The authors report the first independent replication of rs613872, an intronic SNP of TCF4 associated with late-onset Fuchs corneal dystrophy.
Abstract
Purpose.
Fuchs corneal dystrophy (FCD) is an autosomal dominant disease of the corneal endothelium with variable penetrance and expressivity. Recently, rs613872, an intronic variation of TCF4 associated with late-onset FCD, was reported. The present study was undertaken to examine this association in our cohort of FCD patients, to assess the significance of this finding, and to investigate the candidacy of TCF4 in the context of the mapped FCD2 locus.
Methods.
The authors recruited 170 patients with late-onset FCD and 180 age-matched controls. Blood samples were collected, and genomic DNA was extracted. A panel of nine SNPs spanning the entire TCF4 locus was genotyped both on this cohort and on three previously reported FCD2-linked families. The association of an individual SNP with late-onset FCD was evaluated with the Fisher exact test, and the coding exons and exon-intron boundaries of TCF4 were sequenced in 96 affected persons.
Results.
The risk allele G of rs613872 is associated significantly with late-onset FCD (odds ratio, 4.2; P = 4.28 × 10−15) and was present in male and female affected persons without any sex bias, replicating recent findings, though the authors found no apparent correlation with the severity of the disease phenotype. Moreover, the risk allele did not cosegregate with the disease phenotype in any of the three FCD2-linked families. The authors did not identify any pathogenic variants in the coding region of TCF4.
Conclusions.
The authors report the first independent replication of rs613872 conferring risk of late-onset FCD. Their data suggest that this risk factor is likely independent of the FCD2 locus, whose causality remains unknown.
Fuchs corneal dystrophy (FCD) is a heritable, progressive disease of the corneal endothelium and a leading cause of corneal transplantation in the United States.1,2 Generally, disease onset begins in the fifth decade of life and progresses slowly over the next two to three decades.2–4 Phenotypically, FCD is characterized by the presence of corneal guttae, histologically defined as drop-like excrescences of Descemet membrane, the collagen-rich basal lamina of the corneal endothelium.4,5
FCD is a genetically heterogeneous disorder that exhibits an autosomal dominant mode of inheritance with variable penetrance and expressivity. The rare form of early-onset FCD is causally associated with mutations in COL8A2, whereas the more common late-onset FCD has been localized to four loci—FCD1, FCD2, FCD3, and FCD4—on chromosomes 13, 18, 5, and 9, respectively.6–10 In addition, heterozygous loss of function mutations in SLC4A11 have been identified in sporadic and familial cases of late-onset FCD.11,12 Similarly, mutational analysis and subsequent functional evaluations have shown that missense changes in TCF8 lead to late-onset FCD, with evidence for genetic interaction between this locus and FCD4 that modifies the severity of the disorder.10
TCF8 encodes a zinc finger transcription factor that regulates gene expression by repressing or activating target genes.13,14 Sobrado et al.15 demonstrated recently that the expression of TCF8 is regulated by another transcription factor protein, TCF4, suggesting that TCF4 and TCF8 are elements of a common pathway relevant to the pathomechanism of late-onset FCD. TCF4 encodes a helix-loop-helix protein present in most eukaryotic organisms, with important roles in essential developmental processes.16–18 The TCF4 locus maps in the critical interval of FCD2 on chromosome 18q. Recently, an intronic TCF4 SNP, rs613872, was identified through a genome-wide study to be associated with late onset-FCD.19 Given this report and the implication of TCF8 in the pathogenicity of FCD, we investigated the role of TCF4 in the etiology of late-onset FCD.
Materials and Methods
Patients and Control Subjects
Our cohort consisted of 170 patients with sporadic FCD and 180 ethnically matched control subjects. All participants underwent detailed ophthalmic evaluation that included slit lamp biomicroscopy. Affectation status and disease severity were determined with the scale proposed by Krachmer et al.2 Positive disease status was indicated if the patient had a minimum Fuchs Krachmer grading score of 1, which represented 12 or more central nonconfluent guttae, in at least one eye. The inclusion criteria for control subjects consisted of a minimum age of 58 years and no signs or symptoms of FCD on examination using a slit lamp biomicroscope. The study protocol was approved by the Joint Committee on Clinical Investigation at the Johns Hopkins University School of Medicine and was in accordance with the Declaration of Helsinki and with HIPAA regulations. Written informed consent was obtained from all study participants. A sample of approximately 10 mL blood sample was collected from each study participant. DNA was extracted with a purification kit (Gentra Puregene Blood Kit; Qiagen, Santa Clara, CA).
Genotype Analyses
Polymerase chain reaction (PCR) was performed in 5-μL volumes containing 10 ng genomic DNA, 2.5 μL SNP genotyping master mix (TaqMan; Applied Biosystems, Foster City, CA), and 0.125 μL genotyping assay mix (TaqMan; Applied Biosystems). Reactions for all 9 SNPs were amplified independently in a thermocycler (9700; Applied Biosystems). The cycling parameters consisted of 2-minute incubation at 50°C and denaturation at 95°C for 10 minutes, followed by 40 cycles of 10 seconds at 95°C and 1-minute elongation at 72°C with a final 10-minute extension at 72°C. Amplified products were analyzed for the enrichment of specific alleles (ABI 7900HT Sequence Detection System; Applied Biosystems).
Statistical Analysis
Hardy-Weinberg equilibrium of the genotypic frequencies among control subjects was calculated with plink software (http://pngu.mgh.harvard.edu/∼purcell/plink/summary.shtml).20 The Fisher exact test was performed to test the allelic and genotypic associations of all the SNPs using plink algorithms with an alternative hypothesis that the true odds ratio is not equal to 1. These statistical tests were independently confirmed using R software (http://www.r-project.org/). Linear combinations of the regression estimates were performed to compare the age-severity relationships from the two groups using another statistical analysis package (STATA version 11.0; StataCorp, College Station, TX).21,22
Sequencing Analysis
Primer pairs for TCF4 were designed using the primer3 program; primer sequences and annealing temperatures are available on request. PCR products were analyzed on a 2% agarose gel and purified by ethanol precipitation. The PCR primers for each exon were used for bidirectional sequencing using reaction mix (BigDye Terminator Ready; Applied Biosystems), according to the manufacturer's instructions. Sequencing products were precipitated and resuspended in 10 μL formamide (Applied Biosystems) and denatured at 95°C for 5 minutes. Sequencing was performed (ABI PRISM 3100 Automated Sequencer; Applied Biosystems), and sequencing results were assembled with ABI sequencing analysis software (PRISM version 3.7; Applied Biosystems) and analyzed (SeqScape software; Applied Biosystems).
Results
To understand the genetic basis of FCD, we ascertained a cohort of patients with sporadic and familial FCD. To replicate the recently reported association of intronic TCF4 alleles with FCD,19 we chose 170 affected persons (62 men, 108 women). Additionally, we collected samples from 180 ethnically matched control DNA subjects (82 men, 98 women) who had no signs of guttae and were thus negative for FCD. The mean Krachmer grade of the affected persons was 2.97 (range, 1–6) and their mean age was 65 years (range, 32–97 years), slightly younger than that of the control subjects (mean, 72 years; range, 58–93 years). Demographic characteristics of the study participants are shown in Table 1.
Table 1.
Clinical Characteristics of the Study Subjects
| Case Subjects |
Control Subjects | |||
|---|---|---|---|---|
| Combined | Male | Female | ||
| Average age ± SD, y | 66.25 ± 12.97 | 62.47 ± 12.81 | 68 ± 12.70 | 71.99 ± 7.52 |
| Age range, y | 32–97 | 32–90 | 32–97 | 58–93 |
| Krachmer grade, average | 3.66 ± 1.76 | 3.58 ± 1.85 | 3.70 ± 1.73 | NA |
| Krachmer grade, range | 1–6 | 1–6 | 1–6 | NA |
We genotyped nine SNPs (rs1261078, rs41483647, rs11152369, rs2958182, rs613872, rs658977, rs581653, rs17089907, and rs2646965) in 350 participants (170 patients, 180 control subjects). The alleles for all nine SNPs were in Hardy-Weinberg equilibrium in the control subjects (data not shown). Among these SNPs, the G allele of rs613872 showed significant association in our patients, with P = 4.28 × 10−15 (Table 2). Additionally, the C allele of rs11152369 was marginally enriched in our patients, with P = 1.80 × 10−2 (Table 2). The minor alleles of rs2958182, rs658977, and rs581653 were present in slight excess in our control subjects, whereas the alleles of the remaining five SNPs were equally distributed among patients and control subjects (Table 2). In addition, we did not find any evidence of association of rs10490775 in our patients; the PTPRG locus on chromosome 3 (data not shown), suggesting either that it does not contribute to late-onset FCD or that the overall allele effect is small and not readily detectable in all populations (because of insufficient power).
Table 2.
Distribution of SNP Alleles in Case and Control Subjects with Late-Onset FCD
| SNP | SNP Coordinates | Allele | Allele Count |
P | Odds Ratio | |
|---|---|---|---|---|---|---|
| Cases | Controls | |||||
| rs1261078 | chr18:52,866,791 | A | 0.906 | 0.875 | 0.2268 | 1.374 |
| G | 0.094 | 0.125 | ||||
| rs41483647 | chr18:52,901,607 | A | 0.874 | 0.856 | 0.5092 | 1.166 |
| G | 0.126 | 0.144 | ||||
| rs11152369 | chr18:53,066,328 | A | 0.885 | 0.936 | 0.0231 | 0.527 |
| C | 0.115 | 0.064 | ||||
| rs2958182 | chr18:53,149,021 | A | 0.247 | 0.325 | 0.0242 | 0.682 |
| T | 0.753 | 0.675 | ||||
| rs613872 | chr18:53,210,302 | G | 0.388 | 0.130 | 4.28 × 10−15 | 4.217 |
| T | 0.612 | 0.870 | ||||
| rs658977 | chr18:53,213,887 | A | 0.112 | 0.175 | 0.0180 | 0.594 |
| G | 0.888 | 0.825 | ||||
| rs581653 | chr18:53,215,739 | A | 0.900 | 0.825 | 0.0043 | 1.907 |
| T | 0.100 | 0.175 | ||||
| rs17089907 | chr18:53,302,088 | C | 0.953 | 0.961 | 0.7096 | 0.820 |
| T | 0.047 | 0.039 | ||||
| rs2646965 | chr18:53,692,716 | A | 0.629 | 0.633 | 0.9376 | 0.983 |
| C | 0.371 | 0.367 | ||||
FCD is believed to be more prevalent in women than in men.3 Therefore, we asked whether the presence of the G allele of rs613872 is more prevalent in female patients. We found no evidence for gender-specific enrichment. The risk allele was equally distributed among the male and female patients (rs613872, P = 0.36) and was not associated with an increased risk for the disease phenotype. Similarly, alleles of rs41483647, rs11152369, rs2958182, rs1319637, rs658977, and rs581653 were present in patients without gender bias (data not shown).
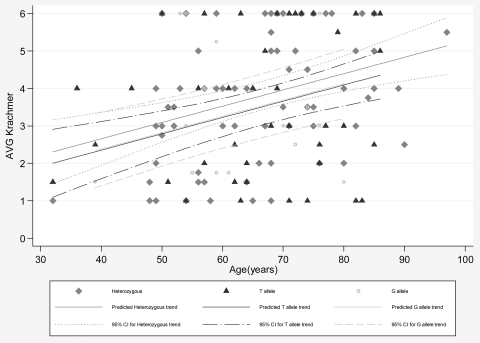
Next, we asked whether the G allele of rs613872 is associated with an increased risk for a severe disease phenotype. We examined the age-severity profiles of the patients homozygous for the G allele of rs613872 and compared them with affected heterozygous carriers and patients homozygous for the major allele using a linear regression model adjusted for age, the estimated difference between different groups. As shown in Figure 1, the estimated difference between affected heterozygous carriers and patients homozygous for the T allele was 0.302 (P = 0.33), the difference between patients homozygous for G allele and patients homozygous for T allele was 0.031 (P = 0.95), and the difference between the heterozygous carriers and patients homozygous for the G allele was −0.27 (P = 0.57).
Figure 1.
Comparison of the age severity profiles of affected persons homozygous for the G allele, heterozygous affected persons, and affected persons homozygous for the T allele. The estimated difference between affected heterozygous carriers and affected persons homozygous for the T allele was 0.302 (P = 0.337), the difference between affected persons homozygous for G allele and affected persons homozygous for T allele was 0.031 (P = 0.951), and the difference between the heterozygous carriers and affected persons homozygous for the G allele is −0.271 (P = 0.57).
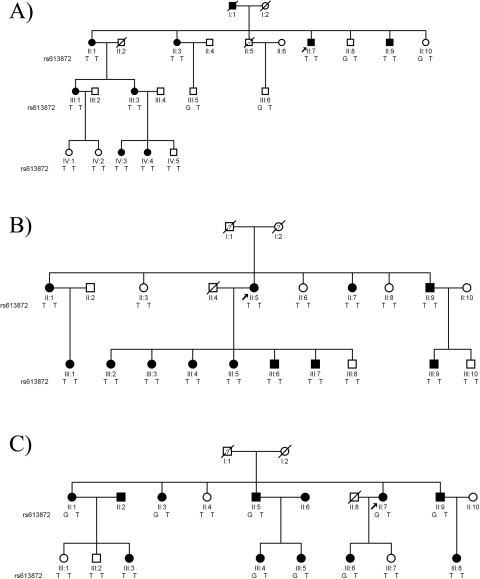
FCD2 has proven to be the most common locus in our familial cohort, with approximately 40% our large families mapping to chromosome 18q under an autosomal dominant model. To investigate the possible contribution of TCF4 to these families, we examined the segregation of the risk allele in three large families localized to FCD2 with significant LOD scores. In each case, and not surprising given the frequency of the G allele in the population, we found no evidence in which this variant could explain the disorder. In family MO the risk allele was present in all but two affected persons. Although this might argue multi-locus transmission as reported recently in another FCD family,10 in family PA the risk allele was absent from all persons tested (affected or unaffected), whereas in family HU the risk allele was present in only two affected persons and one unaffected person (Fig. 2).
Figure 2.
Pedigree drawings of families HU, PA, and MO with alleles of rs613872. Squares: men; circles: women; filled symbols: positive affected status; empty symbols: negative affected status; diagonal line through a symbol: deceased person; arrow next to a symbol: proband; question mark in a symbol: disease status unknown.
Given that rs613872 is present in the intronic region of TCF4 and that we recently identified pathogenic lesions in TCF8, another transcription factor implicated in the pathogenicity of both FCD and PPCD, we investigated whether causal mutations in TCF4 contribute to the genetic load of late-onset FCD. Therefore, we sequenced the entire coding region, including the exon-intron boundaries of TCF4 in 96 patients; however, we found no rare alleles except for known and reported SNPs.
Discussion
Recently, Baratz et al.19 reported rs613872 associated with late-onset FCD. We report the first independent replication of the association study implicating rs613872 in the pathogenicity of late-onset FCD in our cohort of 350 subjects of Northern European descent. Although our data confirm significant association with the disease phenotype, the fact is that the risk allele does not segregate with the disease phenotype in three FCD2 families (indeed, it is completely absent from family PA, which, alone generates a significant LOD score of 3.25 for FCD2). In addition, we did not identify any rare allele in the coding regions of TCF4. Taken together, these results suggest that rs613872 represents a susceptibility locus for late-onset FCD and is likely independent of FCD2 and that the causality of FCD2 remains unknown. The possibility remains that the TCF4 association might tag distant rare haplotypes elsewhere on 18q. However, given the known biological involvement of TCF4 with a bona fide FCD gene, TCF8, a more parsimonious explanation is that the TCF4 intronic association and FCD2 locus are genetically independent.
Heterozygous missense and nonsense mutations in TCF4 have been identified in patients with Pitt-Hopkins syndrome, a debilitating disorder characterized by mental retardation, wide mouth and distinctive facial features, and intermittent hyperventilation followed by apnea.23 SNP rs613872 resides in an intronic region that spans more than 100 kb, and its proximity to the flanking exons prompted us to investigate all conserved elements in the immediate neighborhood. We did identify an element approximately 500 bases 3′ of rs613872 that is conserved across mammalian genomes. However, when this sequence was examined for regulatory signatures with rVista (http://rvista.dcode.org/), it failed to identify any candidate enhancer or promoter motifs.
Together with the recent report by Baratz et al.,19 these data represent an interesting paradox wherein the most common late-onset FCD locus, FCD2, under Mendelian criteria and the most common susceptibility allele maps within a few Mb of each other. The combinatorial segregation analysis of the TCF4 risk allele and ancestral recombinants found in our FCD2-linked pedigrees potentially point to two independent genetic effects. However, we cannot formally exclude the possibility that multiple haplotypes at TCF4 that might be too rare to capture by genome-wide association study might, in fact, be tagging long-range alleles in the FCD2 region. Ultimately, genotyping of TCF4 in additional cohorts, especially from different ethnic backgrounds, or the identification of strong, rare, mutations that can explain FCD in the FCD2-linked families will be required to address the observed linkage and association signals.
Acknowledgments
The authors thank all the family members for their participation in this study.
Footnotes
Supported by National Eye Institute Grant R01EY016835 (JDG), Kwok Research Fund (JDG), and by Wilmer Biostatistics Core Grant R01EY01765 (data analysis). NK is a Distinguished Brumley Professor. JDG is a Margaret C. Mosher Professor of Ophthalmology.
Disclosure: S.A. Riazuddin, None; E.J. McGlumphy, None; W.S. Yeo, None; J. Wang, None; N. Katsanis, None; J.D. Gottsch, None
References
- 1. Lorenzetti DW, Uotila MH, Parikh N, Kaufman HE. Central cornea guttata: incidence in the general population. Am J Ophthalmol. 1967;64:1155–1158 [PubMed] [Google Scholar]
- 2. Krachmer JH, Purcell JJ, Jr, Young CW, Bucher KD. Corneal endothelial dystrophy: a study of 64 families. Arch Ophthalmol. 1978;96:2036–2039 [DOI] [PubMed] [Google Scholar]
- 3. Fuchs E. Dystrophia epithelialis corneae. Graefes Arch Clin Exp Ophthalmol. 1910:478–508 [Google Scholar]
- 4. Vogt A. Weitere Ergebnisse der Spaltlampenmikroskopie des vordern Bulbusabschnittes. Arch Ophthalmol. 1921:63–113 [Google Scholar]
- 5. Bergmanson JP, Sheldon TM, Goosey JD. Fuchs' endothelial dystrophy: a fresh look at an aging disease. Ophthalmic Physiol Opt. 1999;19:210–222 [DOI] [PubMed] [Google Scholar]
- 6. Gottsch JD, Zhang C, Sundin OH, Bell WR, Stark WJ, Green WR. Fuchs corneal dystrophy: aberrant collagen distribution in an L450W mutant of the COL8A2 gene. Invest Ophthalmol Vis Sci. 2005;46:4504–4511 [DOI] [PubMed] [Google Scholar]
- 7. Gottsch JD, Sundin OH, Liu SH, et al. Inheritance of a novel COL8A2 mutation defines a distinct early-onset subtype of Fuchs corneal dystrophy. Invest Ophthalmol Vis Sci. 2005;46:1934–1939 [DOI] [PubMed] [Google Scholar]
- 8. Sundin OH, Jun AS, Broman KW, et al. Linkage of late-onset Fuchs corneal dystrophy to a novel locus at 13pTel-13q12.13. Invest Ophthalmol Vis Sci. 2006;47:140–145 [DOI] [PubMed] [Google Scholar]
- 9. Sundin OH, Broman KW, Chang HH, Vito EC, Stark WJ, Gottsch JD. A common locus for late-onset Fuchs corneal dystrophy maps to 18q21.2-q21.32. Invest Ophthalmol Vis Sci. 2006;47:3919–3926 [DOI] [PubMed] [Google Scholar]
- 10. Riazuddin SA, Zaghloul NA, Al-Saif A, et al. Missense mutations in TCF8 cause late-onset Fuchs corneal dystrophy and interact with FCD4 on chromosome 9p. Am J Hum Genet. 2010;86:45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vithana EN, Morgan PE, Ramprasad V, et al. SLC4A11 mutations in Fuchs endothelial corneal dystrophy. Hum Mol Genet. 2008;17:656–666 [DOI] [PubMed] [Google Scholar]
- 12. Riazuddin SA, Vithana EN, Seet LF, et al. Missense mutations in the sodium borate co-transporter SLC4A11 cause late onset Fuchs corneal dystrophy. Hum Mutat. 2010;31:1261–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Higashi Y, Moribe H, Takagi T, et al. Impairment of T cell development in deltaEF1 mutant mice. J Exp Med. 1997;185:1467–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chamberlain EM, Sanders MM. Identification of the novel player deltaEF1 in estrogen transcriptional cascades. Mol Cell Biol. 1999;19:3600–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sobrado VR, Moreno-Bueno G, Cubillo E, et al. The class I bHLH factors E2–2A and E2–2B regulate EMT. J Cell Sci. 2009;122:1014–1024 [DOI] [PubMed] [Google Scholar]
- 16. Murre C, McCaw PS, Vaessin H, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544 [DOI] [PubMed] [Google Scholar]
- 17. Jan YN, Jan LY. HLH proteins, fly neurogenesis, and vertebrate myogenesis. Cell. 1993;75:827–830 [DOI] [PubMed] [Google Scholar]
- 18. Zhuang Y, Cheng P, Weintraub H. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2–2, and HEB. Mol Cell Biol. 1996;16:2898–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baratz KH, Tosakulwong N, Ryu E, et al. E2–2 protein and Fuchs's corneal dystrophy. N Engl J Med. 2010;363:1016–1024 [DOI] [PubMed] [Google Scholar]
- 20. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rabe-Hesketh S, Skrondal A. Multilevel and Longitudinal Modeling Using Stata. Chicago: Stata Press; 2005 [Google Scholar]
- 22. Hamilton LC. Statistics with STATA: Updated for Version 9. North Scituate, MA; Duxbury Press; 2006 [Google Scholar]
- 23. Amiel J, Rio M, de Pontual L, et al. Mutations in TCF4, encoding a class I basic helix-loop-helix transcription factor, are responsible for Pitt-Hopkins syndrome, a severe epileptic encephalopathy associated with autonomic dysfunction. Am J Hum Genet. 2007;80:988–993 [DOI] [PMC free article] [PubMed] [Google Scholar]