Abstract
HIV-1 drug resistance is a major clinical problem. Resistance is evaluated using in vitro assays measuring the fold change in IC50 caused by resistance mutations. Antiretroviral drugs are used at concentrations above IC50, however, and inhibition at clinical concentrations can only be predicted from IC50 if the shape of the dose–response curve is also known. Curve shape is influenced by cooperative interactions and is described mathematically by the slope parameter or Hill coefficient (m). Implicit in current analysis of resistance is the assumption that mutations shift dose–response curves to the right without affecting the slope. We show here that m is altered by resistance mutations. For reverse transcriptase and fusion inhibitors, single resistance mutations affect both slope and IC50. For protease inhibitors, single mutations primarily affect slope. For integrase inhibitors, only IC50 is affected. Thus, there are fundamental pharmacodynamic differences in resistance to different drug classes. Instantaneous inhibitory potential (IIP), the log inhibition of single-round infectivity at clinical concentrations, takes into account both slope and IC50, and thus provides a direct measure of the reduction in susceptibility produced by mutations and the residual activity of drugs against resistant viruses. The standard measure, fold change in IC50, does not correlate well with changes in IIP when mutations alter slope. These results challenge a fundamental assumption underlying current analysis of HIV-1 drug resistance and suggest that a more complete understanding of how resistance mutations reduce antiviral activity requires consideration of a previously ignored parameter, the dose–response curve slope.
Keywords: HIV/AIDS, pharmacology, virology, highly active antiretroviral therapy, evolution
With suboptimal treatment, drug-resistant HIV-1 evolves rapidly (1–7). Resistance results from mutations introduced by the error-prone HIV-1 reverse transcriptase (RT) (8–10). Treatment with inadequately suppressive regimens and/or problems with adherence allow additional cycles of replication and selection of resistant variants. For some antiretroviral drugs, single amino acid substitutions in the drug target produce high-level resistance (3, 11). Some mutations confer cross-resistance within a drug class (12–15). Many reduce enzyme or protein function, thereby decreasing viral fitness (16–19).
At the molecular level, resistance often results from mutations that interfere with drug binding to the target enzyme or protein (20–23). Additional mechanisms may also contribute. For example, resistance to zidovudine involves mutations that promote excision (24–26). Although the molecular mechanisms of resistance are well studied, the pharmacodynamics of resistance are less well understood. Resistance is typically measured as a change in IC50 (Table S1) relative to WT virus (27–30). Antiretroviral drugs are used at concentrations above IC50, however, and inhibition at clinical concentrations can only be predicted from IC50 if the shape of the dose–response curve is known. The shape is influenced by cooperative interactions and is described mathematically by the slope parameter or Hill coefficient (m) (31, 32). Certain drugs, notably nonnucleoside RT inhibitors (NNRTIs) and protease inhibitors (PIs), have cooperative dose–response curves with high slopes even though they target enzymes that are univalent with respect to the inhibitor (33). These high slopes may reflect a unique form of intermolecular cooperativity operative when multiple copies of a drug target participate in a given step in the virus life cycle (33). High slopes allow for extraordinarily high-level inhibition at concentrations above IC50.
In all basic pharmacodynamic models, including the Hill equation, the median effect equation (32), and the sigmoidal maximum effect (Emax) model (34), m has an exponential relationship to drug effect (31). Thus, slope is an important determinant of antiviral activity. Implicit in current analysis of resistance is the assumption that mutations shift dose–response curves to the right without affecting slope. The effects of resistance mutations on slope have never been described, however. If a mutation increases IC50 and decreases m, it may cause more resistance than is anticipated from a consideration of IC50 alone. Thus, the effects of mutations on slope must be understood. Here, we measure these effects and demonstrate that consideration of slope provides a unique way to understand the effects of resistance mutations.
Results
Dose–Response Curves for Antiretroviral Drugs Against Resistant Viruses.
Inhibition caused by a drug can be expressed as the fraction of single-cycle infection events affected by the drug (fa) or the fraction that remains unaffected (fu = 1 − fa) and is determined by the drug concentration D, IC50, and m according to the median effect equation (32, 35):
or
We studied single mutations that confer at least partial resistance according to the International AIDS Society-USA Drug Resistance Mutations Group and the Stanford University HIV Drug Resistance Database (36–38). Importantly, we analyzed resistance using a single-round infectivity assay because multiround assays distort m (39). Primary CD4+ T lymphoblasts were used as target cells because they mimic the principal target cells for HIV-1 in vivo. Assays were done in 50% (vol/vol) human serum to account for protein binding and with preincubations of target cells with nucleoside RT inhibitors (NRTIs) to allow concentrations of the active triphosphate forms of these drugs to reach steady state (SI Methods).
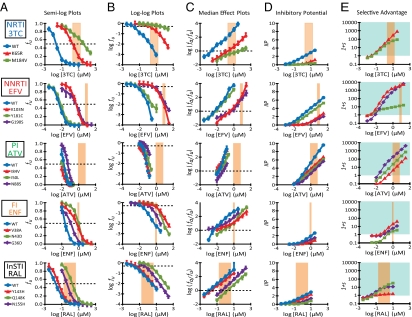
Fig. 1 shows dose–response curves for representative drugs from five classes. For each drug, Fig. 1A shows a standard semilog dose–response curve (fu vs. log D). Resistance mutations shift curves to the right and increase the IC50. For example, the K65R and M184V mutations in RT shift the curve for the NRTI lamivudine (3TC) substantially to the right (Fig. 1A, 3TC). Interestingly, single mutations associated with resistance to the PI atazanavir (ATV) cause only minor shifts and do not increase IC50 by more than 10-fold (Fig. 1A, ATV). A problem with semilog plots shown is that differences in antiviral activity at higher drug concentrations are obscured as fu approaches 0. Log-log dose–response curves (Fig. 1B) provide a better indication of antiviral activity at high drug concentration and reveal the impact of slope. If a mutation lowers m, inhibition achieved by the drug may be dramatically reduced. This is illustrated for M184V (Fig. 1B, 3TC), which increases IC50 but also reduces m such that increases in 3TC concentration cause a much less dramatic fall in fu than for WT. To determine IC50 and m, we used the median effect model (Eq. 2) to linearize dose–response curves (Fig. 1C). It is possible to directly determine m from the slope of median effect plots. The effect of M184V on slope is readily evident (Fig. 1C, 3TC). We have previously shown that PIs have steep slopes (33), and this is clear in the plot for ATV against WT (Fig. 1C, ATV). Some PI mutations reduce slope. For example, I84V, I50L, and N88S caused significant decreases in m (P = 0.0042, P = 0.029, and P = 0.016, respectively). Interestingly, mutations conferring resistance to the integrase strand transfer inhibitor (InSTI) raltegravir (RAL) did not significantly alter m.
Fig. 1.
Analysis of HIV-1 drug resistance mutations requires a consideration of dose–response curve slope. (A) Standard semilog dose–response curves for representative drugs from five different classes of antiretrovirals. Drugs were tested for inhibition of WT (blue curves) and mutant viruses bearing the indicated drug resistance mutations in a single-round infectivity assay. The fraction of infection events that remain unaffected (fu) by the indicated D is shown. Error bars represent SE. The peach-shaded region indicates the clinical concentration range of the relevant drug. fu = 0.5 (dotted line) indicates IC50 for each drug. Note that the D axis for 3TC and RAL is shifted by 1 log relative to the other drugs. (B) Log-log plots of the dose–response curves from A. IC50 for each drug can be determined from the points where the curves intersect log fu = −0.3 (dotted line). Error bars represent SE. (C) Median effect plots of the dose–response curves from A. IC50 for each drug can be determined from the points where the curves intersect log (fa/fu) = 0 (dotted line). The m value is the actual slope of the median effect plot. Error bars represent SE. (D) IIP for WT and mutant viruses at the indicated D. IIP was computed using Eq. 3 and the measured IC50 and m. (E) Selective advantage (written as 1 + s, where s is the selection coefficient) is the ratio of the infectivity of a preexisting mutant virus to the infectivity of WT at the indicated D. The blue shading indicates regions where mutant viruses have selective advantage. ENF, enfuvirtide.
Inhibition of replication at a given drug concentration can be predicted using the median effect equation, given IC50 and m (33). Inhibition can be expressed as instantaneous inhibitory potential (IIP), the number of logs by which single-round infection events are reduced at a clinically relevant drug concentration:
IIP plots for the selected resistance mutations are shown in Fig. 1D. As previously described (33), IIP values for NNRTIs and PIs are greater than the IIP values for drugs from other classes because of higher m values for these classes. Resistance mutations reduced IIP at clinical concentrations, but residual activity against resistant virus varied dramatically for different drugs and mutations. Importantly, drugs with high IIP values for WT [efavirenz (EFV) and ATV] retained more activity against some resistant viruses than drugs from other classes had against WT. For example, N88S in protease is considered to cause high-level ATV resistance (37), but the IIP of ATV against this mutant is still higher than the IIP of 3TC, enfuvirtide (ENF), or RAL toward WT.
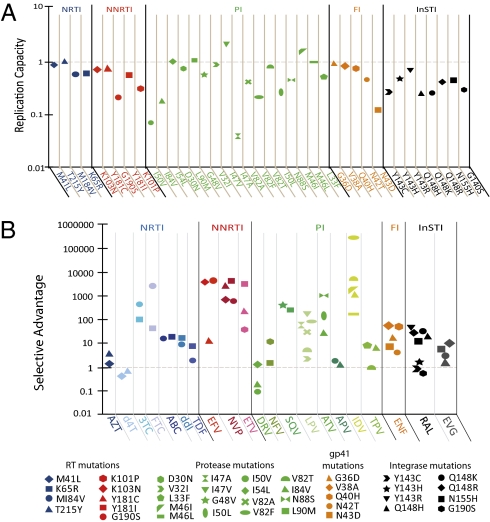
With suboptimal suppression, mutants with selective advantage over WT evolve. Selective advantage is the ratio of infectivity of a preexisting mutant to infectivity of WT at a given D. It can be estimated by multiplying the ratio fu (mutant)/fu (WT) at that D by the replication capacity, the fractional infectivity relative to WT in the absence of drug. Selective advantage takes into account reductions in fitness caused by the mutation. The selective advantage profiles for specific mutants are shown in Fig. 1E. Selective advantage >1 indicates that the mutant will prevail. In the clinical concentration range, most mutants had a selective advantage over WT, but the degree varied widely.
Effect of Resistance Mutations on IC50 and m.
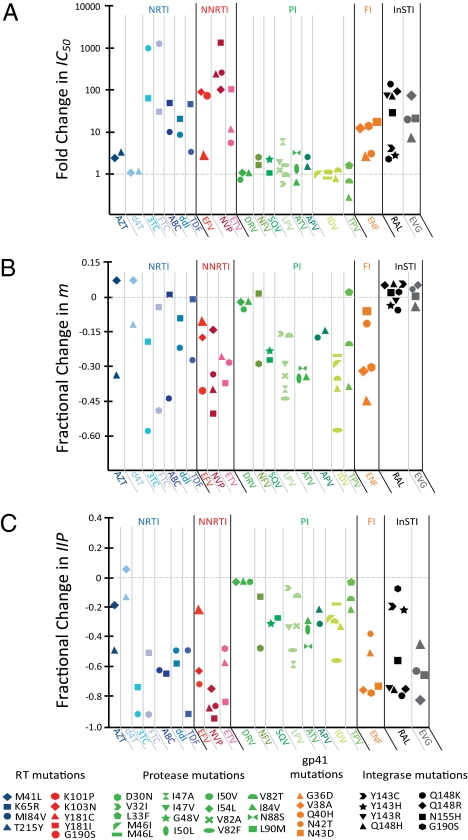
The above analysis was extended to major resistance mutations affecting licensed drugs from five classes. Fig. 2A shows the fold change in IC50 for each single mutant. Fig. 2B shows the fractional change in m relative to WT. Most NRTI, NNRTI, and fusion inhibitor mutations decreased m but increased IC50. In contrast, single PI mutations caused only relatively minor shifts in IC50, whereas m decreased. Interestingly, InSTI mutations resulted in an inverse pattern in which m values were preserved but IC50 increased (Fig. 2 A and B). Thus, depending on drug class, there were fundamental differences in the way resistance mutations affected the dose–response curves.
Fig. 2.
Effect of single drug resistance mutations on IC50 (A), m (B), and IIP (C). IC50 and m were determined for each mutant using median effect plots (Eq. 2) as previously described (33). The fractional change in IIP was computed as: 1 − (IIPmutant /IIPWT). IIP values for WT and mutant viruses were computed using Eq. 3 and the reported D at Cmax (33). Drugs are grouped by class: NRTIs (blue shades), NNRTIs (red shades), PIs (green shades), fusion inhibitor (orange), and InSTIs (black and gray). Within each class, mutations are indicated by the shape of the symbol. ABC, abacavir; APV, amprenavir; AZT, zidovudine; d4T, stavudine; DRV, darunavir; ENF, enfuvirtide; ETV, etravirine; EVG, elvitegravir; FI, fusion inhibitor; FTC, emtricitabine; LPV, lopinavir; NFV, nelfinavir; NVP, nevirapine; SQV, saquinavir; TPV, tipranavir.
To illustrate the effects of slope, we looked for mutations that altered m but not IC50. The V82F mutation in protease does not affect IC50 for indinavir (IDV) but does decrease m (Fig. 2 A and B). As shown in Fig. S1, this change results in a marked decrease in IDV susceptibility concentrations above IC50. This is most evident in a median effect plot. Extrapolation into the clinical concentration range reveals that the change in m alone can give a 5-log decrease in inhibition at minimum plasma concentration.
To extend the generality of these observations, we examined clinical isolates bearing the common resistance mutation M184V or K103N (Fig. S2). Isogenic viruses lacking the relevant mutation were generated by reverting the relevant codon to WT. As expected, M184V caused a large decrease in the slope of the 3TC dose–response curve, whereas K103N caused a moderate decrease in the slope of the EFV dose–response curve. Thus, the principles described here are not unique to laboratory strains.
Effect of Resistance Mutations on IIP.
Because changes in m can have a major effect on antiviral activity, we compared IIP values at the peak plasma concentration (Cmax) for each drug between WT and mutant viruses (Fig. 2C). Most resistance mutations decreased IIP compared with WT, albeit to varying extents. Interestingly, PI mutations caused substantial reductions in IIP even though they had only minor effects on IC50. This reflects the fact that IIP takes m, as well as IC50, into consideration.
The effect of the common M184V mutation on susceptibility to tenofovir disoproxil fumarate (TDF) illustrates the potential clinical significance of these concepts. M184V is thought to cause minimal resistance or even hypersusceptibility to TDF (40–43). As shown in Fig. S3A, there is little difference in IC50 between WT and M184V; however, the log-log plot (Fig. S3B) reveals a shallower slope for the M184V mutant, indicating substantial resistance to TDF at the high end of the clinical concentration range. This resistance would not be evident in assays that report only changes in IC50. At high concentrations, both WT and mutant viruses are inhibited. As is discussed below, the results of several clinical trials are consistent with the notion that M184V causes TDF resistance. Similar observations were made with clinical isolates carrying M184V (Fig. S3C).
Fold Change in IC50 Correlates Poorly with Fractional Change in IIP When Mutations Affect m.
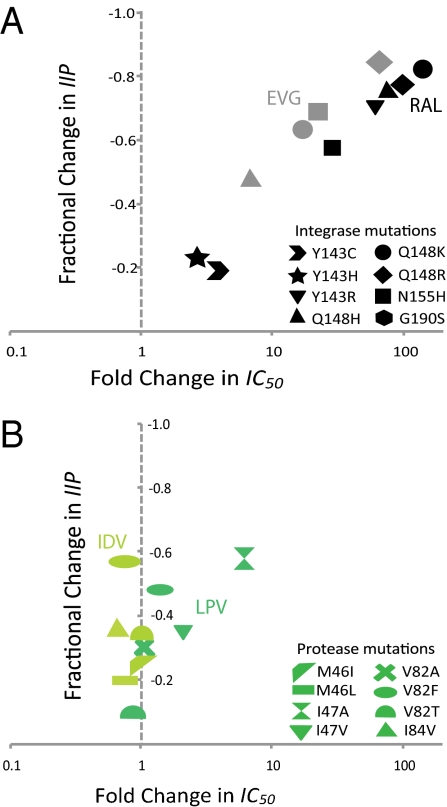
Our results indicate that the analysis of drug resistance is compromised by failure to include slope when this parameter is altered by a mutation. For integrase mutants, there were no significant fractional changes in m compared with WT (+0.064 to −0.058; P > 0.05). In contrast, PI mutants show up to a 0.578 fractional decrease in m (P < 0.05) (Fig. 2). Thus, PI resistance cannot be accurately assessed by considering IC50 alone. Fig. 3 demonstrates a lack of correlation between fold change in IC50 and fractional change in IIP when a mutation significantly changes m. For InSTI mutations, fractional change in IIP correlates well with fold change in IC50 because m is not altered (correlation coefficient = 0.99) (Fig. 3A). On the other hand, a poor correlation coefficient (0.35) was obtained for PI mutants (Fig. 3B). Hence, using fractional change in IIP ensures that the effects of a change in m are accounted for.
Fig. 3.
Correlation of fractional change in IIP with fold change in IC50. (A) Relationship between fractional change in IIP Cmax and fold change in IC50 for integrase mutants with respect to RAL and elvitegravir (EVG). (B) Relationship between fractional change in IIP Cmax and fold change in IC50 for protease mutants with respect to LPV and IDV. LPV, lopinavir.
Residual IIP Against Drug-Resistant Viruses.
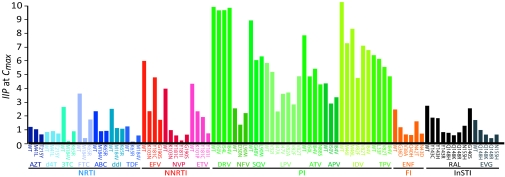
Because IIP is simply the number of logs by which a drug reduces in single-round infectivity, it can be used to compare the expected antiviral activity of different drugs against different viruses under clinical conditions. Fig. 4 displays the predicted IIP of each drug against WT and mutant viruses at Cmax based on m and IC50 measured in primary cells. The reduction in IIP produced by single mutations varies dramatically. Certain drugs, notably the PIs, retain substantial IIP against single mutants, consistent with the clinical observation that PI resistance generally requires multiple mutations. To some extent, differences in residual IIP reflect the tremendous differences in antiviral activity of different drug classes caused by differences in m.
Fig. 4.
Residual IIP against resistant viruses. Bars indicate the IIP against WT and viruses carrying the indicated single drug resistance mutations at Cmax. ABC, abacavir; APV, amprenavir; AZT, zidovudine; d4T, stavudine; DRV, darunavir; ENF, enfuvirtide; ETV, etravirine; EVG, elvitegravir; FI, fusion inhibitor; FTC, emtricitabine; LPV, lopinavir; NFV, nelfinavir; NVP, nevirapine; SQV, saquinavir; TPV, tipranavir.
Effect of Resistance Mutations on Replication Capacity and Selective Advantage.
We also measured the replication capacity of each viral clone in the absence of drug by comparing the infectivity of standardized amounts of WT and mutant viruses in primary cells (Fig. 5A). As expected, most single mutations reduced replication capacity. The effects observed were consistent with but of lower magnitude than effects observed in multiround assays (44, 45). Single mutations in protease produced varied effects on replication capacity. The L90M, I47V, and M46I mutants exhibited higher replication capacity than WT, consistent with a previous study (46). The protease mutants I50V and I47A had highly impaired replication capacity (<0.1) relative to WT. Overall, these results suggest that common resistance mutations impair replication capacity by up to 10-fold in a single cycle.
Fig. 5.
Effect of mutations on replication capacity and selective advantage. (A) Replication capacity was determined as the ratio of the infectivity of resistant virus to the infectivity of WT. (B) Selective advantage of viruses bearing single resistance mutations in the presence of the indicated drugs at Cmax. ABC, abacavir; APV, amprenavir; d4T, stavudine; DRV, darunavir; EFV, efavirenz; ENF, enfuvirtide; ETV, etravirine; EVG, elvitegravir; FTC, emtricitabine; LPV, lopinavir; NFV, nelfinavir; NVP, nevirapine; SQV, saquinavir; TPV, tipranavir.
Changes in replication capacity must be considered in assessing the selective advantage conferred by a mutation and are incorporated in the calculation of selective advantage described above. Fig. 5B shows the selective advantage of all mutants studied relative to WT at Cmax. Most single mutants had a 10- to 10,000-fold selective advantage over WT. For the thymidine analog stavudine and the PI darunavir, single mutants had little selective advantage, consistent with the observation that resistance to these drugs involves accumulation of multiple mutations (47, 48).
Analysis of Resistance Mutations Using Primary Cells vs. Transformed Cell Lines.
The above results were obtained using primary CD4+ T cells, which mimic the principal target cells for HIV-1 in vivo. Clinical methods for determining drug resistance phenotype rely on transformed cell lines, some of which are nonlymphoid (29). The use of cell lines facilitates high-throughput analysis and eliminates the donor-to-donor variability that can be seen in primary cell assays. Although IC50 may vary slightly from donor to donor in our primary CD4+ T-cell assay, the effect of resistance mutations on slope is relatively constant (Fig. S4). Nevertheless, primary cells are not optimal for high-throughput analysis; therefore, we compared dose–response curves in primary CD4+ T lymphoblasts, a transformed human T-cell line (Jurkat), and the human embryonic kidney cell line (293T) used in current clinical phenotypic assays (29) (Figs. S5 and S6). In all three cell types, M184V decreased the slope of the 3TC dose–response but there were differences in the extent of inhibition and the degree of slope change, particularly with the nonlymphoid 293T cells. Interestingly, the TDF effect described in Fig. S3 was not evident in either transformed line. Analysis of resistance in primary cells may reveal effects not evident in cell lines, but whether this advantage outweighs the difficulty, cost, and variability inherent in primary cell assays requires further study.
Discussion
IC50 is used as a measure of the potency of antiretroviral drugs. Resistance is typically expressed as the fold change in IC50 relative to WT. Clinical concentrations of drugs used are typically well above IC50, however. Therefore, determining inhibition at clinical concentrations requires a method for extrapolating from IC50 to higher concentrations. IC90 is sometimes used (46, 49); however, like IC50, it represents only a single point on the dose–response curve. However, m describes how inhibition changes with drug concentration, and is thus critical for understanding drug effects at clinical concentrations. Previous analyses of resistance have ignored m or assumed that mutations do not alter it. Here, we show that some single resistance mutations cause changes in slope that dramatically affect the amount of inhibition produced by drugs.
The importance of slope is illustrated by the M184V mutation and its effect on the response to TDF. This mutation reduces the slope such that in some regions of the dose–response curve, the mutant virus is resistant to TDF despite minimal change in IC50. Our results may explain the unexpected failure of a TDF, 3TC, and abacavir regimen (50). M184V was detected as the main mutation in most patients failing this regimen. It has been suggested that failure may have been attributable to viruses with K65R in addition to M184V (51, 52). In many failing patients, however, this double mutant was not detected. A simpler explanation is that M184V alone confers partial resistance to TDF. M184V also decreases slope of the didanosine (ddI) dose–response curve, which may explain the unexpected failure of TDF, 3TC, and ddI.*
Our results also show that some single resistance mutations leave substantial residual IIP. Because IIP takes into account alterations in both IC50 and m, it provides a unique way to quantitate how much antiviral activity a drug retains against mutant virus. We previously showed that the high slope values of the PIs and the NNRTIs result in high IIP (33). Not surprisingly, single mutations do not abolish all this antiviral activity. The PI class retains the highest residual IIP values in the presence of single mutations. Measuring the fraction of IIP remaining against a mutant virus provides an intuitive numerical value that is more quantitative than the current system of categorizing resistance as low, intermediate, or high level, based on fold change in IC50. Fold change in IC50 ignores m and is potentially boundless, resulting in demarcations of resistance that are not consistent across drug classes. Because fractional decrease in IIP uses a finite intuitive numerical value incorporating m, we propose that it may be a more useful way to compare the extent of resistance.
Our results also have implications for mechanisms of drug action. InSTI mutations affected IC50 but not m. Increases in IC50 can be attributed to decreased affinity, resulting in the need for higher drug concentrations for inhibition. Biochemical analyses of integrase mutants indicate that the off-rate is remarkably increased relative to WT (53). This finding, coupled with our observation of a constant slope close to 1, indicates that integrase inhibitors affect WT and mutant viruses by blocking a noncooperative reaction. At low multiplicity of infection, the critical reaction is mediated by a single molecular complex per cell consisting of a single integrase tetramer bound to the ends of the complete linear cDNA.
In contrast, single PI mutations decrease slope but have minor effects on IC50. Protease mutations are classified by position relative to the active site: substrate cleft mutations, protease flap mutations, and those at other conserved residues (23, 54). All single mutations studied exhibited the same pattern, however, showing modest change in IC50 but decreased slope. The comparatively small fold change in IC50 caused by protease mutations has been observed previously (55). We hypothesize that the reduction in m may reflect the fact that PIs block a cooperative process in which multiple copies of protease participate in virion maturation (33). The loss in enzyme efficiency caused by mutations affects the number of protease molecules needed to complete maturation before irreversible decay processes intervene.
As we and others have pointed out (33, 56–58), IIP is only one of several factors that determine the magnitude and durability of viral suppression by antiretroviral drugs. Other factors include drug half-life, distribution, toxicity and tolerability, drug interactions, and genetic barriers to resistance. A study by Henrich et al. (57) shows that IIP and inhibitory quotient have only modest correlations with clinical trial outcomes as measured using intent-to-treat analysis after 48 wk. This is expected, because many factors in addition to intrinsic antiviral activity determine clinical outcome. In addition, it is becoming clear that many regimens have sufficient inhibitory potential to suppress viral replication completely. In this situation, clinical outcome will be dominated by other factors. These caveats also apply to the residual IIP against resistant viruses. Furthermore, residual IIP only indicates the extent of inhibition a drug exerts on mutant virus and does not indicate how well this mutant virus is able to replicate and compete with other variants, including WT. Selective advantage profiles account for changes in replication capacity and indicate the relative probability that one mutant will be selected for at various drug concentrations. The observed high residual IIPs of some drugs, notably the PIs, suggest that these viral variants may nevertheless be adequately suppressed despite their higher selective advantage.
Taken together, these results demonstrate that a consideration of dose–response curve slope is important for assessing resistance. Values of replication capacity, IC50, m, and IIP obtained in our primary cell system may differ from those obtained with other isolates and with assays using cells lines. For simplicity, we studied single mutations, but the concepts discussed are grounded in fundamental laws of pharmacology and, with further development, can be applied to more complex patterns of resistance. It will be of interest to determine how m and IC50 are altered by complex patterns of mutations that arise in some patients, such as the thymidine analog mutations; the Q151M-complex; and those seen with PI resistance, including Gag-cleavage site mutations.
Methods
Detailed materials and methods are provided in SI Methods.
Dose–response curves for anti–HIV-1 drugs were obtained using an NL4-3 construct (33, 59) bearing single resistance mutations in a single-round infectivity assay. CXCR4-pseudotyped WT viruses encoding GFP in the env gene were used to infect primary CD4+ T lymphoblasts. Patient isolates bearing the M184V and K103N mutations were cloned into the NL4-3 backbone, and viruses generated from these were used in the single-round assay.
Infectivity was quantified by flow cytometry, and fu was calculated as %GFP+ cells in the presence of drug normalized by %GFP+ cells without drug. Using Eq. 2, dose–response curves were linearized, and IC50, m, and IIP were determined as previously described (33).
Supplementary Material
Acknowledgments
We thank Joel Blankson for his helpful review of this manuscript. This work was supported by National Institutes of Health Grant AI081600 and by the Howard Hughes Medical Institute.
Footnotes
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018360108/-/DCSupplemental.
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
*Jemsek J, Hutcherson P, Harper E, Poor virologic responses and early emergence of resistance in treatment naive, HIV-infected patients receiving a once-daily triple nucleoside regimen of didanosine, lamivudine, and tenofovir DF, 11th Conference on Retroviruses and Opportunistic Infections, February 8–11, 2004, San Francisco, CA (abstract 51).
References
- 1.Larder BA, Darby G, Richman DD. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science. 1989;243:1731–1734. doi: 10.1126/science.2467383. [DOI] [PubMed] [Google Scholar]
- 2.Coffin JM. HIV population dynamics in vivo: Implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 3.Wei X, et al. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 4.Clavel F, Hance AJ. HIV drug resistance. N Engl J Med. 2004;350:1023–1035. doi: 10.1056/NEJMra025195. [DOI] [PubMed] [Google Scholar]
- 5.Hirsch MS, et al. International AIDS Society-USA. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Top HIV Med. 2008;16:266–285. [PubMed] [Google Scholar]
- 6.Shafer RW, Schapiro JM. HIV-1 drug resistance mutations: An updated framework for the second decade of HAART. AIDS Rev. 2008;10:67–84. [PMC free article] [PubMed] [Google Scholar]
- 7.Gardner EM, Burman WJ, Steiner JF, Anderson PL, Bangsberg DR. Antiretroviral medication adherence and the development of class-specific antiretroviral resistance. AIDS. 2009;23:1035–1046. doi: 10.1097/QAD.0b013e32832ba8ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Preston BD, Poiesz BJ, Loeb LA. Fidelity of HIV-1 reverse transcriptase. Science. 1988;242:1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 9.Roberts JD, Bebenek K, Kunkel TA. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 10.Mansky LM, Temin HM. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tisdale M, Kemp SD, Parry NR, Larder BA. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci USA. 1993;90:5653–5656. doi: 10.1073/pnas.90.12.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richman DD. Susceptibility to nucleoside analogues of zidovudine-resistant isolates of human immunodeficiency virus. Am J Med. 1990;88(5BSuppl):8S–10S. doi: 10.1016/0002-9343(90)90414-9. [DOI] [PubMed] [Google Scholar]
- 13.Boyer PL, Currens MJ, McMahon JB, Boyd MR, Hughes SH. Analysis of nonnucleoside drug-resistant variants of human immunodeficiency virus type 1 reverse transcriptase. J Virol. 1993;67:2412–2420. doi: 10.1128/jvi.67.4.2412-2420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Race E, Dam E, Obry V, Paulous S, Clavel F. Analysis of HIV cross-resistance to protease inhibitors using a rapid single-cycle recombinant virus assay for patients failing on combination therapies. AIDS. 1999;13:2061–2068. doi: 10.1097/00002030-199910220-00008. [DOI] [PubMed] [Google Scholar]
- 15.Miller V, Larder BA. Mutational patterns in the HIV genome and cross-resistance following nucleoside and nucleotide analogue drug exposure. Antivir Ther. 2001;6(Suppl 3):25–44. [PubMed] [Google Scholar]
- 16.Back NK, et al. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 17.Goudsmit J, de Ronde A, de Rooij E, de Boer R. Broad spectrum of in vivo fitness of human immunodeficiency virus type 1 subpopulations differing at reverse transcriptase codons 41 and 215. J Virol. 1997;71:4479–4484. doi: 10.1128/jvi.71.6.4479-4484.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Croteau G, et al. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J Virol. 1997;71:1089–1096. doi: 10.1128/jvi.71.2.1089-1096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quiñones-Mateu MEAE. HIV-1 fitness: Implications for drug resistance, disease progression, and global epidemic evolution. In: Kuiken C, et al., editors. HIV Sequence Compendium 2001. Los Alamos, NM: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 2001. pp. 134–170. [Google Scholar]
- 20.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 21.Ding J, et al. Structure of HIV-1 RT/TIBO R 86183 complex reveals similarity in the binding of diverse nonnucleoside inhibitors. Nat Struct Biol. 1995;2:407–415. doi: 10.1038/nsb0595-407. [DOI] [PubMed] [Google Scholar]
- 22.Das K, et al. Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant. J Mol Biol. 1996;264:1085–1100. doi: 10.1006/jmbi.1996.0698. [DOI] [PubMed] [Google Scholar]
- 23.Mahalingam B, Louis JM, Hung J, Harrison RW, Weber IT. Structural implications of drug-resistant mutants of HIV-1 protease: High-resolution crystal structures of the mutant protease/substrate analogue complexes. Proteins. 2001;43:455–464. doi: 10.1002/prot.1057. [DOI] [PubMed] [Google Scholar]
- 24.Arion D, Kaushik N, McCormick S, Borkow G, Parniak MA. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): Increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry. 1998;37:15908–15917. doi: 10.1021/bi981200e. [DOI] [PubMed] [Google Scholar]
- 25.Meyer PR, Matsuura SE, So AG, Scott WA. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc Natl Acad Sci USA. 1998;95:13471–13476. doi: 10.1073/pnas.95.23.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer PR, Matsuura SE, Mian AM, So AG, Scott WA. A mechanism of AZT resistance: An increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol Cell. 1999;4:35–43. doi: 10.1016/s1097-2765(00)80185-9. [DOI] [PubMed] [Google Scholar]
- 27.Japour AJ, et al. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. The RV-43 Study Group, the AIDS Clinical Trials Group Virology Committee Resistance Working Group. Antimicrob Agents Chemother. 1993;37:1095–1101. doi: 10.1128/aac.37.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hertogs K, et al. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob Agents Chemother. 1998;42:269–276. doi: 10.1128/aac.42.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petropoulos CJ, et al. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2000;44:920–928. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qari SH, et al. Comparative analysis of two commercial phenotypic assays for drug susceptibility testing of human immunodeficiency virus type 1. J Clin Microbiol. 2002;40:31–35. doi: 10.1128/JCM.40.1.31-35.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill AV. The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J Physiol. 1910;40:iv–vii. [Google Scholar]
- 32.Chou TC. Derivation and properties of Michaelis-Menten type and Hill type equations for reference ligands. J Theor Biol. 1976;59:253–276. doi: 10.1016/0022-5193(76)90169-7. [DOI] [PubMed] [Google Scholar]
- 33.Shen L, et al. Dose-response curve slope sets class-specific limits on inhibitory potential of anti-HIV drugs. Nat Med. 2008;14:762–766. doi: 10.1038/nm1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holford NH, Sheiner LB. Understanding the dose-effect relationship: Clinical application of pharmacokinetic-pharmacodynamic models. Clin Pharmacokinet. 1981;6:429–453. doi: 10.2165/00003088-198106060-00002. [DOI] [PubMed] [Google Scholar]
- 35.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 36.Rhee SY, et al. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31:298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shafer RW. Rationale and uses of a public HIV drug-resistance database. J Infect Dis. 2006;194(Suppl 1):S51–S58. doi: 10.1086/505356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson VA, et al. Update of the drug resistance mutations in HIV-1: December 2009. Top HIV Med. 2009;17:138–145. [PubMed] [Google Scholar]
- 39.Ferguson NM, Fraser C, Anderson RM. Viral dynamics and anti-viral pharmacodynamics: Rethinking in vitro measures of drug potency. Trends Pharmacol Sci. 2001;22:97–100. doi: 10.1016/s0165-6147(00)01615-1. [DOI] [PubMed] [Google Scholar]
- 40.Wainberg MA, et al. In vitro selection and characterization of HIV-1 with reduced susceptibility to PMPA. Antivir Ther. 1999;4:87–94. doi: 10.1177/135965359900400205. [DOI] [PubMed] [Google Scholar]
- 41.Naeger LK, Margot NA, Miller MD. Increased drug susceptibility of HIV-1 reverse transcriptase mutants containing M184V and zidovudine-associated mutations: Analysis of enzyme processivity, chain-terminator removal and viral replication. Antivir Ther. 2001;6:115–126. [PubMed] [Google Scholar]
- 42.Whitcomb JM, Parkin NT, Chappey C, Hellmann NS, Petropoulos CJ. Broad nucleoside reverse-transcriptase inhibitor cross-resistance in human immunodeficiency virus type 1 clinical isolates. J Infect Dis. 2003;188:992–1000. doi: 10.1086/378281. [DOI] [PubMed] [Google Scholar]
- 43.Wolf K, et al. Tenofovir resistance and resensitization. Antimicrob Agents Chemother. 2003;47:3478–3484. doi: 10.1128/AAC.47.11.3478-3484.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cong ME, Heneine W, García-Lerma JG. The fitness cost of mutations associated with human immunodeficiency virus type 1 drug resistance is modulated by mutational interactions. J Virol. 2007;81:3037–3041. doi: 10.1128/JVI.02712-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quiñones-Mateu ME, Arts EJ. Fitness of drug resistant HIV-1: Methodology and clinical implications. Drug Resist Updat. 2002;5:224–233. doi: 10.1016/s1368-7646(02)00123-1. [DOI] [PubMed] [Google Scholar]
- 46.Mammano F, Trouplin V, Zennou V, Clavel F. Retracing the evolutionary pathways of human immunodeficiency virus type 1 resistance to protease inhibitors: Virus fitness in the absence and in the presence of drug. J Virol. 2000;74:8524–8531. doi: 10.1128/jvi.74.18.8524-8531.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watkins T, Resch W, Irlbeck D, Swanstrom R. Selection of high-level resistance to human immunodeficiency virus type 1 protease inhibitors. Antimicrob Agents Chemother. 2003;47:759–769. doi: 10.1128/AAC.47.2.759-769.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molla A, et al. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med. 1996;2:760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 49.Bangsberg DR, Kroetz DL, Deeks SG. Adherence-resistance relationships to combination HIV antiretroviral therapy. Curr HIV/AIDS Rep. 2007;4:65–72. doi: 10.1007/s11904-007-0010-0. [DOI] [PubMed] [Google Scholar]
- 50.Gallant JE, et al. ESS30009 Study. Early virologic nonresponse to tenofovir, abacavir, and lamivudine in HIV-infected antiretroviral-naive subjects. J Infect Dis. 2005;192:1921–1930. doi: 10.1086/498069. [DOI] [PubMed] [Google Scholar]
- 51.Khanlou H, Yeh V, Guyer B, Farthing C. Early virologic failure in a pilot study evaluating the efficacy of therapy containing once-daily abacavir, lamivudine, and tenofovir DF in treatment-naive HIV-infected patients. AIDS Patient Care STDS. 2005;19:135–140. doi: 10.1089/apc.2005.19.135. [DOI] [PubMed] [Google Scholar]
- 52.Underwood MR, et al. Sensitivity of phenotypic susceptibility analyses for nonthymidine nucleoside analogues conferred by K65R or M184V in mixtures with wild-type HIV-1. J Infect Dis. 2009;199:84–88. doi: 10.1086/595296. [DOI] [PubMed] [Google Scholar]
- 53.Dicker IB, et al. Biochemical analysis of HIV-1 integrase variants resistant to strand transfer inhibitors. J Biol Chem. 2008;283:23599–23609. doi: 10.1074/jbc.M804213200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ala PJ, et al. Molecular basis of HIV-1 protease drug resistance: Structural analysis of mutant proteases complexed with cyclic urea inhibitors. Biochemistry. 1997;36:1573–1580. doi: 10.1021/bi962234u. [DOI] [PubMed] [Google Scholar]
- 55.Rhee SY, et al. HIV-1 protease mutations and protease inhibitor cross-resistance. Antimicrob Agents Chemother. 2010;54:4253–4261. doi: 10.1128/AAC.00574-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.MacArthur RD. Instantaneous inhibitory potential and inhibitory quotient show a modest association with virologic outcome: Is either a useful surrogate for clinical drug efficacy? Clin Infect Dis. 2010;51:99–100. doi: 10.1086/653431. [DOI] [PubMed] [Google Scholar]
- 57.Henrich TJ, Ribaudo HJ, Kuritzkes DR. Instantaneous inhibitory potential is similar to inhibitory quotient at predicting HIV-1 response to antiretroviral therapy. Clin Infect Dis. 2010;51:93–98. doi: 10.1086/653430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen L, Siliciano RF. Achieving a quantitative understanding of antiretroviral drug efficacy. Clin Infect Dis. 2010;51:1105–1106, author reply 1106–1107. doi: 10.1086/656688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang H, et al. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J Virol. 2004;78:1718–1729. doi: 10.1128/JVI.78.4.1718-1729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.