparum and conservation of amino acid sequences with P.yoelii revealed that the start sites of some of the annotated genes are predicted falsely. The actual gene may start from a position further downstream. Some very large annotated genes seem to represent two or more genes. Indeed, exact information on full-length cDNAs supported by physical full-length cDNA clones is indispensable for precise annotation of the correct gene structures. For further information regarding genes for which revision of the annotation should be necessary, please refer to our database (http://fullmal.ims.u-tokyo.ac.jp/annotation); the details of this issue will be described elsewhere (J. Watanabe, M. Sasaki, Y. Suzuki and S. Sugano, in preparation). Expansion of comparative analysis to genome sequences along with full-length cDNA of other apicomplexan organisms will be also useful for investigations of evolution and for analysis of the pathogenicity of respective parasites.
Acknowledgments
ACKNOWLEDGEMENTS
We thank DYNACOM Co., Ltd for providing experienced technical assistance. Nucleotide sequences and gene predictions were downloaded from PlasmoDB (http://plasmoDB.org). This database has been constructed and maintained by a Grant-in-Aid for Publication of Scientific Research Results from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Gardner M.J., Hall,N., Fung,E., White,O., Berriman,M., Hyman,R.W., Carlton,J.M., Pain,A., Nelson,K.E., Bowman,S. et al. (2002) Genome sequence of the human malaria parasite Plasmodium falciparum. Nature, 419, 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Florens L., Washburn,M.P., Raine,J.D., Anthony,R.M., Grainger,M., Haynes,J.D., Moch,J.K., Muster,N., Sacci,J.B., Tabb,D.L. et al. (2002) A proteomic view of the Plasmodium falciparum life cycle. Nature, 419, 520–526. [DOI] [PubMed] [Google Scholar]
- 3.Hall N., Pain,A., Berriman,M., Churcher,C., Harris,B., Harris,D., Mungall,K., Bowman,S., Atkin,R., Baker,S. et al. (2002) Sequence of Plasmodium falciparum chromosomes 1, 3–9 and 13. Nature, 419, 527–531. [DOI] [PubMed] [Google Scholar]
- 4.Gardner M.J., Shallom,S.J., Carlton,J.M., Salzberg,S.L., Nene,V., Shoaibi,A., Ciecko,A., Lynn,J., Rizzo,M., Weaver,B. et al. (2002) Sequence of Plasmodium falciparum chromosomes 2, 10, 11 and 14. Nature, 419, 531–534. [DOI] [PubMed] [Google Scholar]
- 5.Hyman R.W., Fung,E., Conway,A., Kurdi,O., Mao,J., Miranda,M., Nakao,B., Rowley,D., Tamaki,T., Wang,F. et al. (2002) Sequence of of Plasmodium falciparum chromosome 12. Nature, 419, 534–537. [DOI] [PubMed] [Google Scholar]
- 6.Lasonder E., Ishihama,Y., Andersen,J.S., Vermunt,A.M., Pain,A., Sauerwein,R.W., Eling,W.M., Hall,N., Waters,A.P., Stunnenberg,H.G. et al. (2002) Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature, 419, 537–542. [DOI] [PubMed] [Google Scholar]
- 7.Le Roch K.G., Zhou,Y., Blair,P.L., Grainger,M., Moch,J.K., Haynes,J.D., De la Vega,P., Holder,A.A., Batalov,S., Carucci,D.J. et al. (2003) Discovery of gene function by expression profiling of the malaria parasite life cycle. Science, 301, 1503–1508. [DOI] [PubMed] [Google Scholar]
- 8.Watanabe J., Sasaki,M., Suzuki,Y. and Sugano,S. (2001) FULL-malaria: a database for a full-length enriched cDNA library from human malaria parasites, Plasmodium falciparum. Nucleic Acids Res., 29, 70–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki Y. and Sugano,S. (2003) Construction of a full-length enriched and a 5′-end enriched cDNA library using the oligo-capping method. Methods Mol. Biol., 221, 73–91. [DOI] [PubMed] [Google Scholar]
- 10.Florea L., Hartzell,G., Zhang,Z., Rubin,G.M. and Miller,W. (1998) A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res., 8, 967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kent W.J. (2002) BLAT—the BLAST-like alignment tool. Genome Res., 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlton J.M., Angiuoli,S.V., Suh,B.B., Kooij,T.W., Pertea,M., Silva,J.C., Ermolaeva,M.D., Allen,J.E., Selengut,J.D., Koo,H.L. et al. (2002) Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature, 419, 512–519. [DOI] [PubMed] [Google Scholar]
- 13.Huang X., Miller,W., Schwartz,S. and Hardison,R.C. (1992) Parallelization of a local similarity algorithm. Comput. Appl. Biosci., 8, 155–165. [DOI] [PubMed] [Google Scholar]