Abstract
IL-6/Jak2 signaling is viewed critical for persistent Stat3 activation in cancer. However, IL-6-induced Stat3 activity is transient in normal physiology. Here we identify a mechanism important for persistent Stat3 activation in tumor cells and the tumor microenvironment. We show that sphingosine-1-phosphate receptor 1 (S1PR1), a G-protein-coupled receptor for lysophospholipid sphingosine-1-phosphate (S1P), is elevated in Stat3-positive tumors. Stat3 is a transcription factor for the S1pr1 gene. Enhanced S1pr1 expression activates Stat3 and upregulates Il6 gene expression, thereby accelerating tumor growth and metastasis. Conversely, silencing S1pr1 in tumor cells or immune cells inhibits tumor Stat3 activity, tumor growth and metastasis. S1P/S1PR1-induced Stat3 activation is persistent, in contrast to transient Stat3 activation by IL-6. S1PR1 activates Stat3 in part by upregulating Jak2 tyrosine kinase activity. We demonstrate that Stat3-induced S1pr1 expression, as well as S1P/S1PR1 pathway, is important for persistent Stat3 activation in cancer cells and the tumor microenvironment and for malignant progression.
Aberrant IL-6-Jak-Stat3 signaling in cancer cells has emerged as an important mechanism for cancer initiation, development and progression1–9. In addition to its direct importance to tumor cells, a major role of paracrine and autocrine IL-6/Stat3 signaling in facilitating tumor progression and inflammatory cell-mediated transformation has been demonstrated1,6–8,10,11. However, in normal physiology, IL-6/gp130 induced Jak/Stat3 signaling is tightly regulated, due to the existence of negative feedback mechanisms for gp130 signaling and Jak family tyrosine kinases12,13. Although a constellation of growth factors and cytokines can stimulate Stat3 activity, which could have synergistic effects on prolonging Stat3 activation, many growth factors, cytokines and other factors-induced Stat3 activity in tumors requires the IL-6-Jak2 signaling pathway6. These observations collectively raise the critical question of how Stat3 remains persistently activated in cancer, especially in tumor cells in which Stat3 activation is mainly driven by IL-6 and in tumor stromal non-transformed cells such as myeloid cells, where IL-6-induced Stat3 activation plays a critical role for tumor initiation and progression.
An ideal model to study this crosstalk between tumor cells and tumor stromal cells is the B16 tumor, which display relatively low levels of Stat3 activity in cell culture, but is greatly enhanced in vivo, at least in part through IL-611,14. Our PCR-based microarrays using B16 tumor-derived myeloid cells indicated that, among several known Stat3 downstream genes, including IL-6, IL-1β and VEGF15–17, the largest difference in gene expression levels between Stat3+/+ and Stat3−/− tumor myeloid cells was S1pr1, also known as Edg1 (Supplementary Table 1).
The S1PR1 gene encodes one of the G-protein-coupled receptors for sphingosine-1-phosphate (S1P), a biologically active metabolite of sphingolipid18,19. A critical role of S1P-S1PR1 in lymphocyte egress and chemotaxis20–23, cell proliferation/survival, and tumor angiogenesis/metastasis has been shown24–26, However, it remains poorly defined at the molecular level how S1PR1 mediates such complex biological responses18,19. Our findings here provide new insights into the downstream molecular events of S1P/S1PR1 signaling and identify a mechanism for persistent Stat3 activation in tumors.
RESULTS
Stat3 activity elevates S1pr1 expression in tumors
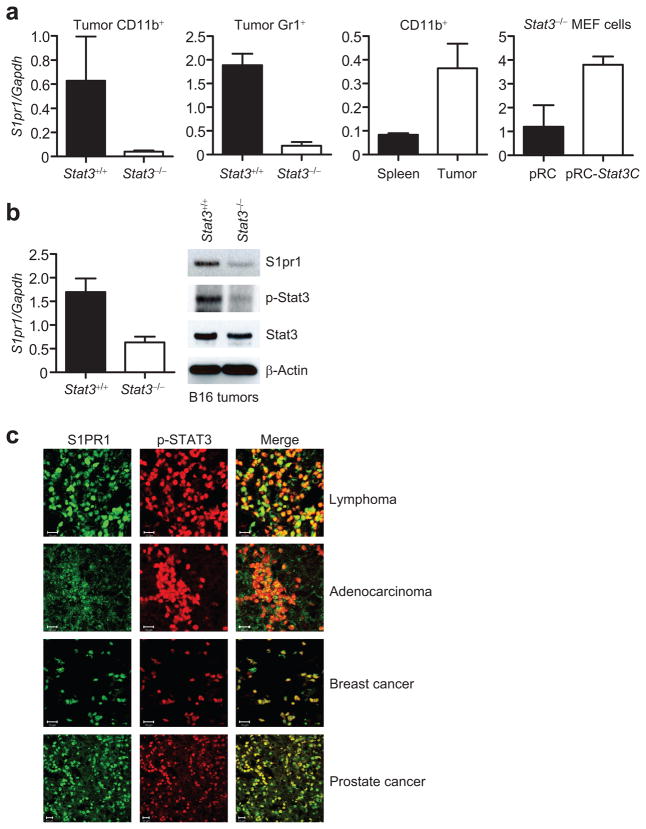
Targeted gene ablation of Stat3 in the myeloid compartment reduces tumor growth, and also Stat3 activity in the entire tumor14,27, including tumor cells (Supplementary Fig. 1a). Real-time PCR using various populations of tumor myeloid cells indicated that ablating Stat3 inhibited S1pr1 expression (Fig. 1a, panels 1 and 2), confirming our PCR-based microarray analysis (Supplementary Table 1). S1pr1 expression was elevated in tumor-derived myeloid cells relative to their normal splenic myeloid cells (Fig. 1a, panel 3). Introducing constitutively-active Stat3 mutant, Stat3C28, into Stat3−/− MEF cells increased S1pr1 expression (Fig. 1a, panel 4). Furthermore, B16 tumors in mice with Stat3−/− myeloid cells displayed reduced S1pr1 expression, at both the RNA and protein levels, compared to B16 tumors with Stat3+/+ myeloid cells (Fig. 1b). However, expression of S1pr2 and S1pr3 was not upregulated by Stat3 in tumor-infiltrating immune cells nor in whole tumors (Supplementary Fig. 1b).
Figure 1.
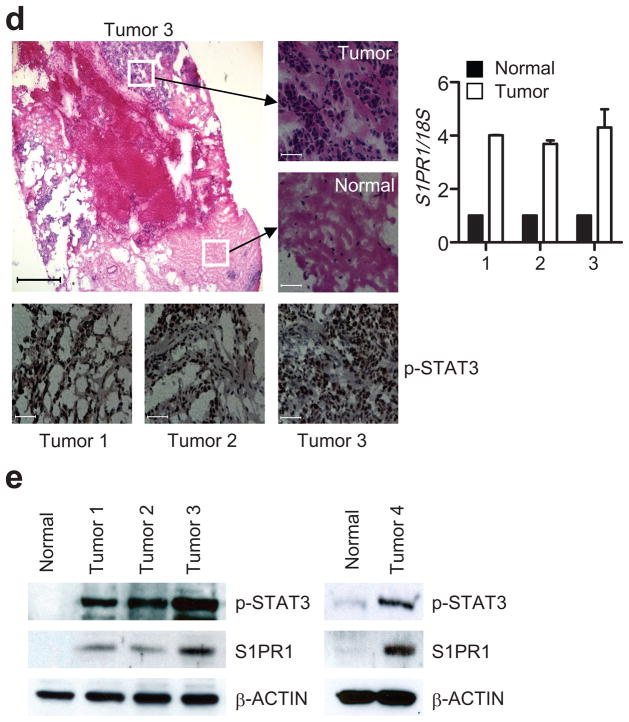
Stat3 activity in tumors promotes S1pr1 expression. (a) Quantification of S1pr1 mRNA expression by real-time RT-PCR in B16 tumor-infiltrating CD11b+ and Gr1+ myeloid cells in mice with Stat3+/+ and Stat3−/− hematopoietic cells, also in tumor vs. splenic CD11b+ cells, and in Stat3−/− MEF cells transfected with indicated plasmids. Data represent means ± s.d. (n = 3). (b) Real-time RT-PCR and Western blotting measuring S1pr1 mRNA and protein expression in whole B16 tumors. Data represent means ± s.d. (n = 3). (c) Immunofluorescent staining showing co-expression of p-STAT3 and S1PR1 in human tumor tissue sections. Scale bar, 10 μm. (d) Measurement of S1PR1 mRNA levels in normal vs. malignant human breast tissues. Top panels: H&E staining to identify normal vs. malignant patches in the breast tissue sections; lower panels, IHC to detect p-STAT3. Scale bars; H&E, 500 μm; IHC, 100 μm. Right panel: quantification of S1PR1 mRNA levels by real-time RT-PCR using RNAs prepared from malignant and normal patches of human breast tumor sections. Values from normal tissue areas are designated as 1, means ± s.d. (n = 2). (e) Western blotting showing S1PR1 protein levels in normal vs. tumor breast tissues. Human breast tumor sections from the same tumors (1–3) were used for both RNA and protein preparations.
Immunohistochemical analyses indicated that expression of S1PR1 and phosphorylated STAT3 (p-STAT3) overlaped in diverse human tumor tissues (Fig. 1c and Supplementary Fig. 2a). The elevated expression of S1PR1 was confirmed in areas with mostly malignant tumor cells vs. those with mainly non-malignant cells of human breast tissue sections by real-time RT-PCR (Fig. 1d). We also performed H&E and p-STAT3 staining on the consecutive tissue sections to determine normal vs. malignant areas and STAT3 activity, respectively (Fig. 1d). We then employed Western blotting to confirm the correlation between STAT3 activity and S1PR1 expression in human breast cancer tissues (Fig. 1e). Detection of reduced S1PR1 protein levels by knocking down S1PR1 in a human cancer cell line supported that the antibody to S1PR1 specifically detects S1PR1 by Western blotting (Supplementary Fig. 2b).
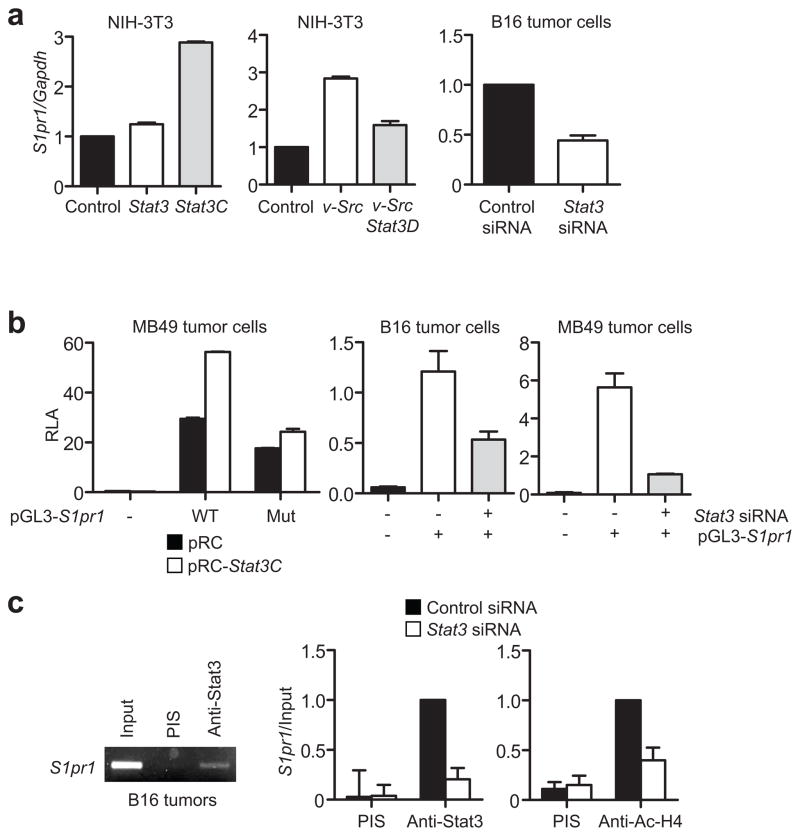
We next determined whether Stat3 might be a transcriptional activator of the S1pr1 gene. Upregulating Stat3 activity in NIH-3T3 fibroblasts by transfecting a vector encoding Stat3C, or v-Src that activates Stat329, or co-transfecting v-Src with a dominant-negative mutant, Stat3D, showed that Stat3 activity modulated S1pr1 mRNA expression (Fig. 2a, left and middle). siRNA knockdown of Stat3 in B16 tumor cells also lowered S1pr1 mRNA (Fig. 2a, right). MB49 tumor cells were transfected with S1pr1 promoter/luciferase reporter vector (pGL3-S1pr1. WT). Co-expression of Stat3C with the S1pr1 reporter construct increased S1pr1 promoter activity, which was abrogated by mutating the Stat3 DNA-binding site in the S1pr1 promoter (positioned at −588; pGL3-S1pr1. Mut) or by co-transfecting with Stat3 siRNA (Fig. 2b).
Figure 2.
Stat3 is a direct transcriptional activator of S1pr1. (a) Real-time RT-PCR showing S1pr1 transcription in cells with various levels of Stat3 activity. S1pr1 mRNA levels in NIH-3T3 cells transfected with indicated expression vectors or in tumor cells with Stat3 siRNA. Results are means ± s.d. (n = 3). Values from control vector or control siRNA are designated as 1. (b) Luciferase assay to analyze the effect of Stat3 activity on S1pr1 promoter activity. WT = wild-type pGL3-S1pr1; Mut = pGL3-S1pr1 with mutated Stat3-binding site. Relative Luciferase Activity (RLA) was calculated as a ratio of Firefly and Renilla luciferase activity. Results are shown as means ± s.e.m. of RLA performed in triplicates. Data represent one of 3 experiments. (c) ChIP assays showing Stat3 binding to the S1pr1 promoter. The chromatins were prepared from either B16 tumors, or B16 tumor cells cultured in medium supplemented with tumor supernatant derived from C-4 mouse melanoma cells transfected with control or Stat3 siRNA. PCR (left) or quantitative real-time PCR (middle and right) using primers for S1pr1 promoter region showing immunoprecipitated DNA fragments. Data in the middle and right were first calculated as [DNA amount in antibody-specific IP – DNA in PIS IP]/[DNA in input], and then compared with the data from antibody-specific IP sample, which is designated as 1.
We next carried out chromatin immunoprecipitation (ChIP) assays to assess whether Stat3 directly binds to the S1pr1 promoter. Stat3 protein bound to the S1pr1 promoter in B16 tumors (Fig. 2c, left), and in cultured B16 tumor cells treated with conditioned medium supplemented with C-4 melanoma tumor cell supernatant, which induces Stat3 activation14. Stat3 knockdown by siRNA abrogated Stat3 binding to the S1pr1 promoter and reduced association of acetylated histone H4 with the promoter, indicating the requirement of Stat3 for S1pr1 promoter activity (Fig. 2c, middle and right). Furthermore, STAT DNA-binding activity to the S1PR1 promoter in human T cells correlated with STAT3 activity (Supplementary Fig. 3).
S1pr1 promotes tumor progression by activating Stat3
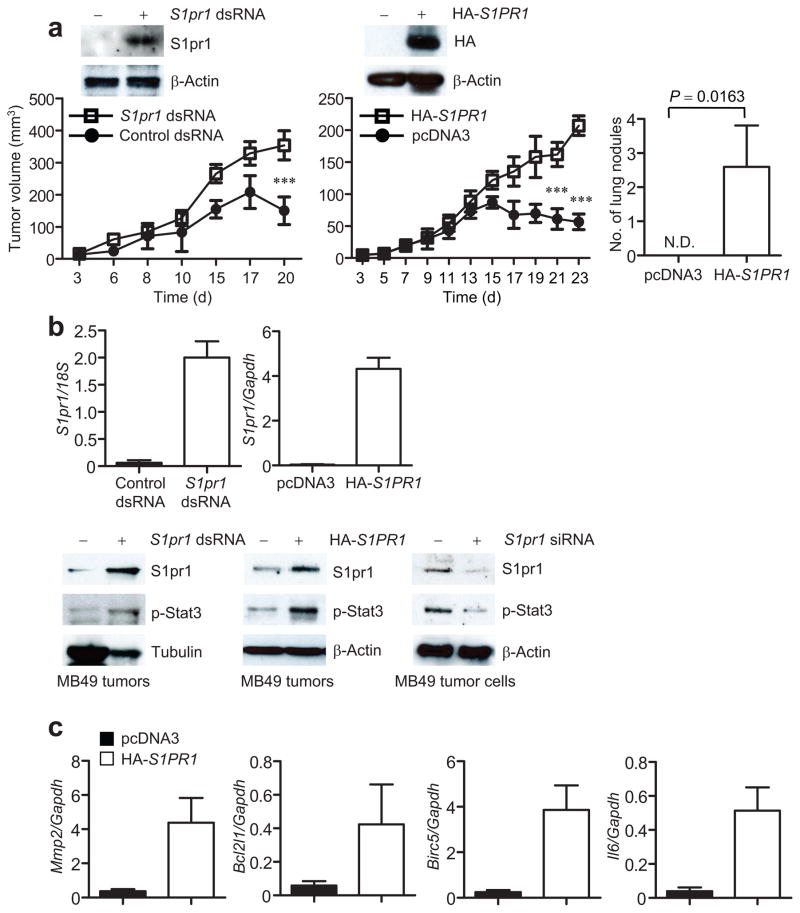
Although silencing S1pr1 in tumor endothelial cells by siRNA or neutralizing S1P with an antibody in mouse tumor models inhibits tumor growth24,26, whether elevated S1pr1 expression in tumor cells directly promotes tumor growth remained to be determined. To increase endogenous S1pr1 expression in tumor cells, we transfected MB49 mouse bladder tumor cells with a double-stranded 21-mer RNA. The selected double-stranded RNA is complementary to the S1pr1 promoter and can increase endogenous S1pr1 expression (Supplementary Fig. 4a). The ability of certain double-stranded short RNAs (dsRNA) derived from promoter regions that specifically activate gene expression has been described30. MB49 tumors do not grow progressively in female mice because these tumor cells express a male antigen31. Increasing endogenous S1pr1 expression in tumor cells by the dsRNA allowed the tumors to grow progressively in both female and male mice (Fig. 3a, left and Supplementary Fig. 4b). To confirm this finding, we transiently transfected into MB49 tumor cells a plasmid vector expressing the human S1PR1 gene, which increased S1PR1 expression (Fig. 3a, right top). We then challenged mice with both control and S1PR1-expressing MB49 tumor cells. Results from these experiments confirmed that increasing S1PR1 expression in tumor cells promoted both tumor growth and metastasis (Fig. 3a). We also confirmed the tumor-promoting effects of S1PR1 in the B16 tumor model (Supplementary Fig. 4c).
Figure 3.
Overexpression of S1pr1 in tumor cells promotes tumor progression and activates Stat3. (a) S1PR1 expression in tumor cells on tumorigenesis in mice. Insets in top panels: Western blotting showing increased expression of S1pr1 in MB49 tumor cells by either dsRNA or plasmids encoding S1PR1. Left, Tumor growth was measured at the indicated time points; means ± s.e.m., n = 6. Right, numbers of spontaneous lung metastatic nodules in mice with control and S1PR1-expressing MB49 tumors. N.D., not detected; means ± s.e.m., n = 6.*, P<0.1.***, P<0.001. (b) Effects of S1pr1 expression on Stat3 activity. Top panels: quantification of S1pr1 mRNA levels by real-time RT-PCR in tumors consisting of both tumor cells and stromal cells including immune cells; means ± s.d., n = 3. Lower left and middle panels: Western blotting showing S1pr1 and p-Stat3 protein levels in MB49 tumors. Lower right panel: quantification of S1pr1 and p-Stat3 protein levels by Western blotting in tumor cells transfected with control or S1pr1 siRNA. (c) Real-time RT-PCR showing RNA expression levels of several Stat3 target genes in MB49 tumors with or without elevated S1pr1 expression; means ± s.d., n = 3.
Analysis of MB49 tumors showed that enhancing S1pr1 expression elevated S1pr1 at both RNA and protein levels, which led to an increase in p-Stat3 levels (Fig. 3b). To confirm that S1pr1 expression affects Stat3 activity in tumor cells, we performed the converse experiment by transfecting MB49 tumor cells with siRNA to silence S1pr1, which inhibited Stat3 activity (Fig. 3b, bottom right). Furthermore, growing MB49 tumors with or without increased S1PR1 expression showed a marked difference in RNA levels of several well-established Stat3 downstream genes (Fig. 3c).
Tumor cell S1pr1 impacts Stat3 in stromal cells
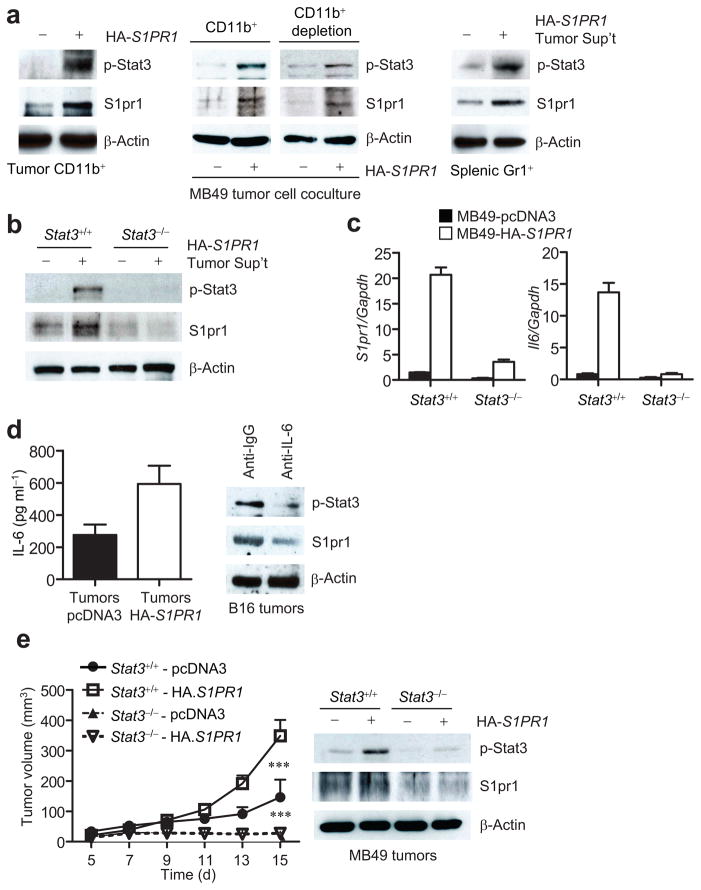
To determine whether Stat3 activation induced by increased S1PR1 expression in tumor cells affected its activity in tumor stromal cells, we examined S1pr1 and p-Stat3 expression in myeloid cells isolated from MB49 tumors formed by transplanting tumor cells with or without increased S1PR1 expression. CD11b+ myeloid cells derived from tumors with increased S1PR1 expression displayed high levels of both S1pr1 and p-Stat3 (Fig. 4a, left).
Figure 4.
Elevated S1PR1 expression in tumor cells increases Stat3 activation and S1pr1 expression in tumor-infiltrating myeloid cells. (a) Western blotting showing p-Stat3 and S1pr1 levels in tumor-infiltrating CD11b+ cells from MB49 tumors (left); in splenic CD11b+ and CD11b− (CD11b+ depleted) subsets co-cultured with MB49 tumor cells expressing either control or S1PR1 vectors (middle); in Gr1+ cells incubated in medium with ex vivo cultured supernatant of S1PR1-expressing MB49 tumors. (b) Western blotting to quantify S1pr1 and p-Stat3 levels in Stat3+/+ and Stat3−/− splenocytes cultured in media supplemented with the same tumor supernatants as in (a). (c) Real-time RT-PCR showing RNA expression levels of S1pr1 and Il6 in Stat3+/+ and Stat3−/− myeloid cells exposed to medium supplemented with tumor supernatants transfected with either control (black bars) or S1PR1-expressing (white bars) vectors, means ± s.e.m., n = 3. (d) Left, ELISA to determine IL-6 production by ex vivo cultured tumor cells from indicated tumors; means ± s.e.m., n = 3. Right, Western blotting of S1pr1 and p-Stat3 in ex vivo cultured B16 tumors incubated with control and IL-6 neutralizing antibody. (e) Left, growth curve of MB49 tumors in mice with Stat3+/+ and Stat3−/− myeloid cells; means ± s.e.m., n = 6. ***, P<0.001. Right, Western blotting measuring p-Stat3 and S1pr1 protein levels in tumors.
We further tested the in vivo finding that S1pr1 expression level could “propagate” from tumor cells to interacting myeloid cells in vitro by co-culturing different myeloid cell populations with both tumor cells and tumor-conditioned supernatants. Data from these experiments indicated that S1pr1 levels in tumor cells increased S1pr1 expression and Stat3 activity in interacting myeloid cells (Fig. 4a, middle). When cultured in the presence of 10% tumor supernatant prepared from S1PR1-expressing MB49 tumor cells, Gr1+ myeloid cells also exhibited enhanced S1pr1 expression and Stat3 activity (Fig. 4a, right). We further determined whether the propagation of S1pr1 upregulation from tumor cells to myeloid cells was Stat3 dependent. Only the S1pr1-positive tumor supernatant was able to upregulate S1pr1 expression in Stat3+/+ but not in Stat3−/− myeloid cells (Fig. 4b).
We next evaluated potential mediators involved in promoting S1pr1-induced Stat3 activation from tumor cells to the tumor stroma. Because the IL-6 cytokine is a major physiological Stat3 activator critical for Stat3-mediated oncogenesis, and given that Stat3 activation increases Il6 gene expression in tumors4,15,32,33, we examined whether increasing S1PR1 expression in tumor cells could impact Il6 expression levels in myeloid cells in vitro. Tumor supernatants from S1PR1-expressing MB49 tumor cells not only increased S1pr1 but also enhanced Il6 expression in myeloid cells, which was Stat3 dependent (Fig. 4c). ELISA confirmed IL-6 protein levels in tumors with and without S1PR1 overexpression (Fig. 4d). Adding IL-6 neutralizing antibody to ex vivo culture of B16 tumors reduced S1pr1 and p-Stat3 levels, demonstrating IL-6 as an important mediator of S1PR1/Stat3 signaling (Fig. 4d).
S1pr1-mediated Stat3 activation is persistent
We also assessed S1pr1/Stat3 co-regulation and Il6 expression in myeloid cells in the tumor milieu. Upon incubation with conditioned media from MB49 tumors, p-Stat3, S1pr1 and IL-6 levels were induced only in S1pr1+/+ but not S1pr1−/− myeloid cells (Supplementary Fig. 5a, middle and right). Ex vivo treatment of tumor-infiltrating immune cells with S1PR1 antagonist, W146, inhibited Stat3 activity (Supplementary Fig. 5b). To determine whether S1pr1-mediated tumorigenesis in vivo depends on Stat3 activity in myeloid cells, we implanted control and S1PR1 expressing MB49 tumor cells subcutaneously into mice with Stat3+/+ and Stat3−/− hematopoietic cells. Without Stat3 in hematopoietic cells, elevated S1PR1 expression in tumor cells failed to promote tumor growth (Fig. 4e). Taken together, these data suggested the reciprocal regulation of S1pr1 and Stat3 in tumor cells and the tumor microenvironment.
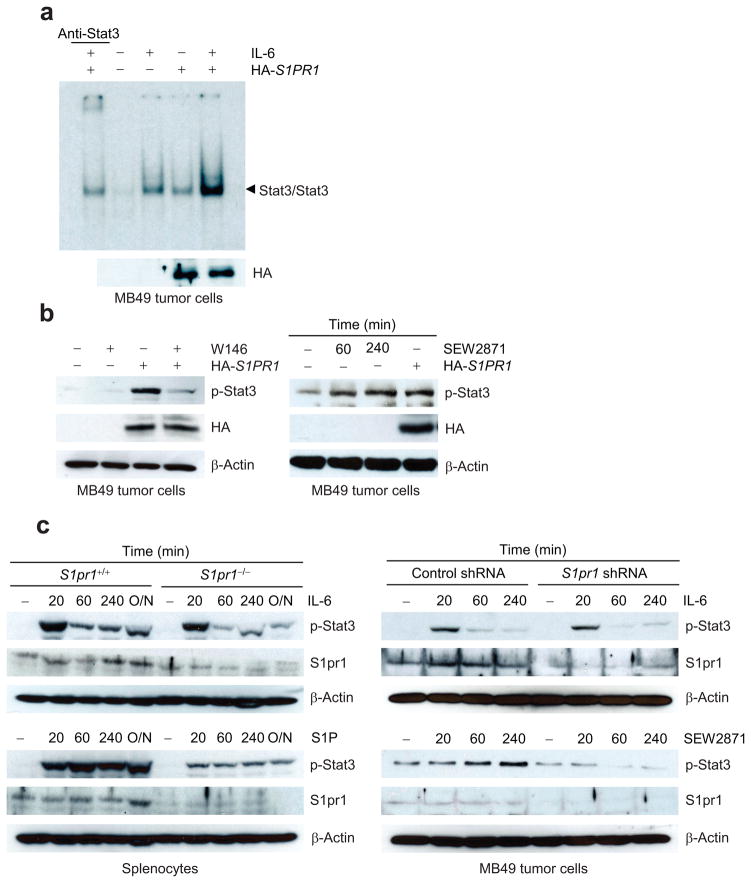
Further characterizing S1PR1 expression-induced Stat3 activation by Stat3 DNA-binding experiments (EMSA) showed that increasing S1PR1 expression led to Stat3 activation without additional stimuli (Fig. 5a). Adding W146 into cultured S1PR1-expressing MB49 tumor cells abolished Stat3 activation (Fig. 5b, left). We also examined whether S1P-S1PR1 signaling activates Stat3. Endothelial cells are known to express S1pr1 and respond to S1P26. Following treatment with S1P, cells from an immortalized mouse endothelial cell line, which displayed low but detectable endogenous Stat3 activity17 and S1pr1 expression, exhibited an induction of Stat3 activation (Supplementary Fig. 6a, left). Increasing endogenous S1pr1 level by its agonist, SEW2871, also increased p-Stat3 in MB49 tumor cells (Fig. 5b, right). S1P-induced Stat3 activation was more pronounced in the S1PR1-expressing tumor cells than in their counterparts without elevated S1PR1 expression (Supplementary Fig. 6a, right).
Figure 5.
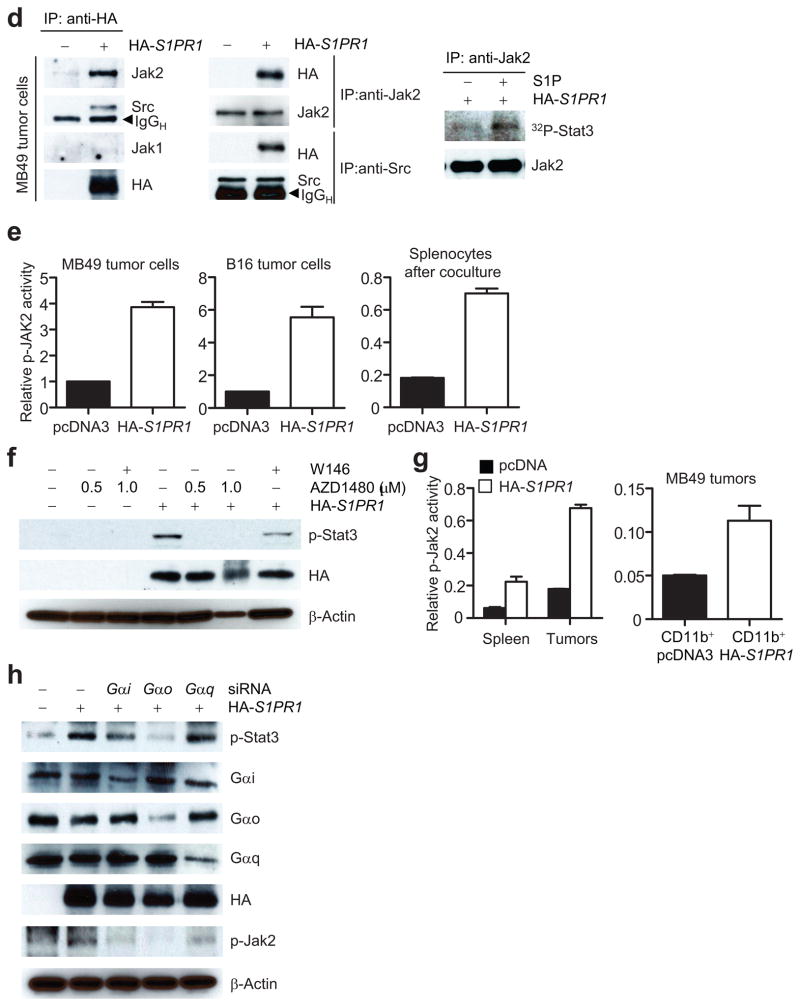
S1P/S1PR1-induced Stat3 activation is persistent and mediated by Jak2. (a) EMSA measuring Stat3 DNA-binding activity in MB49 cells treated by IL-6. (b) Western blotting showing the effects of SEW2871, and W146, on Stat3 activity in MB49 tumor cells transfected with indicated vectors. (c) S1P/S1PR1 signaling induces persistent Stat3 activation. Left, Western blotting comparing p-Stat3 induced by IL-6 or S1P in S1pr1+/+ or S1pr1−/− splenocytes. Right, MB49 tumor cells stably expressing control or S1pr1 shRNA were incubated with either IL-6 or SEW2871. (d) Left, immunoprecipitation by indicated antibodies prior to Western blotting showing S1PR1/Jak2 or Src complexes in control or S1PR1-expressiong MB49 tumor cells. Right, Western blotting showing tyrosyl phosphorylation by Jak2 kinase of recombinant Stat3 protein in vitro using immunoprecipated Jak2 kinase complex from S1PR1-expressing MB49 tumor cells. (e) S1PR1 expression elevates p-Jak2 activity in tumor cells as determined by ELISA-based phosphokinase assay (left and middle). Relative p-Jak2 activity represents the ratio of p-Jak2 in cells expressing S1PR1 over control cells; means ± s.e.m. (n = 3). (f) Assessing the effects of a Jak2 inhibitor on S1PR1-mediated Stat3 activation by Western blotting. (g) ELISA quantifying p-Jak2 in spleens and tumors or tumor-infiltrating CD11b+ myeloid cells. Left, Mice were implanted with indicated MB49 tumor cells. Right, CD11b+ myeloid cells expressing either control or S1PR1 were mixed with MB49 tumor cells before implanting into mice. (h) Western blotting to assess involvement of G protein α subunits for S1PR1/Jak2 signaling.
To further characterize the effects of S1P/S1PR1 vs. IL-6 on Stat3 activation, we treated S1pr1+/+ and S1pr1−/− myeloid cells with either IL-6 or S1P. Stat3 activation was induced upon S1P treatment as early as 20 min and lasted over 18 h, while IL-6-induced Stat3 activation was short lived (Fig. 5c, left). Treating MB49 tumor cells with either IL-6 or SEW2871 generated similar results (Fig. 5c, right). In addition, knocking down S1pr1 in either myeloid cells or tumor cells did not affect IL-6-induced Stat3 activation, while S1PR1 agonists-mediated prolonged Stat3 activation was absent in cells lacking S1pr1 (Fig. 5c).
S1pr1-induced Stat3 activity involves Jak2 kinase
To explore whether Jak2 and Src tyrosine kinases were involved in S1PR1 or S1P/S1PR1-mediated Stat3 activation, we performed immunoprecipitation with an HA-specific antibody to collect HA-tagged S1PR1, followed by Western blot analysis. Results from this experiment indicated S1PR1/Jak2 interaction, which was confirmed by the converse immunoprecipitation using a Jak2 antibody (Fig. 5d, left and middle panels). S1PR1 also interacted with the Src tyrosine kinase together with Jak2, as shown by immunoprecipitation with Src-specific antibody prior to Western blot analysis (Fig. 5d and Supplementary Fig. 6b). In addition, immunoprecipitated Jak2 protein from S1P-stimulated, S1PR1-expressing MB49 tumor cells, had increased kinase activity, as measured by tyrosyl phosphorylation of recombinant Stat3 protein in vitro (Fig. 5d, right). Furthermore, ELISA-based Jak2 phosphokinase assay showed that co-culturing with supernatant collected from S1PR1-expressing tumor cells increased Jak2 kinase activity in both MB49 and B16 tumor cells, and also in splenocytes (Fig. 5e). Blocking Jak2 kinase activity with the Jak2 inhibitor AZD14809 abrogated S1PR1-mediated Stat3 activation in MB49 tumor cells (Fig. 5f). Jak2 activity was also upregulated in tumors harboring elevated S1PR1 (Fig. 5g, left). Additionally, mixing tumor cells with S1PR1-expressing CD11b+ myeloid cells before tumor challenge enhanced Jak2 kinase activity in tumors (Fig. 5g, right).
To further confirm that S1PR1 induces Jak2/Stat3 activation, we silenced the α subunits of G protein using siRNAs against Gα i, o and q in tumor cells overexpressing S1PR1. Consistent with previous findings indicating a requirement of Gα i/o in the S1P/S1PR1 signaling pathway34, and supporting our finding that S1PR1 induces Jak2/Stat3 activation, inhibition of Gαi or Gαo decreased Stat3 activity, and phospho-Jak2 levels, induced by S1PR1 expression in tumor cells (Fig. 5h).
In the case of IL-6 signaling, Jak2 activation induces Stat3-mediated SOCS3 gene expression, which in turn, inhibits Jak2/Stat3 signaling. The question remains whether S1PR1-induced persistent Stat3 activation upregulates SOCS3 expression. In S1PR1 expressing MB49 tumors, SOCS3 mRNA was not elevated, but repressed (Supplementary Fig. 7a). We then examined whether S1PR1 downregulates SOCS3 transcriptional activity by transfecting SOCS3 promoter luciferase construct into MB49 tumor cells together with plasmid containing either pcDNA or HA-S1PR1. Increased S1PR1 expression reduced the promoter activity of SOCS3 (Supplementary Fig. 7b, left). Conversely, blocking S1pr1 by siRNA further enhanced SOCS3 promoter activity in MB49 cells compared to cells expressing control siRNA (Supplementary Fig. 7b, right). IL-6 treatment induced SOCS3 promoter activity was not altered by S1pr1 silencing (Supplementary Fig. 7b, right).
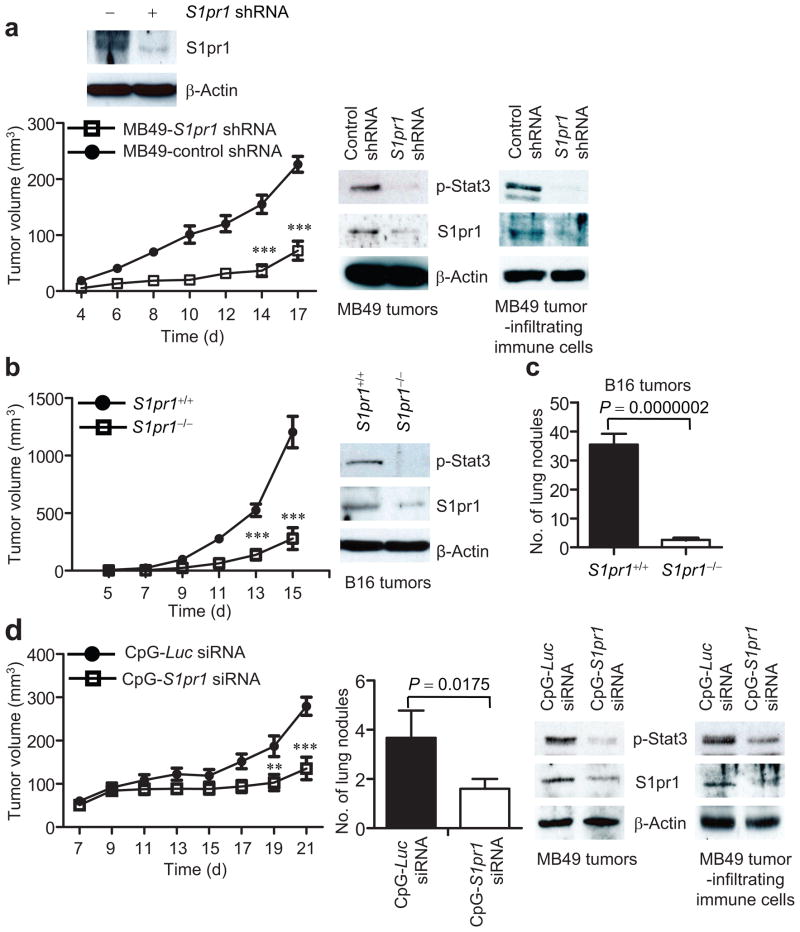
Inhibiting S1pr1 reduces Stat3 activity and tumor progression
We next assessed whether targeting S1PR1 signaling could inhibit Stat3 activity, tumor growth and metastasis. When we implanted into mice MB49 tumor cells stably expressing S1pr1 shRNA, tumor growth, as well as p-Stat3 and S1pr1 expression, was significantly inhibited in tumor and tumor-infiltrating immune cells (Fig. 6a). Furthermore, B16 tumors in mice with S1pr1-deficient myeloid cells showed a marked decrease in tumor Stat3 activity (Fig. 6b). This is consistent with previous studies reporting that ablating Stat3 in myeloid cells affected Stat3 activity in the whole tumor14, including tumor cells (Supplementary Fig. 1a). Lacking S1pr1 alleles in hematopoietic cells also significantly decreased lung metastasis (Fig. 6c).
Figure 6.
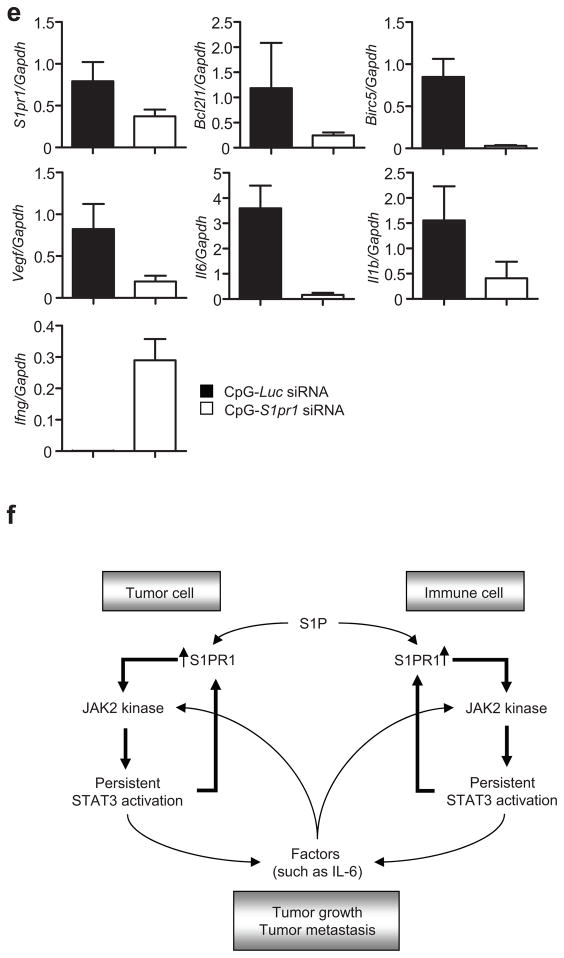
Targeting S1pr1 in either tumor cells or the tumor microenvironment inhibits Stat3 activity, tumor growth and metastasis. (a) Western blotting showing the effect of S1pr1 shRNA on S1pr1 protein expression in MB49 tumor cells (top) and the consequent effects on MB49 tumor growth (lower left, n = 8). Lower right, Western blotting measuring S1pr1 and p-Stat3 expression in MB49 tumors and tumor-associated immune cells. (b) Growth curves to show effects of ablating S1pr1 in hematopoietic cells on tumor growth (left, n = 7). Right, Western blotting to determine Stat3 activity and S1pr1 expression using protein lysates prepared from pooled B16 tumors grown in mice with S1pr1+/+and S1pr1−/− hematopoietic cells. (c) S1PR1/Stat3 signaling and tumor metastasis in vivo. Numbers of lung metastatic nodules in mice with and without S1pr1 in hematopoietic cells receiving systemic B16 tumor cell challenge (n = 10). (d) CpG-S1pr1 siRNA treatment inhibits MB49 tumor progression (left) and numbers of lung metastatic nodules (middle). Western blotting showing the effect of CpG-S1pr1 siRNA in vivo treatment on S1pr1 and p-Stat3 expression in the tumors and tumor-infiltrating immune cells (right). CpG-Luc siRNA = control. Means ± s.e.m., n = 5.*, P<0.1;**, P<0.01;***, P<0.001. (e) Real-time RT-PCR measuring RNA expression of Stat3 downstream genes resulting from targeting S1pr1 in tumors by CpG-S1pr1 siRNA in vivo treatment. Means ± s.d. (n = 3). (f) A proposed model for S1PR1/Stat3 feed-forward loop in both tumor cells and tumor-infiltrating immune cells.
To further evaluate S1pr1 as a molecular target to inhibit Stat3 activity and for cancer therapy, we used an in vivo siRNA delivery approach that links siRNA to a short single-stranded CpG oligonucleotide35. CpG is a ligand for endosomal Toll-like receptor 9, which is mainly expressed in myeloid cells and B cells. Treating mice with established MB49 tumors or B16 tumors with CpG-S1pr1 siRNA conjugate inhibited tumor growth and metastasis, tumor Stat3 activity, and expression of many Stat3 target genes (Fig. 6d,e and Supplementary Fig. 8).
DISCUSSION
Our study has identified a mechanism that facilitates persistent Stat3 activation in cancer, at least in the setting where IL-6/gp130/Jak signaling is the driving force, which is critical for cancer initiation and progression as well as many inflammatory diseases. We show here that S1PR1 expression is elevated in both mouse and human tumors that display persistent Stat3 activity, and also that Stat3 is a direct transcription factor for the S1pr1 gene promoter. Stat3-mediated S1PR1 upregulation in turn, facilitated by S1P and IL-6, contributes to persistent Stat3 activity in both tumor cells and in the tumor microenvironment, thereby promoting malignant progression (Fig. 6f).
Although the importance of persistent Jak2 activation in cancer has become increasingly evident9,36, the underlying mechanisms of continuous Jak2 activation in the tumor microenvironment remain to be further explored. Our findings demonstrate that S1PR1, a G-protein coupled receptor, is capable of activating Stat3, at least in part, through the Jak2 tyrosine kinase. Even though overexpressing a constitutively-activated mutant of Gα subunit, Q205L Gα o protein, in fibroblasts can activate Stat3 through Src kinase37, we show that S1P-S1PR1 signaling in tumor induces Stat3, in part through Gα i/o-mediated Jak2 activation. Src could also contribute to S1PR1-induced Stat3 activation, either directly or indirectly through interacting with Jak2.
We show that S1P/S1PR1 signaling induces persistent Stat3 activation, which is in sharp contrast to the transient nature of Stat3 activation induced by IL-6. Although maintained at low levels in most tissues, S1P level increases under inflammatory conditions and in the tumor microenvironment24,38. Whether Stat3 activity has a regulatory role in S1P levels remains to be determined. Nevertheless, the specificity and activity level of S1P/S1PR1 signaling, which has diverse biological consequences, is in part determined by levels of S1PR119,21,39. By virtue of its ability to directly upregulate S1PR1 expression, Stat3, whose activity levels are modulated by defined cytokines and growth factors, can impact which cell type and when they respond to S1P.
S1PR1 is critical for lymphocyte egress 20–22. It also inhibits TH1-cell cytokines, such as IFN-γ/IL-12, and promotes TH2, including IL-10, and TH17-cell cytokines18,19,40. The critical role of Stat3 in regulating immune responses, in many ways, overlaps with that of S1PR14. Stat3 is also critical for angiogenesis, not only for the production of multiple key angiogenic factors but also for their receptor signaling3–5. Many cytokines and growth factors, such as VEGF, EGF and PDGF, known to interact with S1P/S1PR1 signaling41, are also Stat3 activators and Stat3 in turn promotes their production. Our findings thus link Stat3 signaling directly with the S1P/S1PR1 axis; and their co-regulation in both tumor cells and the tumor stromal cells may have profound biological and therapeutic implications for cancer and other inflammatory diseases.
METHODS
In vivo tumor experiments
Mouse care and experimental procedures were carried out under pathogen-free conditions in accordance with established institutional guidance and approved protocols from the Institutional Animal Care and Use Committee of Beckman Research Institute at City of Hope Medical Center. The generation of Stat3+/+ Stat3−/− mice has been described27. For mice with S1pr1 ablation in hematopoietic cells, we crossed S1pr1flox/flox mice with Mx1-Cre mice.
For subcutaneous MB49 tumor challenge, we injected tumor cells subcutaneously into the flank of female C57BL/6 mice 24 h after transfection with either dsRNA encoding S1pr1 promoter or plasmid expressing HA-tagged S1PR1.
For experimental lung metastasis model, we injected B16 tumor cells intravenously into S1pr1+/+ or S1pr1−/− mice. After 21 days, number of visible metastatic tumor nodules was enumerated in the lung by counting individual nodules.
We killed mice one day after the last tumor measurement in all cases. The detailed methods and sequence for S1pr1 dsRNA and CpG conjugated siRNA are provided in the Supplementary Methods.
Isolation of immune cells
Purification of specific immune subsets and generation of tumor supernatant has been described14,27 and details are provided in the Supplementary Methods.
Sources of human tissues and RNA and protein preparations
We obtained human tissue array slides from the archive of City of Hope Medical Center Pathology Core, and OCT-embedded frozen human breast tumor tissue sections from the Translational Research Core at City of Hope Medical Center, with Institutional Review Board approval. Additional normal and tumor OCT-embedded frozen human breast tissue sections were also purchased from OriGene.
To isolate total RNA from frozen breast tissue sections, we placed the slides on light microscope and while viewing, we gently scraped patches with most either tumor cells or non-malignant cells (confirmed by H&E staining of consecutive sections) with 27 -gauge needle. Tip of the needle was placed into the tube containing lysis buffer for RNA isolation (RNAqueous-Micro kit, Ambion). We used 10 consecutive slides for each RNA preparation.
To extract proteins from human breast cancer tissues, OCT-embedded frozen tissue slides were dried at room temperature for 30 min. Tissues were scraped from the slide and transferred into a tube containing modified RIPA buffer, vortexed briefly, sonicated twice and then subject to three cycles of freezing/thawing in dry ice. After centrifugation, supernatants were collected and stored at −80°C. Five consecutive tissue sections were pooled together for one experiment.
Real-time RT-PCR
Total RNA preparation (mouse tissue or cell lines) and real-time PCR were performed as described35 and details are provided in the Supplementary Methods.
For PCR-based gene profiling in B16 tumor infiltrating CD11b+ myeloid cells, we purchased array plate from SA Bioscience. Data was analyzed according to manufacturer’s instructions.
Immunohistochemical/mmunofluorescent staining and confocal microscopy
IHC staining of OCT-embedded frozen tissue slides were performed as described in the Supplementary Methods.
Immunofluorescent staining and confocal microscopy was performed as described14 and detailed experimental procedures are also provided in the Supplementary Methods.
Promoter activity assay
Promoter activity assay was carried out as described in Supplementary Methods.
Generation of Retroviral S1pr1 Construct and infection
cDNA encoding mouse S1pr1 gene was cloned into MSCV eGFP vector upstream of the internal ribosomal entry site (IRES) site to generate MSCV-S1pr1. Retroviral supernatant was produced using PT-67 packaging cell line (Clontech). We infected CD11b+ myeloid cells (5 × 106 ml−1, 24-well plate) with virus-containing supernatant in the presence of polybrene (8 μg ml−1) using spin infection method (2,450 rpm for 1.5 h).
Chromatin immunoprecipitation (ChIP) assay
We carried out chromatin immunoprecipitation in cells and tissues based on the protocol provided by Millipore-Upstate Biotechnology. Primers used in this study were:5′-TGGACATTCCTGAACTGCTGCT-3′, 3′-TCCGATCCTGGTGAAGCCTTGAAA-5′.
Electrophoretic mobility shift assay (EMSA)
We performed DNA binding assays as previously described1 and details are provided in the Supplementary Methods.
In vitro IP-Kinase assay and phospho-Jak2 ELISA
We used anti-Jak2 antibody to precipitate Jak2 kinase complexes using cell or tissue lysates. Kinase activity was measured in 20 μl reaction buffer containing 1 mM ATP, 10 μCi of 32P-γ-ATP and 0.5 μg of recombinant Stat3 (Active Motif). ELISA-based kinase assay was performed according to supplier’s instruction (Invitrogen).
Statistical analyses
We used two-way ANOVA and Bonferroni post-test to calculate differences in tumor growth; and unpaired t-test to calculate two-tailed P value. P values are shown in figures and legends. Data were analyzed using GraphPad Prism version 4.0 software (GraphPad).
Additional methods
Detailed experimental procedures are described in the Supplementary Methods.
Supplementary Material
Acknowledgments
We thank the Flow Cytometry Core, Light Microscopy Core, Pathology Core, and Translational Research Core, as well as the Animal Facility of the Beckman Research Institute at City of Hope for their superb technical assistance. We would also like to thank P. Chu. of City of Hope Pathology Department for evaluating histology of breast cancer tissue sections, R. L. Proia at US National Institute of Health for generously providing the S1PR1flox/flox mice, V. Poli (University of Turin, Italy) for Stat3−/− MEF cells, T. Ratliff (University of Iowa) for MB49 mouse bladder transitional cell carcinoma cell line, A. Raubitschek (City of Hope Medical Center) for Jurkat human T cell leukemia and Karpas 299 human T cell lymphoma cell lines and S. Priceman of City of Hope for critical reading of this manuscript. This work is funded by National Institutes of Health grant R01CA115815, R01CA115674, R01CA122976 and P50CA107399. Procurement of clinical specimens used in the study was supported by grants from the National Cancer Institute CA 33572 and American Bioscience.
Footnotes
AUTHOR CONTRIBUTIONS
H.Y. and H.L. developed the concept, designed experiments and prepared the manuscript. H.L. also carried out most of experiments, data organization and statistical analysis. J.D, M. Kujawski and Y.L. performed mouse experiments. C.Y. carried out tissue microdissection and RNA preparation. M. Kortylewski performed flow cytometry. Confocal microscopy was performed by A.H. R.J. made conceptual contributions to the studies, particularly S1P/Src and Jak2/Stat3 relationship. G. S and S.F. contributed to the studies for providing clinical relevance of the findings. D.H. synthesized chemical inhibitors that allowed us to perform additional confirmatory experiments.
References
- 1.Catlett-Falcone R, et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10:105–115. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 2.Rebouissou S, et al. Frequent in-frame somatic deletions activate gp130 in inflammatory hepatocellular tumours. Nature. 2009;457:200–204. doi: 10.1038/nature07475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 4.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 6.Gao SP, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bollrath J, et al. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 2009;15:91–102. doi: 10.1016/j.ccr.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Grivennikov S, et al. IL-6 and stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hedvat M, et al. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell. 2009;16:487–497. doi: 10.1016/j.ccr.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ara T, et al. Interleukin-6 in the bone marrow microenvironment promotes the growth and survival of neuroblastoma cells. Cancer Res. 2009;69:329–337. doi: 10.1158/0008-5472.CAN-08-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, et al. IL-17 can promote tumor growth through an IL-6-Stat3 signaling pathway. J Exp Med. 2009;206:1457–1464. doi: 10.1084/jem.20090207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 13.Auernhammer CJ, Bousquet C, Melmed S. Autoregulation of pituitary corticotroph SOCS-3 expression: characterization of the murine SOCS-3 promoter. Proc Natl Acad Sci USA. 1999;96:6964–6969. doi: 10.1073/pnas.96.12.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee H, et al. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell. 2009;15:283–293. doi: 10.1016/j.ccr.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sumimoto H, Imabayashi F, Iwata T, Kawakami Y. The BRAF-MAPK signaling pathway is essential for cancer-immune evasion in human melanoma cells. J Exp Med. 2006;203:1651–1656. doi: 10.1084/jem.20051848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu G, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–2008. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 17.Kujawski M, et al. Stat3 mediates myeloid cell-dependent tumor angiogenesis in mice. J Clin Invest. 2008;118:3367–3377. doi: 10.1172/JCI35213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosen H, Goetzl EJ. Sphingosine 1-phosphate and its receptors: an autocrine and paracrine network. Nat Rev Immunol. 2005;5:560–570. doi: 10.1038/nri1650. [DOI] [PubMed] [Google Scholar]
- 19.Rivera J, Proia RL, Olivera A. The alliance of sphingosine-1-phosphate and its receptors in immunity. Nat Rev Immunol. 2008;8:753–763. doi: 10.1038/nri2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matloubian M, et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 21.Shiow LR, et al. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature. 2006;440:540–544. doi: 10.1038/nature04606. [DOI] [PubMed] [Google Scholar]
- 22.Schwab SR, Cyster JG. Finding a way out: lymphocyte egress from lymphoid organs. Nat Immunol. 2007;8:1295–1301. doi: 10.1038/ni1545. [DOI] [PubMed] [Google Scholar]
- 23.Pappu R, et al. Promotion of lymphocyte egress into blood and lymph by distinct sources of sphingosine-1-phosphate. Science. 2007;316:295–298. doi: 10.1126/science.1139221. [DOI] [PubMed] [Google Scholar]
- 24.Visentin B, et al. Validation of an anti-sphingosine-1-phosphate antibody as a potential therapeutic in reducing growth, invasion, and angiogenesis in multiple tumor lineages. Cancer Cell. 2006;9:225–238. doi: 10.1016/j.ccr.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 25.Spiegel S, Milstien S. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat Rev Mol Cell Biol. 2003;4:397–407. doi: 10.1038/nrm1103. [DOI] [PubMed] [Google Scholar]
- 26.Chae SS, Paik JH, Furneaux H, Hla T. Requirement for sphingosine 1-phosphate receptor-1 in tumor angiogenesis demonstrated by in vivo RNA interference. J Clin Invest. 2004;114:1082–1089. doi: 10.1172/JCI22716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kortylewski M, et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med. 2005;11:1314–1321. doi: 10.1038/nm1325. [DOI] [PubMed] [Google Scholar]
- 28.Bromberg JF, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 29.Yu CL, et al. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 30.Li LC, et al. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci USA. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halak BK, Maguire HC, Jr, Lattime EC. Tumor-induced interleukin-10 inhibits type 1 immune responses directed at a tumor antigen as well as a non-tumor antigen present at the tumor site. Cancer Res. 1999;59:911–917. [PubMed] [Google Scholar]
- 32.Heinrich PC, et al. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem J. 1998;334 (Pt 2):297–314. doi: 10.1042/bj3340297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong Z, Wen Z, Darnell JE., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 34.Okamoto H, et al. EDG1 is a functional sphingosine-1-phosphate receptor that is linked via a Gi/o to multiple signaling pathways, including phospholipase C activation, Ca2+ mobilization, Ras-mitogen-activated protein kinase activation, and adenylate cyclase inhibition. J Biol Chem. 1998;273:27104–27110. doi: 10.1074/jbc.273.42.27104. [DOI] [PubMed] [Google Scholar]
- 35.Kortylewski M, et al. In vivo delivery of siRNA to immune cells by conjugation to a TLR9 agonist enhances antitumor immune responses. Nat Biotechnol. 2009;27:925–932. doi: 10.1038/nbt.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghoreschi K, Laurence A, O’Shea JJ. Selectivity and therapeutic inhibition of kinases: to be or not to be? Nat Immunol. 2009;10:356–360. doi: 10.1038/ni.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ram PT, Horvath CM, Iyengar R. Stat3-mediated transformation of NIH-3T3 cells by the constitutively active Q205L Galphao protein. Science. 2000;287:142–144. doi: 10.1126/science.287.5450.142. [DOI] [PubMed] [Google Scholar]
- 38.Sabbadini RA. Targeting sphingosine-1-phosphate for cancer therapy. Br J Cancer. 2006;95:1131–1135. doi: 10.1038/sj.bjc.6603400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carlson CM, et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 40.Graeler M, Shankar G, Goetzl EJ. Cutting edge: suppression of T cell chemotaxis by sphingosine 1-phosphate. J Immunol. 2002;169:4084–4087. doi: 10.4049/jimmunol.169.8.4084. [DOI] [PubMed] [Google Scholar]
- 41.Igarashi J, et al. VEGF induces S1P1 receptors in endothelial cells: Implications for cross-talk between sphingolipid and growth factor receptors. Proc Natl Acad Sci USA. 2003;100:10664–10669. doi: 10.1073/pnas.1934494100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.