In their natural environments, bacteria often colonize biotic and abiotic surfaces, and the intimate association with biotic surfaces can be a prerequisite for pathogenesis. Once on a surface, cells may remain sessile and simply grow and divide. Alternatively, cells may display active cell movements and translocate across the surface by one of three different mechanisms: swarming, twitching, or gliding (1, 2). Swarming depends on rotating flagella that are randomly distributed on the cell surface and form a bundle that pushes a cell forward (2, 3). Twitching depends on type IV pili, which extend from the cell surface, attach to the surface on which a cell resides, and then retract with a retraction event, pulling a cell forward (1, 4). Whereas swarming and twitching each depend on a cell surface organelle, gliding occurs in the absence of any eye-catching structures, and the molecular mechanism has remained puzzling for years. In PNAS, an important part of the puzzle is solved by the identification of the molecular motor that powers gliding in Myxococcus xanthus (5). Moreover, the finding that this motor moves in a directed manner between subcellular regions has major implications for our understanding of how bacterial cells become spatially organized.
The story begins in 2007 with the astonishing observation that the AglZ protein, which is required for gliding motility in the Gram-negative, rod-shaped bacterium M. xanthus (6), localizes to the leading cell pole as well as to focal adhesion complexes (FACs) that are distributed regularly along the cell body (7). In a moving cell, the FACs remain stationary with respect to the surface on which the cell is translocating and thus indirectly move from the leading to the lagging cell pole. If a cell is immobilized on a surface, the FACs visibly move from one pole to the other. So, in both scenarios, the FACs move in a directed manner from one pole to the other.
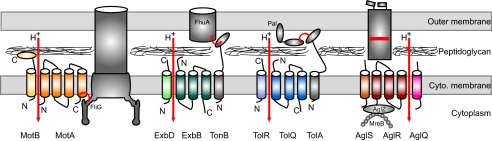
Sun et al. (5) build on these observations and find that tiny beads attached to the surface of immobilized cells also move from the leading to the lagging cell pole. Moreover, the beads colocalize with AglZ-YFP in the FACs, providing evidence that traction force is generated at the sites of the FACs. Using chemical biology, Sun et al. (5) identify the energy source that powers the FACs motor as the H+-gradient over the cytoplasmic membrane. In addition to ATP synthase, three homologous bacterial motors are known to harness the energy of this gradient to generate mechanical force: MotA/MotB (8), ExbB/ExbD (9), and TolQ/TolR (10) (Fig. 1). Armed with this information, Sun et al. (5) searched a collection of M. xanthus gliding mutants (11) and identified the aglQRS locus that encodes one MotA/TolQ/ExbB homolog (AglR) and two MotB/TolR/ExbD homologs (AglQ and AglS) as their candidate for the motor. All three AglQ/AglR/AglS proteins are required for gliding, and according to pull-down assays
Fig. 1.
Schematics of the MotA/MotB, ExbB/ExbD, TolQ/TolR, and AglQ/AglR/AglS proton channels. The membrane topology of the involved proteins is shown in the context of the cell envelope. System specific components are indicated in gray. H+-flux (vertical red arrows) induced interactions that determine the system specific outputs are indicated by red arrows. For simplicity, the H+-flux in the AglQRS system is only shown to involve AglR and AglQ. For simplicity, the interactions between AglQRS and AglZ and between AglZ and MreB are shown as direct. For simplicity, all complexes are shown with 1:1 stoichiometries. The stoichiometry of MotAB is 4:2, with 11 to 12 complexes making up the full stator ring, of ExbBDTonB 7:2:1, of TolQRA 4–6:2:1, and of AglQRS it is not known.
The findings of Sun et al. have implications well beyond gliding motility.
they interact to form a complex. Turning to live-cell imaging, it is shown that AglQ-mCherry localization matches that of AglZ: in moving cells, AglQ-mCherry localizes in a cluster at the leading pole as well as to FACs; and in immobilized cells, AglQ moves from the leading to the lagging cell pole. Genetic inactivation of the H+-channel in the AglQ/AglR/AglS complex blocked gliding as well as trafficking of FACs to the lagging pole but did not interfere with formation of the FACs. Thus, the AglQ/AglR/AglS complex seems to be the component of the FACs that is involved in force generation, and force generation leads to trafficking of the FACs in a directed manner from the leading to the lagging cell pole.
M. xanthus cells occasionally undergo reversals during which cells stop and then resume gliding in the opposite direction, with the old leading pole becoming the new lagging cell pole. During reversals gliding motility proteins relocate between the poles, and the FACs disappear (7, 12). After a reversal, the FACs reappear and now track in the opposite direction, suggesting that they can generate force in at least two directions, depending on the overall leading/lagging pole polarity of a cell.
The findings of Sun et al. (5) have implications well beyond gliding motility. Over the last decade it has become clear that bacterial cells are spatially highly organized (13). In eukaryotic cells, the spatial organization depends on cargo transport by molecular motors that track directionally on filaments, with kinesins and dynein tracking in opposite directions on microtubules and myosins tracking on actin (14). The findings of Sun et al. (5) identify AglQ/AglR/AglS as the first bacterial motor able to move in a directed manner between subcellular regions. Therefore, in a larger perspective, the findings by Sun et al. (5) open up the possibility that similar motors could be involved in organizing bacterial cells spatially by bringing cargo such as proteins, DNA, or mRNA to their individual subcellular addresses.
A challenge for the future will be to understand how the H+-flux through AglQ/AglR/AglS is converted to mechanical force. A comparison with homologous systems is instructive (Fig. 1). The MotA/MotB proteins form an H+-channel in the cytoplasmic membrane, make up the stator part of the flagellar rotary motor, and interact with the FliG protein in the rotor part of the flagellar motor to generate torque, thereby setting up the flagellar rotations (8). MotB contains a domain that fixes MotA/MotB to the peptidoglycan, in that way allowing the complex to function as a stator. Many flagella can rotate both clockwise and counterclockwise, and in both directions rotations depend on the interaction between MotA and FliG. ExbB and ExbD interact with TonB to form a complex in the cytoplasmic membrane (9). H+-flux through ExbB/ExbD induces conformational changes in TonB that are converted to conformational changes in TonB-dependent receptors in the outer membrane, such as FhuA, to energize transport over this membrane. TolQ and TolR together with TolA form a complex in the cytoplasmic membrane that is important for outer membrane integrity and cell division (10). Here, H+-flux through the TolQ/TolR H+-channel induces conformational changes in TolA, which ultimately changes the interaction between TolA and the outer membrane protein Pal (10). Thus, in all three systems the energy from H+-flux is converted to changes in protein conformation that regulate membrane processes. Moreover, in all three systems, the H+-channels MotAB, TolQR, and ExbBD are the motors that harvest the energy from the H+-flux, and this energy is converted to a mechanical output because the motors are hooked up to a partner protein, such as FliG, TolA, or TonB. The AglQ/AglR/AglS system presumably works by a similar mechanism, and the partner protein(s) likely extend from the cytoplasmic membrane to the cell surface, for the proton-flux to be converted to traction force (Fig. 1).
Sun et al.’s article (5) raises many fascinating questions for the future. A critical question is how the FACs move given the presence of peptidoglycan in the periplasm (i.e., does movement of the FACs involve breaking and resealing of peptidoglycan?). FACs are suggested to assemble at the leading pole and disassemble at the lagging pole (7). How does this happen? It was previously suggested the FACs move or even track along the cytoskeletal element formed by the actin-like MreB protein in the cytoplasm (7, 15). An open question is the function of this interaction. Recently, several proteins required for gliding were suggested to be part of FACs (16), and the AgmU protein was proposed to form a helical filament in the periplasm (17). Resolving the function of these proteins in gliding is a formidable challenge. In total, the M. xanthus genome contains eight gene clusters for MotA/TolQ/ExbB and MotB/TolR/ExbD homologs. In addition to aglQRS, the aglXV cluster has been implicated in gliding motility (11, 17). It will be interesting to determine how many different gliding motors M. xanthus contains.
Gliding motility systems in different bacteria are very different (1). For instance, the gliding proteins in Flavobacterium johnsoniae are not present in M. xanthus, suggesting that mechanisms for gliding evolved independently several times (1). In the case of M. xanthus, the identification of AglQ/AglR/AglS as the gliding motor suggests that this gliding machinery evolved by tinkering with spare parts from an existing motor.
Acknowledgments
Our work on motility is supported by The Max Planck Society, the graduate program “Intra- and intercellular transport and communication” funded by the German Research Foundation, and the Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz (LOEWE) Research Center for Synthetic Microbiology.
Footnotes
The author declares no conflict of interest.
See companion article on page 7559.
References
- 1.Jarrell KF, McBride MJ. The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol. 2008;6:466–476. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- 2.Harshey RM. Bacterial motility on a surface: Many ways to a common goal. Annu Rev Microbiol. 2003;57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 3.Kearns DB. A field guide to bacterial swarming motility. Nat Rev Microbiol. 2010;8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craig L, Li J. Type IV pili: Paradoxes in form and function. Curr Opin Struct Biol. 2008;18:267–277. doi: 10.1016/j.sbi.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun M, Wartel M, Cascales E, Shaevitz JW, Mignot T. Motor-driven intracellular transport powers bacterial gliding motility. Proc Natl Acad Sci USA. 2011;108:7559–7564. doi: 10.1073/pnas.1101101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang R, et al. AglZ is a filament-forming coiled-coil protein required for adventurous gliding motility of Myxococcus xanthus. J Bacteriol. 2004;186:6168–6178. doi: 10.1128/JB.186.18.6168-6178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mignot T, Shaevitz JW, Hartzell PL, Zusman DR. Evidence that focal adhesion complexes power bacterial gliding motility. Science. 2007;315:853–856. doi: 10.1126/science.1137223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minamino T, Imada K, Namba K. Molecular motors of the bacterial flagella. Curr Opin Struct Biol. 2008;18:693–701. doi: 10.1016/j.sbi.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Postle K, Kadner RJ. Touch and go: Tying TonB to transport. Mol Microbiol. 2003;49:869–882. doi: 10.1046/j.1365-2958.2003.03629.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhang XY-Z, et al. Mapping the interactions between Escherichia coli tol subunits: Rotation of the TolR transmembrane helix. J Biol Chem. 2009;284:4275–4282. doi: 10.1074/jbc.M805257200. [DOI] [PubMed] [Google Scholar]
- 11.Youderian P, Burke N, White DJ, Hartzell PL. Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner. Mol Microbiol. 2003;49:555–570. doi: 10.1046/j.1365-2958.2003.03582.x. [DOI] [PubMed] [Google Scholar]
- 12.Leonardy S, Freymark G, Hebener S, Ellehauge E, Søgaard-Andersen L. Coupling of protein localization and cell movements by a dynamically localized response regulator in Myxococcus xanthus. EMBO J. 2007;26:4433–4444. doi: 10.1038/sj.emboj.7601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shapiro L, McAdams HH, Losick R. Why and how bacteria localize proteins. Science. 2009;326:1225–1228. doi: 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross JL, Ali MY, Warshaw DM. Cargo transport: Molecular motors navigate a complex cytoskeleton. Curr Opin Cell Biol. 2008;20:41–47. doi: 10.1016/j.ceb.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mauriello EMF, et al. Bacterial motility complexes require the actin-like protein, MreB and the Ras homologue, MglA. EMBO J. 2010;29:315–326. doi: 10.1038/emboj.2009.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nan B, Mauriello EMF, Sun IH, Wong A, Zusman DR. A multi-protein complex from Myxococcus xanthus required for bacterial gliding motility. Mol Microbiol. 2010;76:1539–1554. doi: 10.1111/j.1365-2958.2010.07184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nan B, et al. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc Natl Acad Sci USA. 2011;108:2498–2503. doi: 10.1073/pnas.1018556108. [DOI] [PMC free article] [PubMed] [Google Scholar]