Abstract
Plants deploy intracellular innate immune receptors to recognize pathogens and initiate disease resistance. These nucleotide-binding, leucine-rich repeat (NB-LRR) proteins are activated by pathogen effector proteins that are delivered into the host cell to suppress host defense responses. Little is known about the sites and mechanisms of NB-LRR activation, but some NB-LRR proteins can function inside the plant nucleus. We demonstrate that RPM1 is activated on the plasma membrane and does not relocalize to the nucleus. An autoactive RPM1(D505V) allele that recapitulates key features of normal RPM1 activation also resides on the plasma membrane. There is no detectable relocalization of activated RPM1 to the nucleus. Hindering potential nuclear entry of RPM1-Myc did not affect either its effector-triggered hypersensitive-response (HR) cell death or its disease resistance functions, further suggesting that nuclear translocation is not required for RPM1 function. RPM1 tethered onto the plasma membrane with a dual acylated N-terminal epitope tag retained the ability to mediate HR, consistent with this RPM1 function being activated on the plasma membrane. Plant NB-LRR proteins can thus function at various locations in the cell.
Keywords: plant disease resistance, plant nucleotide-binding leucine-rich repeat receptor immune receptor, type III bacterial effectors
Plants use a two-tiered receptor-based immune system to prevent the invasion of pathogenic microorganisms (1, 2). Plants express transmembrane pattern recognition receptors to detect microbe-associated molecular patterns (MAMPs). This leads to intracellular signaling and transcriptional output responses that can halt the growth of nonpathogens and is termed MAMP-triggered immunity (MTI). Successful pathogens suppress or dampen MTI via delivery of “effector proteins” into the host cell. Hence, effectors are virulence factors. To counter the activities of effector proteins, plants deploy polymorphic intracellular receptors. The largest class of these receptors, composed of nucleotide-binding, leucine-rich repeat (NB-LRR) proteins, has been conserved for several hundred million years and features a structure consisting of N-terminal signaling domains, a conserved central nucleotide-binding site, and C-terminal leucine-rich repeats. Plant NB-LRR proteins are structurally and functionally analogous to animal nucleotide-binding leucine-rich repeat receptor (NLR) innate immune receptors (3, 4). NB-LRR receptors recognize pathogen-encoded effector proteins either directly or indirectly via effector action on a host target. This initiates signal transduction and transcriptional reprogramming, resulting in effector triggered immunity and localized hypersensitive-response (HR) cell death (1, 2).
The functions of NB-LRR domains have been intensively investigated, but the mechanisms of NB-LRR activation and subsequent signaling are not well defined (5). A reasonable model has emerged wherein the N-terminal coiled-coil (CC) or Toll-interleukin-1 receptor (TIR) domain, in conjunction with the LRR domain, inhibits nucleotide exchange and/or hydrolysis in the resting state. Effector recognition, whether direct or indirect, results in an “unfurling” of the molecule, altered intra- and intermolecular interactions, changes in association with cochaperones, enhanced nucleotide turnover, and consequent downstream signaling, which occurs by mechanisms that are largely unknown (6–9).
Preactivation NB-LRR proteins have been localized to multiple subcellular compartments (10). NB-LRR activation has been associated with dynamic relocalization. In some cases, a small fraction of the total NB-LRR pool appears to relocalize to the nucleus, where it is thought to regulate defense gene transcription (10). These examples include both coiled coil (CC) (11–13) and TIR-NB-LRR (14–16) proteins and the RRS1 fusion protein that juxtaposes an NB-LRR protein with a WRKY class DNA binding domain (17). To date, however, there is no generalization regarding site(s) of NB-LRR activation or action.
RPM1 is a CC-NB-LRR protein (18). Inactive, signal-competent RPM1 is a plasma membrane-associated protein (19). RPM1 interacts with another plasma membrane localized protein, RIN4. RPM1 recognizes effector-mediated modifications of RIN4 in the presence of the bacterial type III effector proteins AvrRpm1 or AvrB (20, 21). AvrRpm1 and AvrB are also localized to the host plasma membrane by postdelivery acylation, and this localization is required for their ability to modify RIN4 and, consequently, activate RPM1 (22). RIN4 is required for wild-type levels of RPM1 accumulation at the membrane (20) and is a suppressor of weak RPM1 autoactivity (23). Hence, it is plausible that RPM1 activation occurs on the plasma membrane. Whether activated RPM1 relocalizes to initiate downstream signaling is not clear.
In this study, we determine the localization of an autoactive RPM1 allele that mimics accurately the activation state of wild-type RPM1. Collectively, our results indicate that activated RPM1 remains on the plasma membrane and that nuclear localization of RPM1 is not required for its function. Our results focus attention on how the activation state of RPM1 is transduced into an efficient disease resistance response.
Results
Transient Expression of an MHD Domain RPM1 Mutant Allele Induces Cell Death in Nicotiana benthamiana.
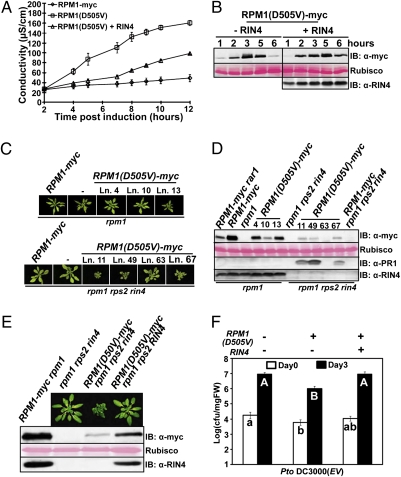
We wanted to address whether RPM1 relocalizes following activation. In principle, pre- and postactivation RPM1 states could coexist in the same cell during activation, due to temporal and spatial differences in signaling mediated by delivery of the relevant effector proteins. We therefore generated an autoactive allele of RPM1 to simplify the analysis of activation and potential consequent relocalization by biochemical methods. Mutations in the conserved MHD motif and Walker B motif can cause autoactivity that is likely achieved by intramolecular conformational rearrangement. We constructed myc-epitope tagged RPM1(D505V) and RPM1(D287A) alleles (Fig. 1A). Each was sufficient to induce cell death, as measured by changes in media conductivity, when transiently expressed in N. benthamiana (Fig. 1 B and C and Fig. S1). Because RPM1(D505V) exhibited stronger autoactivity than RPM1(D287A) (Fig. S1A), it was used for most of the subsequent experiments.
Fig. 1.
The MHD domain mutant RPM1(D505V) induces cell death in N. benthamiana. (A) RPM1 mutants used in this work. Wild-type sequences (Upper) and mutant sequences (Lower) are shown. (B and C) Estradiol-inducible RPM1 alleles were transiently expressed in N. benthamina (OD600 = 0.3). The HR-inducing activity of the RPM1 alleles was measured by ion leakage. (D and E) RPM1 proteins were detected by immunoblot. Samples were collected at 5 h after the induction with 40 μM estradiol and 40 μg of total extract loaded. Rubisco was used for protein loading control. Protein samples from uninfiltrated leaves were used as a control for nonspecific cross-reactivity.
We also reconstructed three previously isolated RPM1 loss-of-function mutants to test their effects in cis on the autoactivity of RPM1 (D505V) (24). The first, RPM1(S43F), is located in the N-terminal coiled-coil domain; the second, RPM1(P105S), is located in a highly variable spacer region; and the third, RPM1(G205E), is in the Walker A motif (P-loop) domain required for ATP binding. As expected, none of these three mutants is autoactive (Fig. 1C). Autoactivity of RPM1(D505V) requires the P-loop motif because it is suppressed in cis in RPM1(G205E/D505V) (Fig. 1B). This result shows that autoactivation of RPM1(D5050V) mimics P-loop–dependent activation of wild-type RPM1 triggered by type III effectors. Similarly, autoactivity of RPM1(D505V) requires the CC domain because it is also suppressed in RPM1(S43F/D505V) (Fig. 1C). By contrast, RPM1(P105S/D505V) retains autoactivity (Fig. 1C), suggesting that the functional requirement for RPM1(P105) occurs before the state in normal RPM1 activation mimicked by RPM1(D5050V). The steady-state abundance of RPM1 (D505V) protein is lower than that of RPM1 or the other mutant alleles (Fig. 1 D and E). Intramolecular suppression of autoactivation restored wild-type accumulation to RPM1(S43F/D505V). Thus, the low levels of RPM1(D505V) protein are consistent with our previous report that the disappearance of RPM1 is correlated with effector-mediated activation and the onset of HR cell death (19). We noted that two common proteasome inhibitors, MG132 and clasto-lactacystin β-lactone, could not stabilize steady-state levels of RPM1(D5050V), suggesting that the disappearance of activated RPM1 is not mediated by the proteasome.
Autoactivity of RPM1(D505V) Can Be Suppressed by RIN4.
RIN4 is formally a negative regulator of RPM1 because RPM1 is weakly autoactive in a rin4 null mutant (23). We co-expressed RIN4 and RPM1(D505V) in N. benthamiana to test the effect of RIN4 on the autoactivity of RPM1(D505V). RIN4 was constitutively expressed from its native promoter, and RPM1(D505V) expression was induced with estradiol 2 d after Agrobacterium infiltration. Our results showed that RIN4 expression delayed and diminished cell death (Fig. 2A). Co-expression of RIN4 also delayed the disappearance of RPM1(D505V) (Fig. 2B).
Fig. 2.
The autoactivity of RPM1(D505V) is sufficient to trigger enhanced disease resistance and can be suppressed by RIN4. (A) Ion leakage induced by transient expression of RPM1(D505V) with or without RIN4 in N. benthamiana. RIN4 is expressed from its native promoter [OD600 = 0.3 for RIN4 and OD600 = 0.3 for RPM1(D505V) or RPM1]. RPM1(D505V) was induced with 40 μM estradiol. (B) RPM1(D505V)-myc expression over a time course after estradiol induction, with or without RIN4 co-expression. (C) Pictures of rosettes from RPM1(D505V)-myc transgenic plants (genetic backgrounds noted at top). The plants were grown under 16-h-long days for 6 wk. (D) Protein expression levels in the transgenic plants. Samples were collected from 4-wk-old plants grown under 9-h short days. (E) Photos and protein expression levels of isogenic RPM1(D505V)-myc line 49 plants with or without RIN4. Line 49 of RPM1(D505V)-myc rpm1 rps2 rin4 (C and D) was crossed with rpm1 rps2 to obtain isogenic RPM1(D505V)-myc rpm1 rps2 plants. (F) Enhanced disease resistance following inoculation with Pto DC3000 of the isogenic plants from E. Seedlings were dip-inoculated with 2.5 × 107 cfu/mL of Pto DC3000. Pair-wise comparison for all means from day 0 or day 3 values were performed with a one-way ANOVA test coupled with Tukey–Kramer HSD with 95% confidence limits.
We confirmed and extended these findings by expressing Myc epitope-tagged RPM1(D505V) from its native promoter [gRPM1(D505V)-Myc] in transgenic Arabidopsis rpm1 or rpm1 rps2 rin4 plants to test the effects of RIN4 on the autoactivity of RPM1(D505V) in Arabidopsis. Use of rpm1 rps2 rin4 as a recipient for these experiments, instead of rpm1 rin4, is necessary to avoid the lethal ectopic autoactivation of RPS2 in the latter genotype (25). Independent transgenic gRPM1(D505V)-Myc rpm1 lines exhibited an essentially wild-type growth phenotype (Fig. 2 C and D) and expressed variable levels of RPM1(D505V). By contrast, independent transgenic gRPM1(D505V) rpm1 rps2 rin4 lines were difficult to recover, severely stunted as homozygous T2 individuals, expressed RPM1 at very low levels, and typically expressed the PR-1 protein, a marker of ectopic basal defense (Fig. 2 C and D). The severity of the dwarf phenotype was correlated with RPM1(D505V) levels in gRPM1(D505V) rpm1 rps2 rin4 T2 transgenic plants (Fig. 2 C and D). These results support our conclusion that RIN4 can also suppress the autoactivity of RPM1(D505V) in transgenic Arabidopsis (Fig. 2D).
RPM1(D505V) levels in rpm1 transgenic lines were lower than wild-type RPM1 from a well-characterized transgenic line, gRPM1-Myc rpm1 (19), and were similar to the expression of the same transgene introgressed into rar1 (Fig. 2D). The RAR1 cochaperone is required to maintain wild-type NB-LRR levels. Its absence results in reduced NB-LRR levels that can drop below a threshold required for function (24, 26). Hence, the level of RPM1-Myc detected in rar1 defines an RPM1 level lower than its functional threshold.
We extended this finding by crossing the dwarfed, PR-1 expressing, single-insertion gRPM1(D505V)-Myc rpm1 rps2 rin4 line 49 transgene (Fig. 2C) into rpm1 rps2. The resulting line had a wild-type phenotype, and the expression level of RPM1(D505V) was higher than that of the same transgene in an rpm1 rps2 rin4 sibling (Fig. 2E). The autoactivity of RPM1(D505V) in rpm1 rps2 rin4 also conferred enhanced basal resistance to the virulent pathogen Pto DC3000, and this was suppressed by the presence of RIN4 (Fig. 2F). We conclude that RIN4 can partially stabilize RPM1(D505V) and fully suppress its autoactivity of RPM1(D505V).
Effector-Mediated Activation of RIN4-Repressed RPM1(D505V).
RPM1(D505V), repressed by RIN4 in the lines shown in Fig. 2 C–E, was reactivated following recognition of AvrRpm1 delivered via the type III secretion system in both HR and growth restriction assays (Fig. S2 A–C). RPM1(D505V) levels in these lines are well below that of wild-type RPM1 (Fig. 2D). We infer that RPM1(D505V) has higher functional efficiency than wild type following effector-mediated activation. The weaker RPM1(D287A) autoactive allele also supported effector-mediated activation, at least for the ability to trigger HR in N. benthamiana (Fig. S1C). Thus, the autoactive RPM1(D505V) allele behaves like wild-type RPM1 with respect to both inhibition by RIN4 and subsequent effector-mediated activation.
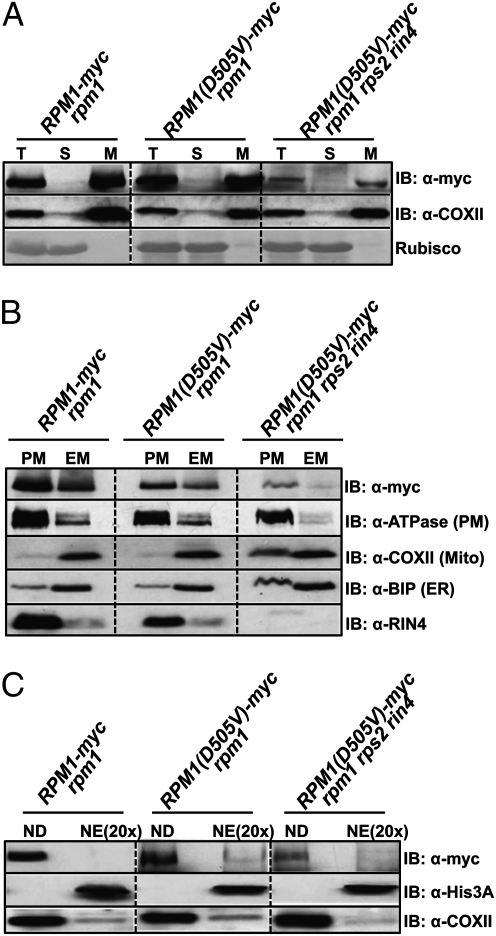
Activated RPM1(D505V) Localizes on the Arabidopsis Plasma Membrane.
We localized RPM1(D505V) in both the presence and the absence of RIN4 in rpm1 or rpm1 rps2 rin4 transgenic plants. We separated extracts into soluble and microsomal membrane fractions by ultracentrifugation (20) (Fig. 3A). These microsomal fractions were further separated into plasma membranes and endomembranes by two-phase partitioning (27). Like wild-type RPM1 (19), RPM1(D505V) is predominantly localized to the plasma membrane (Fig. 3B). It is noteworthy that the autoactive RPM1(D505V) in rpm1 rps2 rin4 also fractionates to the plasma membrane. This result demonstrates that active RPM1 is retained at the plasma membrane.
Fig. 3.
RPM1(D505V) localizes on the plasma membrane in Arabidopsis. (A) RPM1(D505V) resides on microsomal membranes. Total protein (T) was separated into soluble (S) and microsomal membrane (M) fractions by ultracentrifugation. COXII, a mitochondria membrane protein, was used as a membrane protein marker. Rubisco was used as a soluble protein marker. (B) RPM1(D505V) resides on the plasma membrane. The microsomal membrane fraction (M) was further separated into the plasma membrane fraction (PM) and the endomembrane fraction (EM). H+-ATPase, COXII, and Bip were used as plasma membrane, mitochondrial membrane, and endoplasmic reticulum markers, respectively. (C) RPM1(D505V) is not in the nuclear fraction. RPM1(D505V) was present in nuclear depleted (ND) but not in nuclear enriched (NE) fractions. Histone 3A was used as a nuclear protein marker. COXII was used as a non-nuclear marker. Note that NE fractions are loaded at a 20× yield equivalent compared with ND fractions. Three grams of leaves from plants grown under 16-h-long days for 6 wk were used for the experiments. The leaves of RPM1(D505V) rpm1 and RPM1(D505V) rpm1 rps2 rin4 are from line 4 and line 11, respectively (Fig. 2D).
Given recent interest in the relocalization of NB-LRR proteins to the nucleus, we also tested whether RPM1(D505V) relocalized to that compartment. We identified no RPM1(D505V) specifically in our nuclear extracts, even when these were overloaded by 20-fold (Fig. 3C). Overloading by 20-fold means that if 5% of the total RPM1(D505V) were in the nucleus, then that signal would equal the signal detected in the nuclear depleted lanes of Fig. 3C. Thus, our detection limit is significantly less than 5%. These results indicate that it is highly unlikely that activated RPM1(D505V) relocalizes to the nucleus.
We also established that there is no specific fragment processed from N-terminally tagged autoactive T7-RPM1(D505V)-YFP-HA alleles during its activation (Fig. S3). The results validate our results using C-terminal epitope-tagged RPM1(D505V). The results shown in Fig. 3 and Fig. S3 are consistent with the full-length of RPM1(D505V) being the functional molecule.
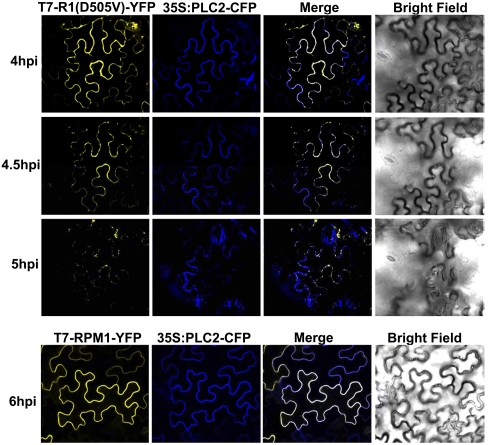
RPM1(D505V) Localizes on the Plasma Membrane of N. benthamiana Epidermal Cells.
We localized the autoactive T7-RPM1(D505V)-YFP-HA and control T7-RPM1-YFP-HA via confocal microscopy. PLC2-CFP was used as a plasma membrane marker (28). The estradiol-induced expression of T7-RPM1(D505V)-YFP-HA caused cell death beginning ∼5 h post induction (Fig. S3A). Consistent with this, dead or dying epidermal cells of N. benthamiana had shrunken shapes under the confocal microscope at this time point (Fig. 4). We observed the localization of RPM1(D505V) in precisely the same cell over time (Fig. 4). RPM1(D505V) was localized predominantly on the plasma membrane at 4 and 4.5 h post induction. Epidermal cell shape was unchanged at these time points. RPM1(D505V) levels were reduced, and the epidermal cell exhibited altered morphology at 5 h post induction. There was some alteration in the pattern of the remaining RPM1(D505V) signal distributed along the plasma membrane, and some redistribution into putative endomembrane structures at the 5-h time point. Because the plasma membrane marker PLC2 colocalized with RPM1(D505V) in the endomembrane structures, and because the translocation of PLC2 occurred only when the cell began to die, these endomembrane structures are likely to be a consequence of cell death, rather than a signaling intermediate. Preactivation T7-RPM1-YFP-HA was stably localized on the plasma membrane throughout this time course.
Fig. 4.
T7-RPM1(D505V)-YFP localizes to the plasma membrane of N. benthamiana epidermal cells. Transient expression of T7-RPM1(D505V)-YFP or T7-RPM1-YFP (OD600 = 0.3) was induced with 20 μM estradiol at 48 h after agrobacteria infiltration. Images were collected at the time after induction noted at left. 35S:PLC2-CFP (OD600 = 0.3) was co-expressed as a plasma membrane marker.
Excluding the Entry of RPM1 into the Nucleus Does Not Block Its Function.
We further confirmed that RPM1(D505V) neither relocalizes to, nor functions in, the nucleus, using nuclear export signal (NES)-tagged RPM1-Myc-NES under the control of the native RPM1 promoter (gRPM1-Myc-NES) (Materials and Methods) (11). We generated stable transgenic gRPM1-Myc-NES rpm1 and gRPM1-Myc-nes rpm1 plants and tested for effector-mediated HR and disease resistance functions over a range of expression levels. The efficiency of RPM1 function in either assay was determined by the RPM1 expression level, and not by the NES or mutant (nes) tag, indicating that the NES sequence does not alter RPM1 function (Fig. S4). Addition of a nuclear localization signal (NLS) onto these NES-tagged molecules affected localization, but did not alter function (Fig. S5). The nuclear localization of NLS-RPM1-GFP-NES was reduced, compared with that of NLS-RPM1-GFP-nes, showing that the NES tag is functional. However, the two proteins exhibited the same efficiency for effector-mediated HR. These data also support the contention that, under these expression conditions, the HR function of RPM1 does not require relocalization to the nucleus.
Plasma Membrane-Tethered RPM1 Retains HR Function in N. benthamiana.
If translocation of RPM1 from the plasma membrane to other subcellular locations is necessary for its function, then a plasma membrane-tethered RPM1 should exhibit reduced function. The first 12 amino acids of CBL1 (calcineurin B-like protein 1) can target proteins to the plasma membrane due to myristoylation on G2 and palmitoylation on C3 (29). We added this 12-amino-acid peptide (i.e., CBL) to the N terminus of several RPM1-Myc derivatives to tether them to the plasma membrane. A mutant CBL (mCBL) that could not be acylated was made as a control (Fig. 5A). We surprisingly found that ∼50% of the RPM1(G205E/D505V) P-loop mutant described above (Fig. 1) was soluble (Fig. 5B), whereas, as noted in Fig. 4, both RPM1 and RPM1(D505V) were strictly membrane localized. We used this result to test whether the CBL tag could retether the soluble RPM1(G205E/D505V) protein to the membrane. CBL, but not mCBL, efficiently tethered the soluble RPM1(G205E/D505V) to the membrane (Fig. 5B). Two-phase partitioning demonstrated that CBL-RPM1(G205E/D505V) was localized to the plasma membrane (Fig. 5D). Both CBL-RPM1 and mCBL-RPM1 were also retained on plasma membrane localization (Fig. 5 C and D).
Fig. 5.
Plasma membrane-tethered RPM1 retains effector-mediated HR function in N. benthamiana. (A). CBL tag contains dual-lipid modification sites. (B) The CBL tag tethers soluble RPM1(G205E/D505V)-Myc to the microsomal membrane following conditional transient expression in N. benthamiama. (C) CBL and mCBL tags do not affect the membrane localization of RPM1-Myc. (D) CBL-tagged proteins localize on the plasma membrane. For B–D, proteins were transiently expressed in N. benthamiana under the control of 35S promoter (OD600 = 0.5). Samples were collected at 40 h after agrobacteria infiltration. (E) The HR function of CBL-tagged RPM1. CBL- or mCBL-tagged RPM1-myc (R1) under the control of 35S promoter, T7-RIN4 (R4) under the control of its native promoter and AvrRpm1-HA (AvrRpm1) under the control of a dexamethasone (Dex)-inducible promoter were co-expressed in N. benthamiana (OD600 = 0.5, 0.3, and 0.05, respectively). AvrRpm1 was induced with 20 μM Dex at 36 h after infiltration. (F) The expression level of the constructs. The protein levels of the tagged RPM1 and RIN4 were detected at 2 h after induction. The protein levels of AvrRpm1 were detected at 6 h after induction.
Constitutive overexpression of CBL-RPM1, but not of mCBL-RPM1 or RPM1, led to weak autoactivity in N. benthamiana compared with effector-activated HR of the same molecules (compare Fig. 5E to Fig. S6). Accumulation of CBL-RPM1 was lower than that of either mCBL-RPM1 or RPM1, consistent with weak autoactivation (Fig. S6). Hence, the weak autoactivity observed with CBL-tagged RPM1, but not with mCBL-tagged RPM1, is likely due to the dual lipid modification rather than to the mere presence of the N-terminal tag. The weak autoactivity of CBL-RPM1 was suppressed by co-expression with RIN4 (Fig. S6), making this molecule a useful tool to test whether tethering RPM1 to the plasma membrane alters effector-mediated activation of HR.
We thus co-expressed in N. benthamiama CBL-RPM1 under the control of the 35S promoter and RIN4 under the control of its native promoter and then conditionally induced AvrRpm1 36 h post infiltration. CBL-RPM1-Myc supported nearly wild-type levels of effector-mediated HR (Fig. 5E). mCBL-RPM1 was less efficient than CBL-RPM1. The functional difference between CBL-RPM1 and mCBL-RPM1 could suggest that the dual-lipid modification of the CBL tag has positive effects on both autoactivity and effector-mediated RPM1-dependent HR, which partially compensates for any negative effects of the N-terminal tag. CBL-RPM1(G205E/D505V) did not support effector-mediated RPM1-dependent HR, indicating that HR requires a functional nucleotide-binding site, but that retethering the largely soluble RPM1(G205E/D505V) to the plasma membrane is insufficient to rescue the P-loop loss-of-function mutation. We conclude that the robust HR function of CBL-RPM1 indicates that plasma membrane-tethered RPM1 retains its HR function.
Discussion
This study was motivated by the question of how NB-LRR proteins, specifically RPM1, are activated by pathogen effectors, or by the activity of pathogen effectors on associated host targets, and whether activation is accompanied by dynamic relocalization of some or all of the active RPM1. Detailed analyses of this kind are still rare for plant NB-LRR proteins, and generalities have proven difficult to establish (5, 10).
Here, we used both transient and stable transgenic expression in transgenic Arabidopsis at native levels to establish that an autoactive RPM1(D505V) allele resides on the plasma membrane and does not obviously relocalize. The autoactivity of this allele is P-loop–dependent, suggesting that its activation proceeds by a mechanism similar to the activation of RPM1 by type III effectors. The autoactivity of RPM1(D505V) was repressed by RIN4 in both experimental systems. RIN4-repressed RPM1(D505V) could still be activated by the type III effector AvrRpm1. Hence, RPM1(D505V) autoactivity is a reasonable proxy for activation of wild-type RPM1. Our cell fractionation and confocal microscopy results indicate that activated RPM1 resides solely on the plasma membrane and is not detectable in the nucleus under conditions where we can easily detect less than 5% of RPM1 protein in the cell. Autoactive alleles in the MHD domain of NB-LRR proteins are likely to be generically useful tools to identify the endogenous partners and regulators of NB-LRR proteins and to sequence activation steps with subcellular localization.
We also tethered RPM1 onto the plasma membrane using a N-terminal dual-acylation sequence. This RPM1 derivative retained the ability to trigger HR in response to AvrRpm1 in the presence of RIN4. Collectively, our results suggest that activated RPM1 resides on the plasma membrane, that effector-mediated activation of RPM1 occurs on the plasma membrane, and, finally, that anchoring RPM1 to the plasma membrane does not significantly alter its HR induction function. Our results are consistent with the conclusion that RPM1 function does not require nuclear translocation for function.
At least two other NB-LRR receptors are likely to act at the plasma membrane. Pit is a CC-NB-LRR protein that is active against rice blast fungus. An analogous MHD domain allele, Pit(D485V), is autoactive in N. benthamiana. Confocal images from rice protoplasts indicated that Pit and Pit(D485D) are plasma membrane-localized (30). Pit interacts with Rac1; Pit(D485V), but not Pit, activated Rac1 (a regulator of reactive oxygen species production and cell death) on the plasma membrane, suggesting that activated Pit also initiates signal transduction on the plasma membrane (30). The plasma membrane-localized CC-NB-LRR protein RPS2 also associates with RIN4 at the membrane, and the relevant type III effector, AvrRpt2, is also likely to be acylated and localized to the plasma membrane, suggesting that the activation of RPS2 also starts on the plasma membrane. Effector-activated RPS2 is stable and localized to a microsomal compartment, although it is not clear whether this is actually the plasma membrane (25, 31).
We report the surprising finding that the P-loop is important for RPM1 localization. We noted that ∼50% of RPM1(G205E/D505V) was soluble and lacked autoactivity, compared with the nearly complete plasma membrane association of both RPM1 and RPM1(D505V). Notably, CBL-RPM1(G205E/D505V) is still a complete loss-of-function allele even though it is tethered to the plasma membrane. Thus, mutations in the RPM1 P-loop affect both localization and function. Consistent with this, the P-loop is required for intramolecular interactions of Rx (32), oligomerization of N (33), and direct interaction of L6 with the AvrL6 effector protein (34). The wide effects of P-loop mutations on NB-LRR function are therefore likely to reflect not only the necessity of nucleotide binding to maintain a functional resting state conformation, but also the correct localization of that optimal conformer for each NB-LRR protein.
RIN4 interacts with the CC domain of RPM1 (20). RIN4 could suppress the autoactivity of RPM1(D505V) either by blocking a required conformational change involving the CC domain or by blocking the interaction of RPM1 with downstream signaling components. The CC domain of RPM1 is critical for function (24). Similarly, the CC domain of the barley powdery mildew A NB-LRR immune receptor MLA interacts with WRKY transcription factors, and a CC dimer is both necessary and sufficient for signaling and required for the interaction of MLA with the WRKY transcription factors (35). These examples provide evidence that the CC domain of NB-LRR immune receptors can provide functions that may change during activation and can demonstrably act in different subcellular contexts. Similar findings are suggested for TIR domains (36) and for the noncanonical N terminus of Prf (37), focusing future work on understanding how these domains act as sensors of both effector-mediated target modification and initiators of downstream signaling.
Animal NLR proteins are also found in diverse subcellular locations, require analogous chaperones to maintain preactivation competence, and can relocalize upon activation to participate in a variety of signaling complexes. The mammalian CIITA NLR protein functions in the nucleus (38–40). NOD2 is present at, and recruits a downstream partner to, the plasma membrane; a mutation in its LRR renders NOD2 cytosolic and nonfunctional (41). NOD1 and NOD2 recruit autophagy components focally to plasma membrane sites of bacterial infection, and a common NOD2 variant associated with Crohn's disease fails to do so (42). These studies reveal that mammalian NLRs localize to different places in the cell before activation, that they can dynamically relocalize, and that their cellular localization influences downstream function. The sum of the data from plant NB-LRR and animal NLR intracellular immune receptors is consistent with the conceptual role of NB-LRR proteins as monitors of cellular homeostasis surveying a wide array of cellular defense machineries across a variety of subcellular addresses (1).
Materials and Methods
Plant Growth Conditions.
Arabidopsis plants used for HR and disease resistance experiments were grown at 24 °C under a 8-h light/16-h dark cycle. Plants for other purposes and N. benthamiana plants were grown in a greenhouse at 24 °C under a 16-h light/8-h dark cycle.
Vectors.
Vectors used in this study were constructed as described in SI Materials and Methods.
Transient Protein Expression in N. benthamiana.
Agrobacterium-mediated transient expression assays in N. benthamiana are described in SI Materials and Methods.
Total Protein Extraction and Primary Antibodies.
Total protein was extracted with extraction buffer [20 mM Tris·HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% SDS, 10 mM DTT]. The supernatant after centrifugation at 9,000 × g for 3 min was used for Western blot. The primary antibodies used were anti-Myc (University of North Carolina, Chapel Hill), anti-GFP (Roche), anti-HA (Roche), anti-T7 (Novagen), anti-COXII (Agrisera), anti–H+-ATPase (Agrisera), anti-Bip (Santa Cruz Biotechnology), anti-RIN4 (20), and anti-histone H3 (Abcam; ab1791). Anti-PR-1 (a gift from X. Dong, Duke University, Durham, NC).
Membrane Fractionation and Isolation of Nuclei.
Two-phase partitioning was performed as previously described (27) with a few modifications as described in SI Materials and Methods. Plant nuclei were isolated with a plant nuclei isolation kit (Sigma-Aldrich). Semipure preparation of nuclei was performed according to the protocol (Sigma-Aldrich).
HR and Disease Resistance Assays.
For conductivity assays, four leaf discs (0.8-cm diameter) were collected and floated in 5 mL of water with three replicates per sample (n = 12) at 2 h after induction with estradiol or dexmethasone. Ion leakage was measured at indicated time points using a conductivity meter (Orion; model 130). For HR assay, the leaves of 5-wk-old plants were infiltrated with Pto DC3000(avrRpm1) at 5 × 107 cfu/mL. The leaves were stained with Trypan blue 6 h after infiltration (43). Bacterial growth was measured as described (44).
Confocal Microscopy.
The localization of T7-RPM1(D505V)-YFP and T7-RPM1-YFP was observed under confocal microscopy (LSM 510 Meta; Carl Zeiss) as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Professors Roger Innes and John McDowell for critical reading, and members of the Dangl/Grant laboratory for helpful discussions. This work was funded by National Science Foundation Arabidopsis 2010 Program Grants IOS-0929410 and DOE DE-FG05-95ER20187 (to J.L.D.). T.K.E. was supported by a National Institutes of Health Training Grant.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104410108/-/DCSupplemental.
References
- 1.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Dodds PN, Rathjen JP. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 3.Ting JP, Willingham SB, Bergstralh DT. NLRs at the intersection of cell death and immunity. Nat Rev Immunol. 2008;8:372–379. doi: 10.1038/nri2296. [DOI] [PubMed] [Google Scholar]
- 4.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: Discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eitas TK, Dangl JL. NB-LRR proteins: Pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol. 2010;13:472–477. doi: 10.1016/j.pbi.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takken FLW, Albrecht M, Tameling WIL. Resistance proteins: Molecular switches of plant defence. Curr Opin Plant Biol. 2006;9:383–390. doi: 10.1016/j.pbi.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 7.van Ooijen G, et al. Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J Exp Bot. 2008;59:1383–1397. doi: 10.1093/jxb/ern045. [DOI] [PubMed] [Google Scholar]
- 8.Takken FL, Tameling WI. To nibble at plant resistance proteins. Science. 2009;324:744–746. doi: 10.1126/science.1171666. [DOI] [PubMed] [Google Scholar]
- 9.Shirasu K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol. 2009;60:139–164. doi: 10.1146/annurev.arplant.59.032607.092906. [DOI] [PubMed] [Google Scholar]
- 10.Caplan J, Padmanabhan M, Dinesh-Kumar SP. Plant NB-LRR immune receptors: From recognition to transcriptional reprogramming. Cell Host Microbe. 2008;3:126–135. doi: 10.1016/j.chom.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 11.Shen QH, et al. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science. 2007;315:1098–1103. doi: 10.1126/science.1136372. [DOI] [PubMed] [Google Scholar]
- 12.Tameling WI, et al. RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell. 2010;22:4176–4194. doi: 10.1105/tpc.110.077461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slootweg E, et al. Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell. 2010;22:4195–4215. doi: 10.1105/tpc.110.077537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burch-Smith TM, et al. A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 2007;5:e68. doi: 10.1371/journal.pbio.0050068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wirthmueller L, Zhang Y, Jones JD, Parker JE. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr Biol. 2007;17:2023–2029. doi: 10.1016/j.cub.2007.10.042. [DOI] [PubMed] [Google Scholar]
- 16.Cheng YT, et al. Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell. 2009;21:2503–2516. doi: 10.1105/tpc.108.064519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tasset C, et al. Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R-mediated immunity in Arabidopsis. PLoS Pathog. 2010;6:e1001202. doi: 10.1371/journal.ppat.1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grant MR, et al. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- 19.Boyes DC, Nam J, Dangl JL. The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci USA. 1998;95:15849–15854. doi: 10.1073/pnas.95.26.15849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackey D, Holt BF, III, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 21.Chung EH, et al. Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe. 2011;9:125–136. doi: 10.1016/j.chom.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nimchuk Z, et al. Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell. 2000;101:353–363. doi: 10.1016/s0092-8674(00)80846-6. [DOI] [PubMed] [Google Scholar]
- 23.Belkhadir Y, Nimchuk Z, Hubert DA, Mackey D, Dangl JL. Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell. 2004;16:2822–2835. doi: 10.1105/tpc.104.024117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tornero P, Chao RA, Luthin WN, Goff SA, Dangl JL. Large-scale structure-function analysis of the Arabidopsis RPM1 disease resistance protein. Plant Cell. 2002;14:435–450. doi: 10.1105/tpc.010393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- 26.Bieri S, et al. RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell. 2004;16:3480–3495. doi: 10.1105/tpc.104.026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsson C, Sommarin M, Widell S. Isolation of highly purified plant plasma membranes and separation of inside-out and right-side-out vesicles. Methods Enzymol. 1994;228:451–469. [Google Scholar]
- 28.Hirayama T, Mitsukawa N, Shibata D, Shinozaki K. AtPLC2, a gene encoding phosphoinositide-specific phospholipase C, is constitutively expressed in vegetative and floral tissues in Arabidopsis thaliana. Plant Mol Biol. 1997;34:175–180. doi: 10.1023/a:1005885230896. [DOI] [PubMed] [Google Scholar]
- 29.Batistic O, Sorek N, Schültke S, Yalovsky S, Kudla J. Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell. 2008;20:1346–1362. doi: 10.1105/tpc.108.058123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawano Y, et al. Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe. 2010;7:362–375. doi: 10.1016/j.chom.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 31.Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 32.Moffett P, Farnham G, Peart J, Baulcombe DC. Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 2002;21:4511–4519. doi: 10.1093/emboj/cdf453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mestre P, Baulcombe DC. Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell. 2006;18:491–501. doi: 10.1105/tpc.105.037234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dodds PN, et al. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci USA. 2006;103:8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maekawa T, et al. Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe. 2011;9:187–199. doi: 10.1016/j.chom.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 36.Bernoux M, et al. Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling and autoregulation. Cell Host Microbe. 2011;9:200–211. doi: 10.1016/j.chom.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gutierrez JR, et al. Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J. 2010;61:507–518. doi: 10.1111/j.1365-313X.2009.04078.x. [DOI] [PubMed] [Google Scholar]
- 38.Cressman DE, Chin KC, Taxman DJ, Ting JP. A defect in the nuclear translocation of CIITA causes a form of type II bare lymphocyte syndrome. Immunity. 1999;10:163–171. doi: 10.1016/s1074-7613(00)80017-5. [DOI] [PubMed] [Google Scholar]
- 39.Zhu XS, et al. Transcriptional scaffold: CIITA interacts with NF-Y, RFX, and CREB to cause stereospecific regulation of the class II major histocompatibility complex promoter. Mol Cell Biol. 2000;20:6051–6061. doi: 10.1128/mcb.20.16.6051-6061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harton JA, Cressman DE, Chin KC, Der CJ, Ting JP. GTP binding by class II transactivator: Role in nuclear import. Science. 1999;285:1402–1405. doi: 10.1126/science.285.5432.1402. [DOI] [PubMed] [Google Scholar]
- 41.Lécine P, et al. The NOD2-RICK complex signals from the plasma membrane. J Biol Chem. 2007;282:15197–15207. doi: 10.1074/jbc.M606242200. [DOI] [PubMed] [Google Scholar]
- 42.Travassos LH, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 43.Koch E, Slusarenko A. Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell. 1990;2:437–445. doi: 10.1105/tpc.2.5.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tornero P, Dangl JL. A high-throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana. Plant J. 2001;28:475–481. doi: 10.1046/j.1365-313x.2001.01136.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.