Summary
Polarized segregation of proteins in T cells is thought to play a role in diverse cellular functions including signal transduction, migration, and directed secretion of cytokines. Persistence of this polarization can result in asymmetric segregation of fate-determining proteins during cell division, which may enable a T cell to generate diverse progeny. Here, we provide evidence that a lineage-determining transcription factor, T-bet, underwent asymmetric organization in activated T cells preparing to divide and that it was unequally partitioned into the two daughter cells. This unequal acquisition of T-bet appeared to result from its asymmetric destruction during mitosis by virtue of concomitant asymmetric segregation of the proteasome. These results suggest a mechanism by which a cell may unequally localize cellular activities during division, thereby imparting disparity in the abundance of cell fate regulators in the daughter cells.
Introduction
After the activation of T cells by antigen-presenting cells, the microtubule organizing center, as well as specific transmembrane and intracellular proteins, rapidly undergo reorganization towards the site of intercellular contact (Monks et al., 1998). This polarized reorganization of T cells has been characterized among naïve and antigen-experienced lymphocytes stimulated in vitro (Huse et al., 2006; Ludford-Menting et al., 2005; Maldonado et al., 2004; Stinchcombe et al., 2006) and in vivo (Azar et al.; Barcia et al., 2006; Beuneu et al., 2010; Friedman et al., 2010; Reichert et al., 2001). The acute polarization and redistribution of proteins subsequent to T cell activation has been suggested to regulate signal transduction and facilitate function, such as directed secretion of cytokines and cytolytic granules (Huse et al., 2006; Stinchcombe et al., 2006).
Polarized segregation of proteins may be evident several hours after activation of naïve T cells (Yeh et al., 2008), and this coalescence may even persist through cell division (Chang et al., 2007). The polarized segregation of proteins during mitosis is reminiscent of an evolutionarily conserved phenomenon known as asymmetric cell division, which allows a single parent cell to give rise to two daughter cells with distinct fates (Betschinger and Knoblich, 2004; Knoblich, 2008; Lechler and Fuchs, 2005). During asymmetric division, key fate determinants are localized to one side of the plane of division, resulting in two daughter cells that inherit different amounts of critical determinants. One such determinant in Drosophila neural stem cells is the transcription factor Prospero, which acts as a binary switch between terminal differentiation and self-renewal (Betschinger and Knoblich, 2004). It has been suggested that a T cell may undergo asymmetric division to give rise to daughter cells that are differentially fated towards the effector and memory lineages (Chang et al., 2007). It remains unknown, however, what determinants are unequally partitioned into the daughter cells of a selected T cell and how their asymmetry is mediated.
Several transcriptional regulators have been implicated in regulating fate decisions of effector and memory T cells (Intlekofer et al., 2005; Joshi et al., 2007; Kallies et al., 2009; Rutishauser et al., 2009; Shin et al., 2009). Genetic evidence suggests that the T-box transcription factor, T-bet, is a critical fate determinant in activated naïve CD8+ T cells, promoting differentiation towards the effector fate while repressing development towards the memory fate (Intlekofer et al., 2005; Joshi et al., 2007). In activated CD4+ T cells, T-bet promotes the T helper 1 (Th1) cell fate while repressing the development of the Th2 and Th17 cell lineages (Hwang et al., 2005; Lazarevic et al., 2010; Szabo et al., 2000; Szabo et al., 2002). Small changes in the amount of these factors can have profound influences on T cell fate (Intlekofer et al., 2005; Joshi et al., 2007; Kallies et al., 2009; Rutishauser et al., 2009; Shin et al., 2009; Szabo et al., 2002).
We now provide evidence that in activated naïve T cells undergoing division, T-bet was asymmetrically partitioned between the daughter cells. Moreover, the mechanism by which T-bet asymmetry was mediated appeared to involve proteasome-dependent degradation specifically during mitosis in the setting of asymmetric distribution of the degradation machinery, the proteasome. The localization of the proteasome was opposite to that of T-bet, such that the daughter cell that received less proteasome acquired more T-bet. This reciprocal partitioning, along with the observation that T-bet asymmetry is prevented by inhibiting its proteasome-dependent degradation, indicates that the asymmetric localization of T-bet and proteasome may be related. Inhibiting the polarized segregation of the proteasome during mitosis, moreover, prevented the asymmetric partitioning of T-bet. Together these findings suggest a mechanism of asymmetric cell division whereby asymmetric localization of the proteasome, and consequently unequal degradation of factors targeted for destruction during mitosis, yields unequal partitioning of key fate determinants to two daughter cells.
Results
Asymmetric partitioning of T-bet during T lymphocyte division
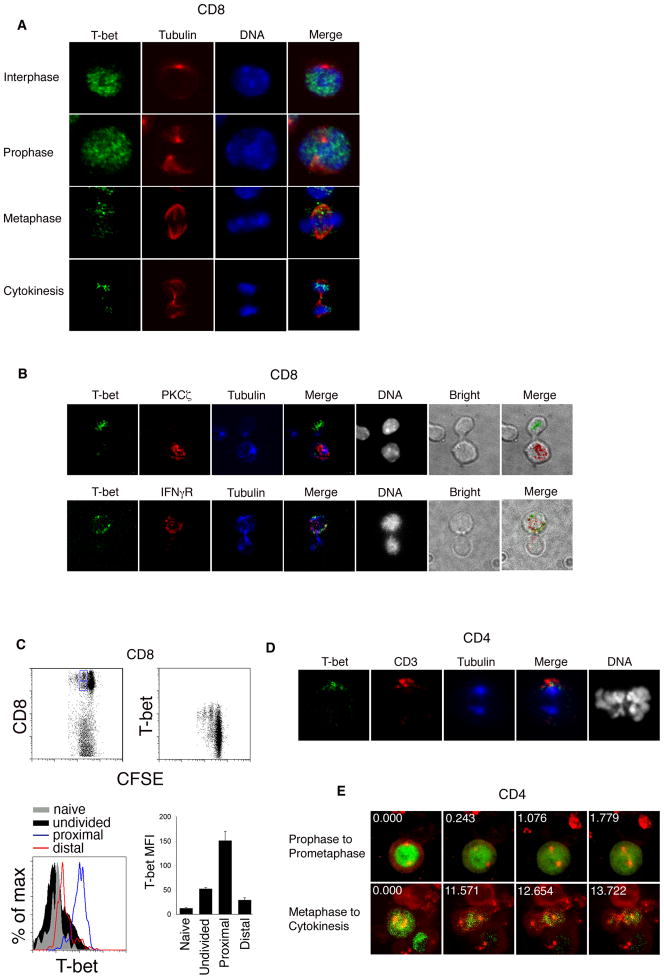
To examine the cellular distribution of T-bet, we employed a model system that has allowed us to examine T cells preparing for their first division in vivo in response to a microbe (Chang et al., 2007). Naïve CD8+ T cells transgenic for the P14 T cell receptor were labeled with a fluorescent dye (CFSE) that allows determination of whether a cell has undergone division. Cells were then adoptively transferred into recipient mice that were infected 24 hours previously with recombinant Listeria monocytogenes bacteria expressing a specific gp33–41 peptide epitope (gp33-L. monocytogenes) recognized by the transgenic T cell receptor. Undivided donor T cells were isolated by flow cytometry at 36 hours after transfer and examined by confocal microscopy. Among activated cells in interphase and prophase, we observed that T-bet was localized in the nucleus (Figure 1A). Among cells in metaphase, we observed a substantial reduction in T-bet signal compared to those in interphase and prophase, suggesting the possibility that T-bet was undergoing degradation prior to cell division. We also observed that among cells in metaphase, T-bet was displaced from the chromatin and localized asymmetrically on one side of the cell. The asymmetry of T-bet persisted into cytokinesis, with unequal amounts of T-bet detected in the conjoined daughter cell pairs (Figure 1B). Based on the preferential partitioning of T-bet into the daughter cell receiving more of the proximal cell marker, interferon-γ receptor (IFNγR), and less of the distal cell marker, Protein Kinase C-zeta (PKCζ (Chang et al., 2007), the greater share of T-bet appeared to be partitioned into the putative proximal daughter cell (Figure 1B).
Figure 1. Asymmetric partitioning of T-bet in dividing T lymphocytes.
(A) Undivided P14 CD8+ TCR transgenic T cells were adoptively transferred into wild-type recipients infected with gp33-L. monocytogenes, harvested at 36h after transfer, and examined by confocal microscopy after staining for T-bet (green), β-tubulin (red), and DNA (blue). Asymmetry of T-bet inheritance was observed in 66% of cells (n=80). (B) Undivided P14 CD8+ TCR transgenic T cells were harvested as in (A) and stained for T-bet (green), PKCζ or IFNγR (red), β-tubulin (blue), and DNA (grayscale). In co-staining experiments where both molecules were asymmetrically inherited, T-bet and the IFNγR were inherited by the same daughter cell in 100% (n=15) of cells, and T-bet and PKCζ were inherited by opposite daughters in 87% (n=14) of cells. (C) CFSE-labeled P14 transgenic CD8+ T cells from recipients infected with gp33-L. monocytogenes were harvested 48h after infection, stained with antibodies to detect CD8 and T-bet, and analyzed by flow cytometry. Putative proximal and distal daughter cells (detected as the second brightest CFSE peak) were gated on the basis of CD8 expression (gates shown in upper left panel). T-bet mean fluorescence intensity (MFI) of the gated proximal (blue histogram), distal (red histogram), naïve (gray filled histogram), and undivided (black filled histogram) populations is shown. Error bars indicate standard error of the mean (SEM). (D) CD4+ T cells were activated in vitro and stained for T-bet (green), CD3 (red), β-tubulin (blue), and DNA (grayscale). In co-staining experiments where both molecules were asymmetrically inherited, T-bet and CD3 were inherited by the same daughter cell in 100% (n=16) of cells. (E) CD4+ T cells were activated in vitro and transduced at 48h with T-bet-GFP and cherry-alpha-tubulin fusion proteins. After 3d, T cells were restimulated in vitro for 24h and imaged by time-lapse confocal microscopy. Prophase cells were imaged through prometaphase (upper panel), and metaphase cells imaged through cytokinesis (lower panel). Asymmetric T-bet partitioning by the daughter cells was observed in 68% of cells (n=23). Relative time (in minutes) is indicated in the upper left-hand corner of each panel. Results are representative of three separate experiments.
We next confirmed that the unequal amounts of T-bet protein acquired by the daughter cells during mitosis persisted after division. We have previously used flow cytometry to distinguish putative proximal and distal daughter populations on the basis of CD8 abundance (Chang et al., 2007). CFSE-labeled P14 transgenic CD8+ T cells were adoptively transferred into recipient mice that were infected 24 hours prior with gp33-L. monocytogenes. At 52 hours post-infection, splenocytes were analyzed by flow cytometry. Examination of T-bet protein amounts revealed greater abundance of T-bet in the putative proximal daughter cells, which expressed higher amounts of CD8, compared to distal daughter cells (Figure 1C). Putative distal daughter cells had higher amounts of T-bet compared to naïve cells and some undivided cells. The amounts of T-bet in the highest-expressing undivided cells appeared to be less than that present in the proximal and distal daughter cells combined, suggesting that re-synthesis of T-bet in the proximal and/or distal daughter cells may follow asymmetric partitioning of pre-existing parent cell T-bet protein during mitosis. As genetic studies have suggested that T-bet drives terminal differentiation of effector T cells while repressing self-renewal of memory CD8+ T cells (Intlekofer et al., 2007; Joshi et al., 2007), asymmetric partitioning of T-bet into the proximal daughter cell is consistent with prior evidence suggesting that the proximal daughter cell gives rise to the effector lineage while the distal daughter cell is the predecessor of the memory lineage (Chang et al., 2007).
In addition to regulating fate decisions in CD8+ T cells, T-bet plays a critical role in the fate choice of CD4+ T cells (Szabo et al., 2000). In a quantitative manner, T-bet promotes Th1 cell differentiation while repressing the development of the Th2 and Th17 cell lineages (Hwang et al., 2005; Lazarevic et al., 2010; Szabo et al., 2000; Szabo et al., 2002). T-bet binds directly to GATA-3 and prevents it from binding to its target DNA (Hwang et al., 2005); T-bet also cooperates with the transcription factor, Runx1, to inhibit the transcription of RORγt (Lazarevic et al., 2010). As with CD8+ T cells, small changes in the amount of T-bet results in profound phenotypic changes. T-bet heterozygous mice, which exhibit only a 50% reduction in T-bet protein relative to wild-type mice (Szabo et al., 2002), exhibit early and dense defects in Th1 cell development and manifest a similar degree of Th2 cell-mediated airway hyperresponsiveness as homozygous T-bet-deficient mice (Finotto et al., 2002; Szabo et al., 2002).
Because of the ability of small differences (50% or less) in the amount for T-bet to alter cell fate and function (Finotto et al., 2002; Intlekofer et al., 2007; Joshi et al., 2007; Szabo et al., 2002), we examined the first daughter cells of CD4+ T cells activated in vitro (Figure S1). We observed a 3.6-fold disparity in T-bet abundance between the T-bet-higher and T-bet-lower daughter cells. The T-bet disparity in the two daughter populations positively correlated with a 3.3-fold greater likelihood to express IFN-γ, and a 4.2-fold more intense IFN-γ signal per expressing cell (Figure S1). Minor disparity in the partitioning of T-bet during the first division of a CD4+ T cell could, thus, influence the subsequent fates of the daughter cells.
To examine the localization of T-bet in dividing CD4+ T cells, we developed a reductionist cell culture-based model system that recapitulated the key features of CD8+ T cell division in vivo. Naïve CD4+ T cells were stimulated with immobilized anti-CD3 and anti-CD28 along with immobilized ICAM1-Fc fusion protein. This approach was taken in order to mimic a polarizing stimulus plus integrin-mediated contact because ICAM1-dependence was one of the defining features of asymmetric T cell division in vivo (Chang et al., 2007) and because immobilized ICAM1-Fc was found to be critical for asymmetric division in vitro (Figure S1). We observed that T-bet was asymmetrically partitioned to the side of the cell that receives more CD3 (Figure 1D). As CD3 is a marker of the immune synapse (Monks et al., 1998), T-bet was partitioned to the side of the cell that is presumed to have been in contact with the stimulus, consistent with the findings in CD8+ T cells activated in vivo (Figure 1B).
To examine the steps leading up to T-bet asymmetry in real time, CD4+ T cells were activated and transduced with retroviruses encoding T-bet-GFP and cherry-alpha-tubulin fusion proteins. Three days later, when the transduced lymphocytes expressing fluorescent fusion proteins were no longer dividing, they were restimulated with immobilized anti-CD3 and ICAM1-Fc fusion protein. Among CD4+ T cells in interphase and prophase, we observed that T-bet was localized in the nucleus, consistent with the staining of endogenous T-bet in CD8+ T cells responding to a microbe in vivo (Figure 1A). During prometaphase, T-bet-GFP began to leak out of the disintegrating nuclear envelope, eventually filling the cytoplasm as it became fully displaced from condensed chromatin in early metaphase (Figure 1E, top panel, and Movie S1). The displacement of T-bet from mitotic chromatin is consistent with the reported behavior of other transcription factors during mitosis, which may be incapable of binding to highly condensed mitotic chromatin (Martinez-Balbas et al., 1995). As metaphase progressed, we observed a decrease in T-bet-GFP fluorescence (Figure 1E, bottom panel, and Movie S2), consistent with the reduction of endogenous T-bet during metaphase in T cells dividing in vivo. As anaphase began and the mitotic spindle began to separate, T-bet-GFP appeared to localize asymmetrically towards one side of the cell, becoming unequally inherited by the incipient daughter cells (Figure 1E, bottom panel, and Movie S2).
T-bet undergoes proteasome-dependent degradation during mitosis
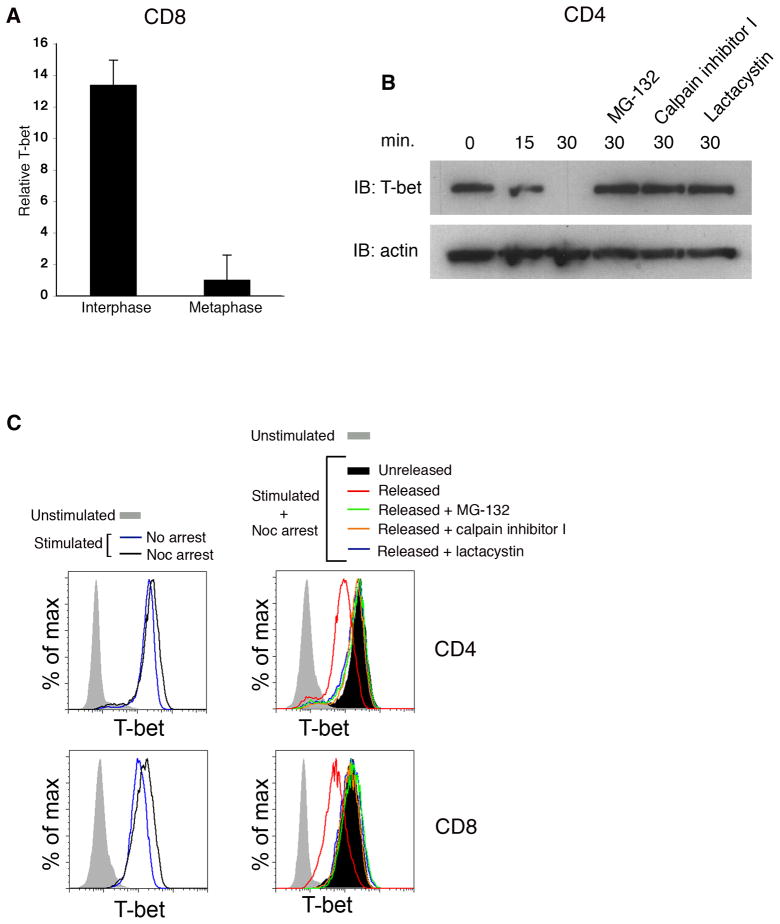
The reduction in T-bet signal observed in cells during metaphase using both static and time-lapse imaging approaches suggested that T-bet might be undergoing degradation just prior to or during its asymmetric localization. Specifically, in experiments where interphase and metaphase blasts were imaged in the same field of view, quantitation of T-bet signal revealed a greater than 90% reduction in metaphase cells compared to interphase cells (Figure 2A). This reduction of T-bet signal was observed in all mitotic T cells, regardless of whether T-bet was partitioned asymmetrically. Using biochemical and flow cytometric approaches, we confirmed that T-bet underwent proteasome-dependent degradation during mitosis. CD4+ or CD8+ T cells were activated in vitro and synchronized with an inhibitor of microtubule polymerization, nocodazole, to enrich for cells reversibly arrested in G2-prometaphase. Cells were then washed free of drug and allowed to progress into metaphase in the presence or absence of proteasome inhibitors. We observed that T-bet appeared to undergo degradation within 30 minutes of release from nocodazole (Figures 2B and 2C). Furthermore, the degradation of T-bet could be prevented by the addition of an inhibitor of proteasome activity (Figures 2B and 2C). Degradation of T-bet was cell cycle-specific, as T-bet underwent degradation after drug washout in cells arrested in G2-M, but not in G1 or S phase (Figure S2).
Figure 2. T-bet undergoes proteasome-dependent degradation during mitosis.
(A) Quantification of T-bet signal in interphase vs. metaphase T cells represented in Figure 1A. T-bet signal was compared between interphase and metaphase blasts imaged in the same field of view (n=61). Error bars indicate SEM. (B) CD4+ T cells were activated in vitro for 24h and then synchronized with nocodazole. Cells were washed free of drug, cultured in vitro with or without the proteasome inhibitors MG-132, calpain inhibitor I, or lactacystin. Cell lysates were prepared at 0, 15, or 30m after nocodazole washout. T-bet and β-actin levels were assessed by immunoblotting. (C) CD4+ or CD8+ T cells were prepared as in (B) and T-bet levels assessed by intracellular staining at 0 and 30m after nocodazole washout. T-bet levels of naïve T cells and activated cells not treated with nocodazole (“freely cycling”) are also shown. Results are representative of three separate experiments.
Asymmetric localization of the proteasome during mitosis
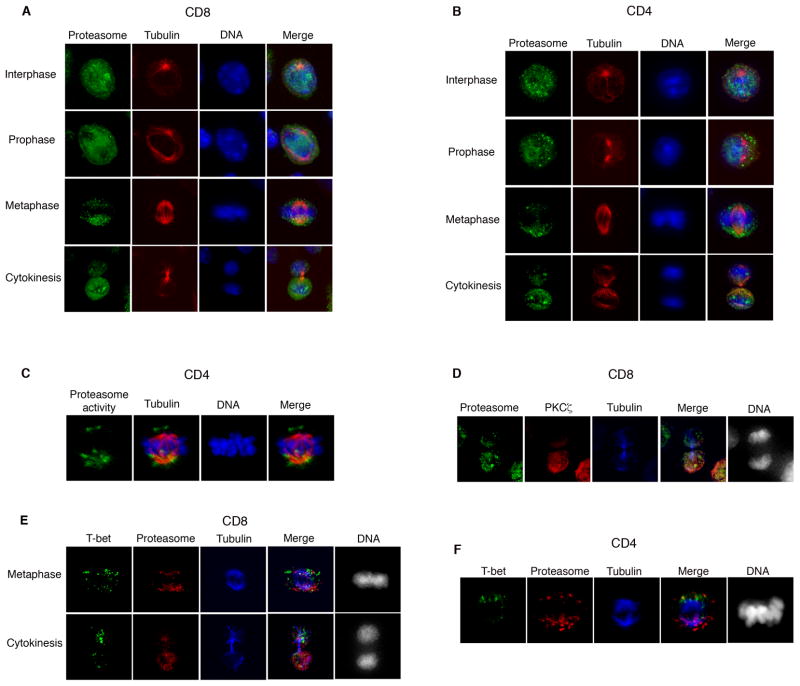
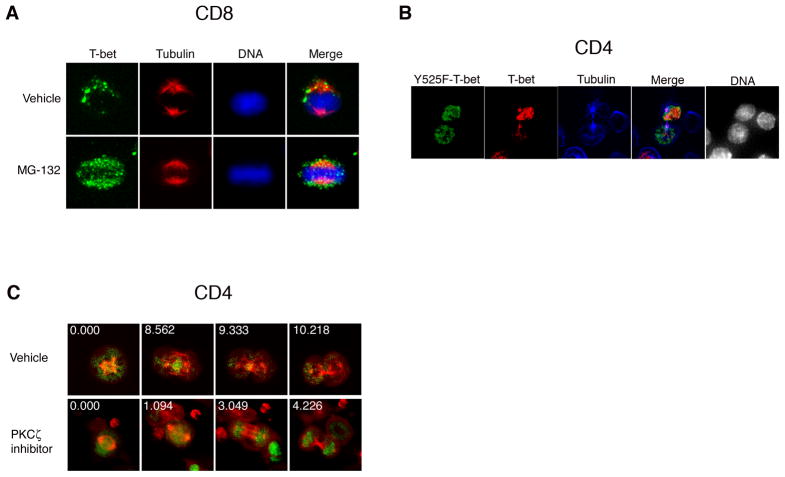
The finding that T-bet underwent degradation specifically during M phase raised the possibility that unequal degradation during mitosis might result in asymmetric partitioning of T-bet into the daughter cells. For asymmetric degradation to occur, however, some component of the destruction process would need to be asymmetrically localized. Consistent with this prediction, examination of activated T lymphocytes dividing in vivo (Figure 3A) and in vitro (Figure 3B) revealed evidence for asymmetry in the localization of the proteasome. During interphase and prophase, the proteasome was localized throughout the cell. During metaphase, however, we observed asymmetric segregation of the proteasome on one side of the lymphocyte, and unequal segregation of the proteasome into daughter cells during cytokinesis. The asymmetric localization of the proteasome, moreover, was confirmed using antibodies to two distinct proteasomal epitopes (Figure S3). Proteasomal asymmetry is not a general feature of cell division, however, as dividing HEK293T cells exhibited equal segregation of the proteasome into the daughter cells (Figure S3).
Figure 3. Unequal proteasomal segregation as a mechanism for asymmetric cell division.
(A)Undivided P14 CD8+ TCR transgenic T cells were harvested as in Figure 1A and stained for the α1 chain of the proteasome 20S subunit (green), β-tubulin (red), and DNA (blue). Asymmetry of proteasome localization was observed in 62% (n=74) of cells. (B) CD4+ T cells were activated in vitro for 28h and stained as in (A). Asymmetry of proteasome localization was observed in 74% (n=125) of cells. (C) CD4+ T cells were activated in vitro for 28h, treated with the proteasome activity probe MVB003 for 2h, and stained for β-tubulin (red), and DNA (blue). Asymmetry of degradative activity was observed in 65% (n=22) of cells. (D) Undivided P14 CD8+ TCR transgenic T cells were harvested as in Figure 1A, and stained for the proteasome 20S α1 subunit (green), PKCζ (red), β-tubulin (blue), and DNA (grayscale). In co-staining experiments where both molecules were asymmetrically inherited, proteasome and PKCζ were inherited by the same daughter cell in 95% of cells (n=34). (E) Undivided P14 CD8+ TCR transgenic T cells were harvested as in Figure 1A, and stained for T-bet (green), proteasome 20S α1 subunit (red), β-tubulin (blue), and DNA (grayscale). In co-staining experiments where both molecules were asymmetrically inherited, proteasome and T-bet were inherited by opposite daughter cells in 90% of cells (n=29). (F) CD4+ T cells were activated as in (B) and stained as in (E). In co-staining experiments where both molecules were asymmetrically inherited, proteasome and T-bet were inherited by opposite daughter cells in 90% of cells (n=14). Results are representative of three separate experiments.
To determine whether asymmetric localization of the proteasome was associated with differential rates of degradation within a mitotic cell, we used the proteasome activity probe, MVB003, which functions as a reporter for degradative activity (Florea et al., 2010). After 24 hours of activation in vitro, T lymphocytes were incubated with the proteasome activity probe and examined by immunofluorescence microscopy. Unequal proteasome activity was observed within mitotic T lymphocytes (Figure 3C), suggesting that both localization and degradative activity of the proteasome were unequal during cell division. In a model wherein asymmetry of T-bet results from unequal degradation by the proteasome, the greater share of T-bet would be predicted to be partitioned into the daughter cell that receives less proteasome. Co-staining experiments using activated CD8+ T lymphocytes dividing in vivo in response to microbe (Figures 3D and 3E) and activated CD4+ T lymphocytes dividing in vitro (Figure 3F) indicated that T-bet was partitioned asymmetrically into the daughter cell that received less proteasome.
The polarity network regulates asymmetry of the proteasome
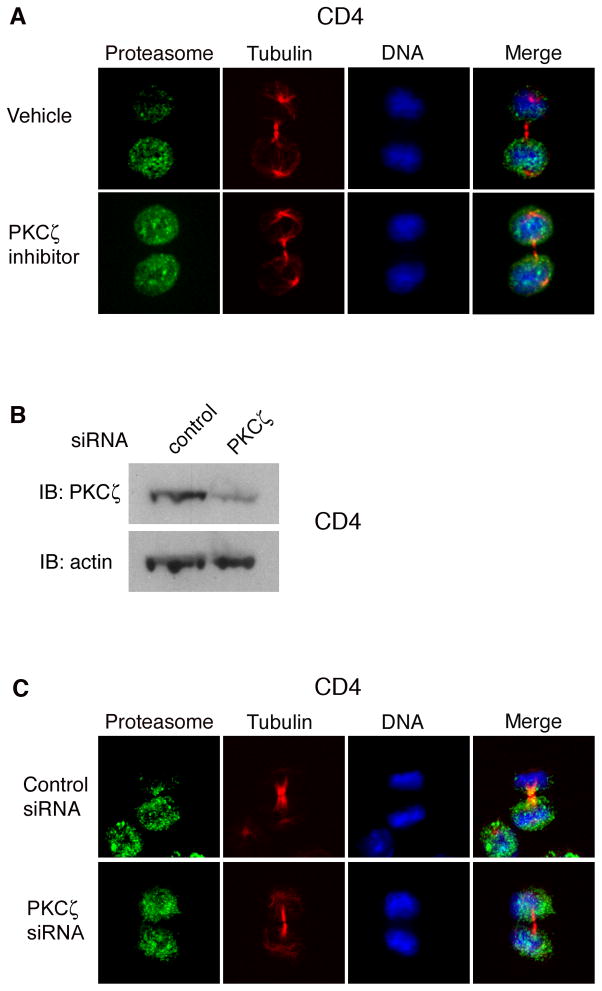
The observation that a conserved network of polarity proteins is involved in T cell migration, polarity, and asymmetric division (Chang et al., 2007; Ludford-Menting et al., 2005; Yeh et al., 2008) raised the possibility that this conserved network might also regulate asymmetry of the proteasome. In particular, the mammalian homologue of atypical PKC (PKCζ), an essential component of a complex containing the partitioning-defective (PAR) proteins Par-3 and Par-6, has been implicated in T cell function (Martin et al., 2005). To determine whether PKCζ might play a role in regulating proteasome asymmetry, we used a pharmacologic inhibitor of PKCζ, the myristolated PKCζ pseudosubstrate, which has been shown to inhibit its kinase activity (Sun et al., 2005). CD4+ T cells were activated in vitro and treated at 24 hours with vehicle or PKCζ inhibitor. We observed that inhibition of PKCζ kinase activity resulted in a loss of proteasomal asymmetry (Figure 4A). Consistent with these results, PKCζ knockdown using a siRNA approach also resulted in a loss of proteasomal asymmetry (Figures 4B and 4C). Inhibition of PKCζ, however, had no effect on PKCζ localization in dividing T cells nor did it affect T-bet amounts (Figure S4). Together these results suggest a role for the conserved polarity network in regulating asymmetry of the proteasome, and consequently, the asymmetry of T-bet.
Figure 4. Asymmetry of the proteasome may depend on the polarity network.
(A) CD4+ T cells were activated in vitro for 28h, treated for 1h with vehicle or a pharmacologic inhibitor of PKCζ, and stained for the proteasomal 20S α1 subunit (green), β-tubulin (red), and DNA (blue). Asymmetry of proteasome localization was observed in 61% (n=25) of vehicle-treated vs. 27% (n=29) of PKCζ inhibitor treated cells (p<0.001). (B) CD4+ T cells were activated in vitro for 48h, and control or PKCζ siRNA was introduced using electroporation. Lysates were prepared 72h later, and PKCζ and β-actin levels assessed by immunoblotting. (C) CD4+ T cells were transfected with control or PKCζ siRNA as in (B). 48h later, cells were restimulated in vitro for 24h and stained with the proteasomal 20S α1 subunit (green), β-tubulin (red), and DNA (blue). Asymmetry of proteasome localization was observed in 63% (n=60) of control transfected vs. 32% (n=62) of PKCζ siRNA transfected cells (p<0.001). Results are representative of two separate experiments.
Phosphorylation of T-bet links its degradation with its asymmetric partitioning
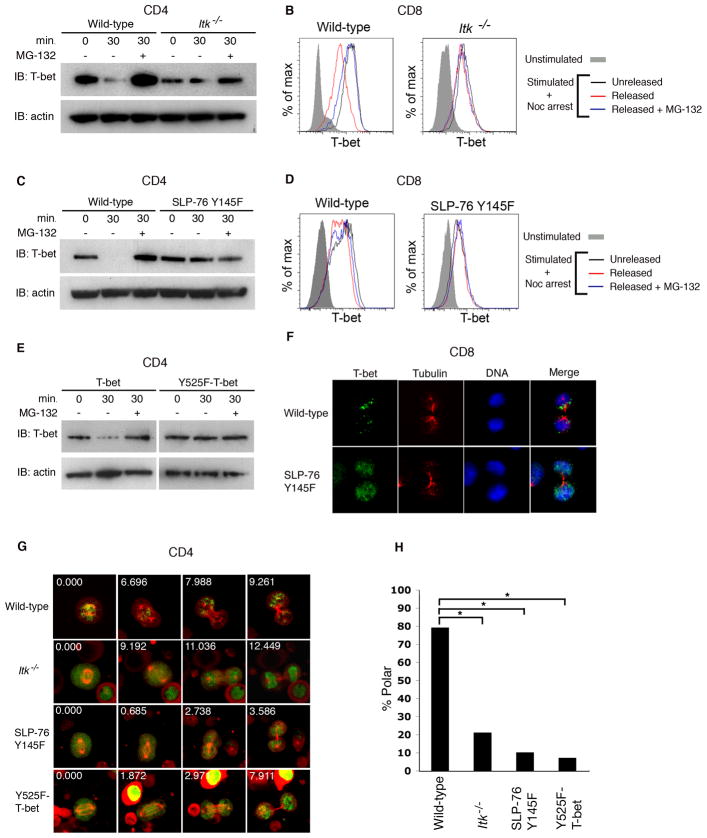
To further explore whether the degradation of T-bet is related to its asymmetric localization, we examined the signals regulating the degradation of T-bet. Tyrosine phosphorylation is a post-translational modification of T-bet that is thought to be critical for mediating its ability to interact with other proteins (Hwang et al., 2005). The inducible T cell kinase, ITK, phosphorylates T-bet at a critical tyrosine residue 525 (Hwang et al., 2005). ITK is activated and recruited to the T cell receptor by the adaptor protein, SLP-76 (Hwang et al., 2005; Jordan et al., 2008). To determine whether tyrosine phosphorylation of T-bet might play a role in targeting it for mitotic degradation, we examined CD4+ and CD8+ T cells from ITK-deficient mice (Hwang et al., 2005) or from mice expressing a tyrosine-to-phenylalanine knock-in mutation in SLP-76 at residue 145 (SLP-76 Y145F) that prevents the activation of ITK (Jordan et al., 2008). In mitotic T cells from SLP-76 Y145F or ITK-deficient mice, T-bet failed to undergo degradation (Figures 5A–D). We also examined cells expressing a mutation of T-bet with a tyrosine-to-phenylalanine substitution at residue 525 (Y525F-T-bet) which prevents it from undergoing phosphorylation (Hwang et al., 2005). CD4+ T cells from T-bet-deficient mice were reconstituted with either wild-type T-bet-GFP or mutant Y525F-T-bet-GFP. We observed that wild-type T-bet, but not mutant Y525F-T-bet, underwent proteasome-dependent degradation during mitosis (Figure 5E). Although both constructs are expressed under the control of retroviral regulatory elements, the general transcriptional inactivity of mitosis may allow us to observe T-bet protein degradation. Together these findings suggest that phosphorylation of T-bet is required for its degradation.
Figure 5. Mutations preventing phosphorylation of T-bet impair its proteasome-dependent degradation and asymmetric partitioning during mitosis.
(A) CD4+ T cells from wild-type or Itk−/− mice were activated in vitro and synchronized with nocodazole as in Figure 2B. After drug washout, cells were cultured with or without MG-132 for 30m. Cell lysates were prepared at 0 or 30m after drug washout. T-bet and β-actin levels were assessed by immunoblotting. (B) CD8+ T cells from wild-type or Itk−/− mice were activated as in (A) and T-bet levels assessed by intracellular staining at 0 or 30m after drug washout. (C) CD4+ T cells from wild-type and SLP-76 Y145F mice were activated and analyzed as in (A). (D) CD8+ T cells from wild-type and SLP-76 Y145F mice were activated and analyzed as in (B). (E) CD4+ T cells from T-bet-deficient mice were transduced with retroviruses encoding wild-type T-bet-GFP or Y525F-T-bet-GFP. After 3d, cells were restimulated for 24h and synchronized with nocodazole and analyzed as in (A). (F) Wild-type or SLP-76 Y145F P14 CD8+ TCR transgenic T cells were adoptively transferred into wild-type recipients infected with gp33-L. monocytogenes, harvested at 36h after transfer, and stained for T-bet (green), β-tubulin (red), and DNA (blue). Asymmetric partitioning of T-bet was observed in 72% (n=21) of wild-type versus 15% (n=26) of SLP-76 Y145F P14 CD8+ T cells (p<0.001). (G) CD4+ T cells from wild-type, Itk−/−, and SLP-76 Y145F mice were transduced with cherry-alpha-tubulin and either T-bet-GFP or Y525F-T-bet-GFP. Cells were imaged as in Figure 1E. (H) Quantification of asymmetric T cell partitioning into daughter cells represented in (G). The number of cells transduced with T-bet-GFP that were examined in each group: wild-type (46), Itk−/− (48), SLP-76 Y145F (29). The number of wild-type cells transduced with Y525F-T-bet-GFP examined was 27. p<0.001 (*). Results are representative of two separate experiments.
If degradation of T-bet is critical for its asymmetry, then defects in phosphorylation that prevent its degradation would also be predicted to disrupt its asymmetry. Mutations that prevent the phosphorylation of T-bet affect its asymmetric partitioning in vivo and in vitro. In mice infected with gp33-L. monocytogenes, CD8+ T cells harboring the SLP-76 Y145F mutation were found to exhibit a loss of T-bet asymmetry compared to wild-type cells (Figure 5F). To further test this hypothesis, CD4+ T cells from wild-type, ITK-deficient, and SLP-76 Y145F mice were transduced with cherry-alpha-tubulin and either wild-type T-bet-GFP or Y525F-T-bet-GFP. In contrast to wild-type cells, dividing cells from ITK-deficient and SLP-76 Y145F mice, as well as those transduced with Y525F-T-bet-GFP, could not support asymmetric partitioning of T-bet (Figures 5G, 5H, and Movies S3-S6). These results suggest that phosphorylation of T-bet, which appears to be required for its degradation, is also necessary for its asymmetric partitioning during mitosis.
Asymmetric localization and function of the proteasome is required for T-bet asymmetry
To further establish a mechanistic link between degradation and asymmetry of T-bet, we treated mitotic T cells with the proteasome inhibitor MG-132. Inhibition of the proteasome resulted in a substantial defect in T-bet asymmetry (Figure 6A), suggesting that degradation of T-bet may be causally linked to its asymmetry. It remains possible, however, that preventing T-bet degradation pharmacologically or through the aforementioned genetic approaches might perturb the asymmetric partitioning of T-bet even if an alternative mechanism were responsible for T-bet asymmetry. To evaluate this possibility, CD4+ T cells from T-bet-deficient mice were simultaneously transduced with wild-type T-bet-cherry and mutant Y525F-T-bet-GFP fusions. We observed that wild-type T-bet, but not Y525F-T-bet, was asymmetrically partitioned into the daughter cells (Figure 6B). This finding supports the hypothesis that unequal degradation underlies T-bet asymmetry: mutant T-bet lacking the ability to be degraded is mislocalized but does not disrupt the ability of wild-type T-bet to be localized unequally, presumably by asymmetric degradation, within the same dividing cell.
Figure 6. Preventing degradation of T-bet disrupts the asymmetric partitioning of T-bet.
(A) Undivided P14 CD8+ TCR transgenic T cells were harvested as in Figure 1A and cultured in vitro with vehicle or MG-132 for 4h prior to staining with T-bet (green), β-tubulin (red), and DNA (blue). (B) CD4+ T cells from T-bet-null mice were transduced with both wild-type T-bet-cherry and Y525F-T-bet-GFP, restimulated in vitro and stained for β-tubulin (blue). Cytokinetic cells expressing both wild-type T-bet (red) and Y525F-Tbet (green) were scored. Asymmetry of wild-type T-bet and Y525F-T-bet was observed in 60% and 9% (n=23) of cells, respectively (p<0.001). (C) CD4+ T cells were transduced as in Figure 1E. After 3d, cells were restimulated for 24h, treated for 1h with vehicle (top panel) or PKCζ inhibitor (bottom panel), and imaged as in Figure 1E. Asymmetric partitioning of T-bet occurred in 82% (n=28) of vehicle-treated versus 14% (n=42) of PKCζ inhibitor-treated cells (p<0.001). Results are representative of two separate experiments.
If localized degradation owing to proteasomal asymmetry were responsible for T-bet asymmetry, inhibiting proteasomal asymmetry would be predicted to disrupt the asymmetric partitioning of T-bet. We observed that loss of proteasomal asymmetry resulting from inhibition of PKCζ was associated with a loss in T-bet asymmetry (Figure 6C). It remains possible that PKCζ may have a direct effect on T-bet asymmetry, in addition to influencing T-bet localization indirectly through its effect on proteasome asymmetry. Inhibiting the activity or asymmetric localization of the proteasome, thus, prevents unequal partitioning of T-bet. Together these results support the hypothesis that localized degradation of T-bet by virtue of proteasome asymmetry may underlie the asymmetric partitioning of T-bet.
Discussion
When a lymphocyte is engaged in an immune response, it must undergo vigorous cell division to amplify its numbers. The progeny of a selected lymphocyte must also adopt new patterns of gene expression representing the spectrum of fates of antigen-experienced cells. Whether the progeny of a single lymphocyte all adopt the same fate or whether the fates of clonally related cells differ has been difficult to establish. Recent studies using single-cell adoptive transfers and cellular barcoding have suggested the possibility that a single naïve cell may give rise to progeny of heterogeneous fates (Gerlach et al., 2010; Schepers et al., 2008; Stemberger et al., 2007). Hypothetically, there are at least two different ways by which sibling cells could adopt dissimilar fates. Cells could be born identically and subsequently receive different signals from their environments, prompting them to diverge in fate. Alternatively, a single cell could unequally transmit information to its daughter cells, causing them to diverge in fate. The evolutionarily conserved process whereby two sibling cells acquire unequal shares of certain determinants is known as asymmetric cell division (Betschinger and Knoblich, 2004; Knoblich, 2008). It has been suggested that a T lymphocyte selected for an immune response undergoes asymmetric division, enabling it to produce progeny of heterogeneous fates (Chang et al., 2007; Oliaro et al., 2010).
In order for a lymphocyte to undergo an asymmetric division it needs to apportion unequal shares of regulatory molecules to its daughter cells. The presence of such determinants at sufficiently high levels should promote the acquisition of one fate, whereas their relative paucity would favor adoption of an alternative fate. In activated CD8+ T cells, the transcription factor T-bet promotes the effector fate at the expense of the memory fate (Intlekofer et al., 2005; Joshi et al., 2007). In activated CD4+ T cells, T-bet promotes the Th1 cell fate while repressing the development of the Th2 and Th17 cell lineages (Hwang et al., 2005; Lazarevic et al., 2010; Szabo et al., 2000; Szabo et al., 2002). These effects of T-bet are highly dose-dependent, as small changes (50% or less) in the abundance of T-bet protein result in profound alterations in CD8+ and CD4+ T cell fate (Intlekofer et al., 2005; Intlekofer et al., 2007; Joshi et al., 2007; Szabo et al., 2002; Finotto et al., 2002). These observations suggest that seemingly small differences in T-bet abundance between the daughter cells of a T cell selected for an immune response would be predicted to influence their subsequent fates.
The present findings suggest that CD8+ and CD4+ daughter T cells that have completed their first division indeed exhibit differences in T-bet abundance. This disparity begins during the single cell stage, as asymmetry of T-bet localization can be observed during mitosis and in the nascent daughter cells even prior to the completion of division. After division, asymmetric segregation of the IFNγ receptor (Chang et al., 2007) could reinforce the pre-existing differences in the amount of T-bet protein between the daughter cells, by virtue of differential IFN-γ signaling resulting in unequal T-bet mRNA expression (Afkarian et al., 2002; Lighvani et al., 2001). Together these distinct mechanisms may function to promote differences in T-bet amounts in the daughter cells.
What signals instruct a dividing cell to asymmetrically apportion T-bet to its daughter cells? Our findings suggest that the tyrosine kinase ITK may be one such critical signal. ITK participates in signalling events downstream of T cell receptor ligation and has a role in developmental and differentiation pathways in T cells (Atherly et al., 2006; Berg, 2007; Gomez-Rodriguez et al., 2009; Siliciano et al., 1992). The present results suggest that another critical function for ITK is to target T-bet for proteasome-dependent degradation during mitosis. In situations where asymmetric partitioning of T-bet is defective, as in ITK-deficient T cells, the failure to exclude T-bet from the distal daughter cell might be predicted to interfere with its ability to become a memory cell. Such a prediction is consistent with recent evidence that suggests a role for ITK in CD8+ memory cell development (Smith-Garvin et al., 2010). Similarly, in a CD4+ T cell, defective T-bet asymmetry by virtue of ITK deficiency might be predicted to result in excess T-bet partitioned to a daughter cell that would have otherwise been fated towards the Th2 cell lineage, thereby precluding it from developing into a Th2 cell. This is consistent with the defect in Th2 cell differentiation that has been observed in ITK-deficient mice (Fowell et al., 1999; Schaeffer et al., 2001).
Signals that solely target T-bet for destruction might not be sufficient to mediate T-bet asymmetry. In order for T-bet to undergo asymmetric inheritance, the signals that target T-bet for proteasome-dependent degradation must seemingly be accompanied by signals that instruct the cell to segregate some component of the degradation machinery asymmetrically during mitosis. Here we provide data suggesting that this segregated component may be the proteasome. Although the signals mediating this asymmetric segregation remain to be extensively evaluated, our initial experiments suggest that the conserved cell polarity network may be involved in mediating this effect, as loss of function of a key member of this family appears to prevent the proteasome from being asymmetrically distributed. Such a mechanism could allow the polarity network, by regulating asymmetry of the degradation machinery, to influence the partitioning of fate determinants that have been targeted for destruction.
The present findings indicate that regulated destruction controlled by distinct localization of the degradation machinery may be a mechanism to allow for the asymmetric partitioning of cell fate determinants. Recent evidence has suggested that regulated degradation can also occur by virtue of polarized segregation of other components of the degradation pathway, such as ubiquitin or even ubiquitinated proteins themselves (Fuentealba et al., 2008; Narimatsu et al., 2009). In this way, distinct mechanisms regulating degradation may function to render unique transcriptional programs between the daughter cells by unequally degrading key transcriptional regulators, such as T-bet or other transcription factors that regulate T cell fate decisions. In addition, it remains possible that other proteins targeted for destruction during mitosis, such as regulators of the cell cycle, proliferation, or homeostasis, could be unequally inherited by the daughter cells owing to proteasome asymmetry. Although the full extent of the disparities mediated by unequal segregation of the proteasome remains to be determined, our findings suggest that proteasome asymmetry may be a novel mechanism to allow for the unequal partitioning of determinants that can influence fate and function in sibling cells.
Experimental Procedures
Mice
All animal work was done in accordance with Institutional Animal Care and Use Guidelines of the University of Pennsylvania. All mice were housed in specific pathogen-free conditions prior to use. Wild-type C57BL/6 and P14 TCR transgenic mice recognizing LCMV peptide gp33–41/Db were used; generation of Tbx21−/− (T-bet-null) mice has been previously described (Intlekofer et al., 2005). The generation of mice expressing a tyrosine-to-phenylalanine knock-in mutation in SLP-76 at residue 145 (Y145F) has been described (Jordan et al., 2008). SLP-76 Y145F P14 TCR transgenic mice were generated by breeding P14 TCR transgenic mice with SLP-76 Y145F mice. Itk−/− mice have been described (Liu et al., 1998). Adoptive transfers and infectious challenges with gp33-Listeria monocytogenes were performed as previously reported (Chang et al., 2007).
T lymphocyte confocal microscopy
Immunofluorescence of T cells was performed as previously described (Chang et al., 2007) using the following antibodies: anti-β-tubulin (Sigma); anti-T-bet, anti-CD3ε (eBioscience); anti-α-tubulin, anti-PKCζ, anti-proteasome 20S α1, anti-proteasome 20S α5, anti-proteasome 19S (Abcam); anti-IFNγR-biotin (BD Bioscience); anti-mouse- and anti-rat Alexa Fluor 488, anti-mouse-, anti-rabbit-, and anti-rat Alexa Fluor 568, anti-rat- and anti-rabbit- Alexa Fluor 647, and streptavidin-conjugated Alexa Fluor 647 (Invitrogen). Hoechst 33258 (Invitrogen) was used to detect DNA. The proteasome activity probe, MVB003, has been previously described (Florea et al., 2010).
Statistical analysis
Asymmetry of cells was summarized as proportions and compared using chi-square or Fisher’s exact test, as appropriate. All statistical tests were two-tailed. P-values of < 0.05 were considered significant.
Acquisition and analysis of T lymphocyte confocal microscopy
Mitotic cells were selected for analysis based on the appearance of tubulin staining, while cells undergoing cytokinesis were identified by dual nuclei and pronounced cytoplasmic cleft by brightfield, and then, secondarily, was the morphology of the other fluorescence channels revealed. Acquisition of image stacks was performed as previously reported (Chang et al., 2007). Using Volocity (Improvision) software, the volume of 3-D pixels (voxels) containing the designated receptor fluorescence was quantified within each hemisphere of mitotic cells, or within each nascent daughter in cytokinetic cells. In mitotic cells, the two hemispheres were delineated using the pattern of tubulin fluorescence to define the poles of the mitotic spindle, with the equator bisecting the line connecting the two poles. In cytokinetic cells, the two nascent daughters were delineated using the pattern of tubulin fluorescence to define the border of each daughter cell. Receptor enrichment in one hemisphere or in one nascent daughter cell greater than 1.5-fold compared to the other hemisphere or daughter cell was considered polarized. All images are depicted using pseudo-colors. In cells labelled with CFSE, the “true” green channel occupied by CFSE fluorescence was not shown. In such cells, anti-tubulin staining was detected using Alexa Fluor 488, which could be resolved in the green channel due to its enhanced brightness relative to CFSE.
Cell culture
CD8+ or CD4+ T cells were purified using the CD8+ or CD4+ T Cell Isolation Kit (Miltenyi), respectively. For microscopy experiments, naïve cells were activated in vitro using immobilized anti-CD3/anti-CD28 and immobilized recombinant ICAM1-Fc fusion protein (R&D Systems), and previously activated cells were restimulated in vitro using immobilized anti-CD3 and immobilized ICAM1-Fc. In certain experiments, cells from T-bet-deficient mice were simultaneously transduced with both wild-type T-bet-cherry and Y525F-T-bet-GFP. In some experiments, after 28 hours of activation, cells were incubated with the proteasome activity probe MVB003 (5μM) for 2 hours prior to harvesting cells for immunofluorescence studies. In certain experiments, an inhibitor of PKCζ, the myristoylated PKCζ mM)(Invitrogen), was added to cells 28 hours after activation for 2 hours prior to harvesting cells for immunofluorescence studies. For biochemistry experiments, naïve cells were activated in vitro using immobilized anti-CD3 and anti-CD28, and previously activated cells were restimulated in vitro using immobilized anti-CD3. Nocodazole (1 mM)(Sigma) was added after 24 hours of stimulation to reversibly synchronize the cells in G2/prometaphase. After 12–16 hours of nocodazole arrest, cells were washed free of nocodazole and then cultured in media alone or with MG-132 (10μM)(Calbiochem), calpain inhibitor I (100nM), or lactacystin (100nM)(Sigma). In other experiments, cells were activated in vitro using immobilized anti-CD3 and anti-CD28 in the presence of mimosine (300 mM), hydroxyurea (200 mM)(Sigma), or nocodazole. After 40 hours, cells were washed free of drug and cultured in media for an additional 30 minutes.
Retroviral constructs
The cherry-alpha-tubulin fusion construct has been previously described (Day et al., 2009). Generation of the MIGR and T-bet-MIGR construct has been described (Mullen et al., 2002). For T-bet-C-terminal-GFP or T-bet-C-terminal-cherry fusion constructs, PCR was performed using Pfx polymerase (Invitrogen) using a forward primer including a BglII site (5′-ATGACAGATCTCCACCATGGGCATCGTGGAGC-3′). For T-bet-GFP, the reverse primer was designed by omitting the stop codon and adding an EcoRI site for in-frame fusion to GFP (5′-ATGACAGAATTCTGTTGGGAAAATAATTATAAAACTGGCCTTC-3′). For T-bet-cherry, the reverse primer was 5′-ATGACAGAATTCGTTGGGAAAATAATTATAAAACTGGCCTTC-3′. The PCR product was digested by BglII and EcoRI (New England Biolabs) and fused in-frame with Cherry in the MIGR retrovirus vector (Shu et al., 2006). The Y525F mutation was introduced using the following primers: forward 5′-ATGACAGATCTCCACCATGGGCATCGTGGAGC-3′ and reverse 5′-ATGACAGAAT TCTGTTGGGAAAATAATTAAAAAACTGGCCTT-3′. The point mutation is underlined. All constructs were confirmed by sequence analysis.
Time-lapse confocal microscopy of activated T lymphocytes dividing in culture
To examine the subcellular localization of T-bet in dividing T cells in real-time, we developed a cell-culture based method to mimic the activation of T cells in vivo by microbe (Chang et al., 2007). CD4+ T cells were purified from wild-type C57BL/6, SLP-76 Y145F, or Itk−/− mice. After in vitro activation with immobilized anti-CD3 and anti-CD28 for 48h, cells were transduced with retroviruses encoding cherry-alpha tubulin and wild-type T-bet-GFP or Y525F-T-bet-GFP. Three days later, when the transduced lymphocytes expressing fluorescent fusion proteins were no longer dividing, cells were restimulated in vitro in a Lab-Tek chambered coverglass (Nunc) using immobilized anti-CD3 and recombinant ICAM1-Fc fusion protein for 24 hours prior to imaging. Immobilized ICAM1-Fc fusion protein was used as ICAM1-dependent interactions between antigen-presenting cells and T cells are required for asymmetric cell division in vivo (Chang et al., 2007). Furthermore, restimulation in the presence of immobilized ICAM1-Fc was found to be necessary for T-bet asymmetry in dividing wild-type cells in culture (Figure S1). Activated interphase cells were generally observed on the bottom of the culture dish where they interacted with anti-CD3 and ICAM1-Fc, but cells generally detached prior to mitosis. Cells were identified in prophase based on tubulin fluorescence and an intact nuclear envelope and imaged through metaphase, or identified in metaphase based on the presence of dual MTOCs and imaged through cytokinesis. 5 Z stack sections were acquired continuously over 30-second intervals using a spinning disk confocal microscope (Olympus). 3-dimensional Z stacks over time were converted into time-lapse movies using Volocity (Improvision). Quantitation was performed as described above. T-bet enrichment in one incipient daughter cell greater than 2-fold compared to the other daughter cell was considered polarized.
Immunoblotting
Cell lysates were prepared in 1% NP40 lysis buffer with the following additives: 0.1M DTT (Roche), protease inhibitor cocktail, sodium vanadate (10mM), NaF (10mM), and PMSF (10mM)(Sigma). Protein was prepared for SDS-PAGE followed by transfer to nitrocellulose membrane. Immunoblotting was performed with the following antibodies: rabbit anti-PKCζ, anti-tubulin-HRP (Abcam), mouse anti-T-bet (eBioscience), anti-mouse- or anti-rabbit-HRP (Cell Signaling), and β-actin-HRP (Sigma).
RNA interference
CD4+ T cells were purified and stimulated in vitro with immobilized anti-CD3/anti-CD28 for 48 hours prior to electroporation with control or PKCζ ON-TARGET SMARTpool siRNA (Thermo Scientific) using a ECM830 Squarewave Electroporator (BTX). Pulses were performed for 10ms at 190 mV. 48 hours after electroporation, cells were analyzed by immunoblotting or restimulated for microscopy studies.
Flow cytometry
Adoptive transfers and infectious challenges with gp33-Listeria monocytogenes were performed as previously reported (Chang et al., 2007). Splenocytes were stained with anti-T-bet-Alexa Fluor 647 (BD Bioscience) or T-bet-eFluor 660 (eBioscience) and anti-CD8 PE (BD Bioscience) and analyzed on a FACS Calibur (BD Bioscience).
Supplementary Material
Acknowledgments
We thank M. Birnbaum for use of the confocal microscope; A. Stout for assistance with time-lapse microscopy; J. Burkhardt for the use of the electroporator; E. Holzbaur, L. Boyd, I. Su, A. Banerjee, S. Gordon, M. Paley, R. Moy, L. Rupp, and K. Baraldi for assistance and discussion. Supported by NIH grants AI042370, AI076458, and AI061699 and the Abramson Family (S.L.R.); NIH grant DK080949 and AGA-FDHN (J.T.C.); NIH grant T32HD007516 (M.L.C.); NIH grants AI37584 and AI46564 (L.J.B.), CA093615 and GM053256 (G.A.K), and AR052802 (M.S.J.); NHMRC (J.O.); Peter MacCallum Cancer Centre, NHMRC, HFSP, and ARC (S.M.R.); and the UCSD Digestive Diseases Research Development Center Grant DK80506. J.T.C. is a Howard Hughes Medical Institute Physician-Scientist Early Career Awardee.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- Atherly LO, Lucas JA, Felices M, Yin CC, Reiner SL, Berg LJ. The Tec family tyrosine kinases Itk and Rlk regulate the development of conventional CD8+ T cells. Immunity. 2006;25:79–91. doi: 10.1016/j.immuni.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Azar GA, Lemaitre F, Robey EA, Bousso P. Subcellular dynamics of T cell immunological synapses and kinapses in lymph nodes. Proc Natl Acad Sci U S A. 2010;107:3675–3680. doi: 10.1073/pnas.0905901107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcia C, Thomas CE, Curtin JF, King GD, Wawrowsky K, Candolfi M, Xiong WD, Liu C, Kroeger K, Boyer O, et al. In vivo mature immunological synapses forming SMACs mediate clearance of virally infected astrocytes from the brain. J Exp Med. 2006;203:2095–2107. doi: 10.1084/jem.20060420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg LJ. Signalling through TEC kinases regulates conventional versus innate CD8(+) T-cell development. Nat Rev Immunol. 2007;7:479–485. doi: 10.1038/nri2091. [DOI] [PubMed] [Google Scholar]
- Betschinger J, Knoblich JA. Dare to be different: asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr Biol. 2004;14:R674–685. doi: 10.1016/j.cub.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Beuneu H, Lemaitre F, Deguine J, Moreau HD, Bouvier I, Garcia Z, Albert ML, Bousso P. Visualizing the functional diversification of CD8+ T cell responses in lymph nodes. Immunity. 2010;33:412–423. doi: 10.1016/j.immuni.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, Longworth SA, Vinup KE, Mrass P, Oliaro J, et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315:1687–1691. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- Day D, Pham K, Ludford-Menting MJ, Oliaro J, Izon D, Russell SM, Gu M. A method for prolonged imaging of motile lymphocytes. Immunol Cell Biol. 2009;87:154–158. doi: 10.1038/icb.2008.79. [DOI] [PubMed] [Google Scholar]
- Finotto S, Neurath MF, Glickman JN, Qin S, Lehr HA, Green FH, Ackerman K, Haley K, Galle PR, Szabo SJ, et al. Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science. 2002;295:336–338. doi: 10.1126/science.1065544. [DOI] [PubMed] [Google Scholar]
- Florea BI, Verdoes M, Li N, van der Linden WA, Geurink PP, van den Elst H, Hofmann T, de Ru A, van Veelen PA, Tanaka K, et al. Activity-based profiling reveals reactivity of the murine thymoproteasome-specific subunit beta5t. Chem Biol. 2010;17:795–801. doi: 10.1016/j.chembiol.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowell DJ, Shinkai K, Liao XC, Beebe AM, Coffman RL, Littman DR, Locksley RM. Impaired NFATc translocation and failure of Th2 development in Itk-deficient CD4+ T cells. Immunity. 1999;11:399–409. doi: 10.1016/s1074-7613(00)80115-6. [DOI] [PubMed] [Google Scholar]
- Friedman RS, Beemiller P, Sorensen CM, Jacobelli J, Krummel MF. Real-time analysis of T cell receptors in naive cells in vitro and in vivo reveals flexibility in synapse and signaling dynamics. J Exp Med. 2010;207:2733–2749. doi: 10.1084/jem.20091201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba LC, Eivers E, Geissert D, Taelman V, De Robertis EM. Asymmetric mitosis: Unequal segregation of proteins destined for degradation. Proc Natl Acad Sci U S A. 2008;105:7732–7737. doi: 10.1073/pnas.0803027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach C, van Heijst JW, Swart E, Sie D, Armstrong N, Kerkhoven RM, Zehn D, Bevan MJ, Schepers K, Schumacher TN. One naive T cell, multiple fates in CD8+ T cell differentiation. J Exp Med. 2010;207:1235–1246. doi: 10.1084/jem.20091175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Rodriguez J, Sahu N, Handon R, Davidson TS, Anderson SM, Kirby MR, August A, Schwartzberg PL. Differential expression of interleukin-17A and −17F is coupled to T cell receptor signaling via inducible T cell kinase. Immunity. 2009;31:587–597. doi: 10.1016/j.immuni.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse M, Lillemeier BF, Kuhns MS, Chen DS, Davis MM. T cells use two directionally distinct pathways for cytokine secretion. Nature Immunology. 2006;7:247–255. doi: 10.1038/ni1304. [DOI] [PubMed] [Google Scholar]
- Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science. 2005;307:430–433. doi: 10.1126/science.1103336. [DOI] [PubMed] [Google Scholar]
- Intlekofer AM, Takemoto N, Kao C, Banerjee A, Schambach F, Northrop JK, Shen H, Wherry EJ, Reiner SL. Requirement for T-bet in the aberrant differentiation of unhelped memory CD8+ T cells. J Exp Med. 2007;204:2015–2021. doi: 10.1084/jem.20070841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- Jordan MS, Smith JE, Burns JC, Austin JE, Nichols KE, Aschenbrenner AC, Koretzky GA. Complementation in trans of altered thymocyte development in mice expressing mutant forms of the adaptor molecule SLP76. Immunity. 2008;28:359–369. doi: 10.1016/j.immuni.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 2009;31:283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Lazarevic V, Chen X, Shim JH, Hwang ES, Jang E, Bolm AN, Oukka M, Kuchroo VK, Glimcher LH. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat Immunol. 2010;12:96–104. doi: 10.1038/ni.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437:275–280. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lighvani AA, Frucht DM, Jankovic D, Yamane H, Aliberti J, Hissong BD, Nguyen BV, Gadina M, Sher A, Paul WE, O’Shea JJ. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc Natl Acad Sci U S A. 2001;98:15137–15142. doi: 10.1073/pnas.261570598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KQ, Bunnell SC, Gurniak CB, Berg LJ. T cell receptor-initiated calcium release is uncoupled from capacitative calcium entry in Itk-deficient T cells. J Exp Med. 1998;187:1721–1727. doi: 10.1084/jem.187.10.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludford-Menting MJ, Oliaro J, Sacirbegovic F, Cheah ET, Pedersen N, Thomas SJ, Pasam A, Iazzolino R, Dow LE, Waterhouse NJ, et al. A network of PDZ-containing proteins regulates T cell polarity and morphology during migration and immunological synapse formation. Immunity. 2005;22:737–748. doi: 10.1016/j.immuni.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Maldonado RA, Irvine DJ, Schreiber R, Glimcher LH. A role for the immunological synapse in lineage commitment of CD4 lymphocytes. Nature. 2004;431:527–532. doi: 10.1038/nature02916. [DOI] [PubMed] [Google Scholar]
- Martin P, Villares R, Rodriguez-Mascarenhas S, Zaballos A, Leitges M, Kovac J, Sizing I, Rennert P, Marquez G, Martinez AC, et al. Control of T helper 2 cell function and allergic airway inflammation by PKCzeta. Proc Natl Acad Sci U S A. 2005;102:9866–9871. doi: 10.1073/pnas.0501202102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Dey A, Rabindran SK, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- Monks CR, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 1998;395:82–86. doi: 10.1038/25764. [DOI] [PubMed] [Google Scholar]
- Mullen AC, Hutchins AS, High FA, Lee HW, Sykes KJ, Chodosh LA, Reiner SL. Hlx is induced by and genetically interacts with T-bet to promote heritable T(H)1 gene induction. Nat Immunol. 2002;3:652–658. doi: 10.1038/ni807. [DOI] [PubMed] [Google Scholar]
- Narimatsu M, Bose R, Pye M, Zhang L, Miller B, Ching P, Sakuma R, Luga V, Roncari L, Attisano L, Wrana JL. Regulation of planar cell polarity by Smurf ubiquitin ligases. Cell. 2009;137:295–307. doi: 10.1016/j.cell.2009.02.025. [DOI] [PubMed] [Google Scholar]
- Oliaro J, Van Ham V, Sacirbegovic F, Pasam A, Bomzon Z, Pham K, Ludford-Menting MJ, Waterhouse NJ, Bots M, Hawkins ED, et al. Asymmetric cell division of T cells upon antigen presentation uses multiple conserved mechanisms. J Immunol. 2010;185:367–375. doi: 10.4049/jimmunol.0903627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert P, Reinhardt RL, Ingulli E, Jenkins MK. Cutting edge: in vivo identification of TCR redistribution and polarized IL-2 production by naive CD4 T cells. J Immunol. 2001;166:4278–4281. doi: 10.4049/jimmunol.166.7.4278. [DOI] [PubMed] [Google Scholar]
- Rutishauser RL, Martins GA, Kalachikov S, Chandele A, Parish IA, Meffre E, Jacob J, Calame K, Kaech SM. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer EM, Yap GS, Lewis CM, Czar MJ, McVicar DW, Cheever AW, Sher A, Schwartzberg PL. Mutation of Tec family kinases alters T helper cell differentiation. Nat Immunol. 2001;2:1183–1188. doi: 10.1038/ni734. [DOI] [PubMed] [Google Scholar]
- Schepers K, Swart E, van Heijst JW, Gerlach C, Castrucci M, Sie D, Heimerikx M, Velds A, Kerkhoven RM, Arens R, Schumacher TN. Dissecting T cell lineage relationships by cellular barcoding. J Exp Med. 2008;205:2309–2318. doi: 10.1084/jem.20072462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Blackburn SD, Intlekofer AM, Kao C, Angelosanto JM, Reiner SL, Wherry EJ. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity. 2009;31:309–320. doi: 10.1016/j.immuni.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ. Novel chromophores and buried charges control color in mFruits. Biochemistry. 2006;45:9639–9647. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- Siliciano JD, Morrow TA, Desiderio SV. itk, a T-cell-specific tyrosine kinase gene inducible by interleukin 2. Proc Natl Acad Sci U S A. 1992;89:11194–11198. doi: 10.1073/pnas.89.23.11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Garvin JE, Burns JC, Gohil M, Zou T, Kim JS, Maltzman JS, Wherry EJ, Koretzky GA, Jordan MS. T-cell receptor signals direct the composition and function of the memory CD8+ T-cell pool. Blood. 2010;116:5548–5559. doi: 10.1182/blood-2010-06-292748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemberger C, Huster KM, Koffler M, Anderl F, Schiemann M, Wagner H, Busch DH. A single naive CD8+ T cell precursor can develop into diverse effector and memory subsets. Immunity. 2007;27:985–997. doi: 10.1016/j.immuni.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JC, Majorovits E, Bossi G, Fuller S, Griffiths GM. Centrosome polarization delivers secretory granules to the immunological synapse. Nature. 2006;443:462–465. doi: 10.1038/nature05071. [DOI] [PubMed] [Google Scholar]
- Sun R, Gao P, Chen L, Ma D, Wang J, Oppenheim JJ, Zhang N. Protein kinase C zeta is required for epidermal growth factor-induced chemotaxis of human breast cancer cells. Cancer Res. 2005;65:1433–1441. doi: 10.1158/0008-5472.CAN-04-1163. [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- Yeh JH, Sidhu SS, Chan AC. Regulation of a late phase of T cell polarity and effector functions by Crtam. Cell. 2008;132:846–859. doi: 10.1016/j.cell.2008.01.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.