Abstract
In recent years, considerable progress has been made in understanding the molecular mechanisms underlying olfaction in insects. Because of the diverse nature of the gene families involved, this process has largely relied on genomic data. As a consequence, studies have focused on a small subset of species with extensive genomic information. For Lepidoptera, a large order historically crucial to olfactory research, this circumstance has mostly limited advances to the domesticated species Bombyx mori, with some progress in the noctuid Heliothis virescens based on a nonpublic partial genome database. Because of the limited behavioral repertoire and nonexistent ecological importance of Bombyx, molecular data on the tobacco hornworm Manduca sexta are of utmost importance, especially with regards to its position as a classical olfactory model and its complex natural behavior. Here we present the use of transcriptomic and microarray data to identify members of the main olfactory gene families of Manduca. To assess the quality of our data, we correlate information on expressed receptor genes with detailed morphological data on the antennal lobe. Finally, we compare the expression of the near-complete transcript sets in male and female antennae.
Sensory assessment of the environment is crucial to survival and reproduction and therefore directly affects the fitness of animals. In insects, receptors for sensory modalities, such as olfaction, taste, mechanosensation, and thermo- and hygrosensation, are situated in the antenna. The antennal olfactory sense is of special importance to insects, as it provides information on the chemical quality of, e.g., food sources or oviposition sites, and is also used in intraspecific communication via pheromones. Because of their highly specific and sensitive olfactory sense and extraordinarily complex chemosensory behavior, lepidopteran species have been widely used in the field of insect olfaction. For example, the first identified pheromone, bombykol, is the main pheromone component of the silk moth, Bombyx mori. Moths also played an important role in physiological studies, both at the olfactory periphery and the central nervous levels (1, 2). Because the tobacco hornworm, Manduca sexta, reflective of natural ecosystems shows far more complex behavior compared with the domesticated Bombyx, it was the focus of many studies and led to important advances in the field (e.g., refs. 3–5).
In our effort to analyze the antennal transcriptome of M. sexta, we focused on gene families that have been implicated in olfaction: odorant binding proteins (OBPs), chemosensory proteins (CSPs), sensory neuron membrane proteins (SNMPs), odorant degrading enzymes (ODEs), odorant receptors (ORs), gustatory receptors (GRs), and ionotropic receptors (IRs; refs. 6–9). The key players are the OR proteins (10). ORs are embedded in the dendrites of olfactory sensory neurons (OSNs) in the antennae and—to a lesser extent—the maxillary palps. ORs are seven-transmembrane domain receptors with inverted membrane topology in comparison with other G protein coupled receptors (11). Insect ORs are not related to vertebrate ORs (11). ORs apparently share a common ancestor with GRs (12), which can also be expressed in neurons in the insect antennae (13, 14). ORs supposedly function as dimers (11, 15), with a conserved protein acting as an ion channel (originally called DOr83b in Drosophila but now with the universal name ORCO; refs. 16 and 17) and a variable partner (OrX) that apparently determines ligand specificity (18).
Besides OR-based detection of odorants, recent discoveries have implicated IRs as being involved in odor detection as well (7). IRs are relatives of ionotropic glutamate receptors (iGluR) with atypical binding domains that are conserved across protostome lineages and therefore far more ancient than ORs (19). The IR family contains a conserved subgroup, the “antennal,” presumably olfactory IRs (19). Both IR- and OR-expressing OSN populations expressing the same receptor innervate the same spherical structures, called “glomeruli,” in the antennal lobe (AL), the first olfactory neuropil of the insect brain. All neurons expressing the same receptor target the same glomerulus (7, 20–23). The total number of glomeruli in the AL is thus related to the total number of functionally distinct OSN populations and therefore to the number of receptor genes (8).
Because of the large sequence diversity exhibited by olfaction-related genes across insects (10, 24–27), identification by purely homology-based methods has been of limited use. Therefore, recent research in the molecular field of moth olfaction has largely been restricted to Bombyx due to the availability of genomic data (27–32).
Here we report the analysis of an antennal transcriptome of Manduca. Assessment of the data based on Gene Ontology (GO) indicated enrichment not only in signal detection and transduction, but also in enzymatic activity. Additionally, we identified extensive sets of representatives of all gene families involved in olfaction. Using the correlation between the total number of antennal receptor genes and olfactory glomeruli, we estimated the success of our approach by comparison with a newly constructed AL map. Furthermore, we analyzed sexually dimorphic gene expression by using microarrays to identify genes potentially involved in sex-specific behavior, such as oviposition or pheromone detection. Our results demonstrate the feasibility of the approach and provide the basis for a deeper understanding of lepidopteran olfaction in the context of complex behavior.
Results
Initial Sequencing.
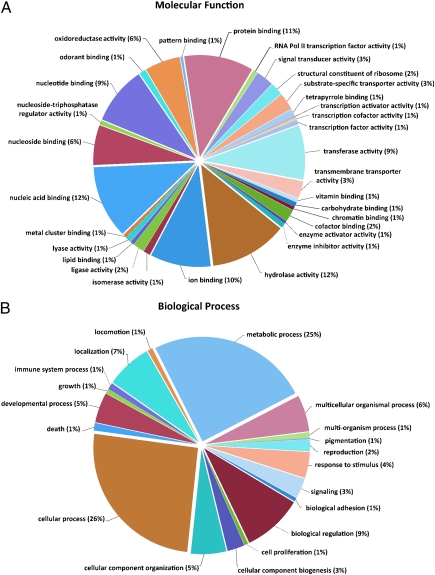
Transcriptomic sequence data were generated by using a normalized antennal cDNA library and GS/FLX 454 technology. In total, we acquired 66 million bases of sequence data in 275,949 ESTs. After assembly, resulting in 22,934 contigs, GO annotation was used for an initial assessment of the transcriptome (Fig. 1). GO annotation associates analyzed transcripts with terms from hierarchical vocabularies describing, e.g., molecular function, to allow metaanalyses of gene populations (33). Besides basic cell functions, GO terms connected to olfaction (e.g., “odorant binding”) and signal transduction (e.g., “response to stimulus,” “signal transducer activity”) were well represented. Additionally, we found strong representation of terms connected to enzymatic activity (for example, “hydrolase activity,” 12%; “transferase activity,” 9%). In comparison with other tissues, an inordinate number of transcripts (Manduca midgut transcriptome with 49% of the data without BLAST hit) have no associated GO terms (15,803 out of 22,934 contigs, ~69%), potentially representing orphan genes.
Fig. 1.
GO analyses of Manduca antennal transcriptome data. GO analysis of Manduca sequences corresponding to 7,131 contigs, as predicted for their involvement in molecular functions (A) and biological processes (B), is shown. Data are presented as level 3 GO categorization for molecular function and level 2 GO categorization for biological process. Classified gene objects are depicted as percentages (in brackets) of the total number of gene objects with GO assignments.
Nonreceptor Olfactory Gene Families.
For a more detailed analysis of the transcriptome, we focused on putative members of olfactory gene families, starting with OBP-coding genes. OBPs are small, globular proteins comprising 140–220 amino acids (9, 34) that are characterized by a pattern of six conserved cysteines (35, 36). In addition to the 12 OBPs reported for Manduca (37–40), we were able to identify 6 unique putative OBP-coding transcripts (Fig. S1), 3 of which seemingly include the full coding sequence. All non-full-length transcripts encode overlapping but distinct OBP domains, establishing them as parts of independent genes. The total amount of putative OBP-coding genes in Manduca is 18, compared with 20 reported for Bombyx.
A second group of proteins present in the sensillum lymph are the CSPs (6). Although they exhibit a similar expression in the antenna to OBPs, CSPs are not related. They are generally more conserved and differ in tertiary structure, with four cysteines forming two disulfide bridges in a configuration distinct from OBPs (6). Thus far, five CSPs have been reported for Manduca (38). We identified a total of 14 additional CSP-coding gene fragments in the antennal transcriptome, 4 of which encode full-length proteins (Fig. S2). The total number (21 CSPs) exceeds the number reported for Bombyx (16 CSPs; ref. 41).
A third class of nonreceptor proteins described in M. sexta is sensory marker neuron proteins, CD36-class proteins expressed in the antenna (SNMPs; ref. 9). Although SNMP-1 is expressed in pheromone sensitive OSNs and necessary for pheromone detection (42), SNMP-2 is expressed by the support cells surrounding these neurons (43). Both described Manduca SNMPs (44) were represented in our data.
Receptor Coding Transcripts.
The fourth protein family of interest is the family of IRs. IRs show extended sequence variability in comparison with other gene families but share a distinct structure (7). Generally, IRs are subdivided into diverse, antennal, and the IR25a/8a subgroup, with the latter two involved in olfaction in the antenna (19). In the genome of Bombyx, 11 putative antennal IRs and 2 pseudogenes have been identified (19). Additionally, 12 homologous IRs have been identified from an antennal EST database of the noctuid Spodoptera littoralis (45). We were able to identify six transcripts encoding homologous receptors in Manduca. All of them show clear homology to antennal IRs in Bombyx (Fig. S3).
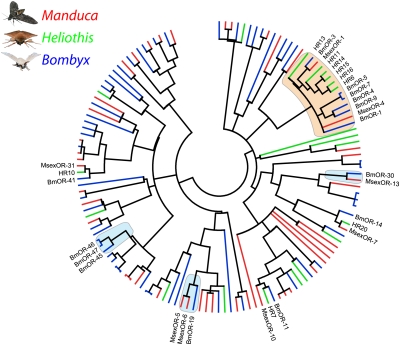
Central to odorant detection are ORs. Because of their high sequence variability, the most common method to identify OR-coding genes is to analyze genomic sequence databases. For Manduca, the coreceptor (MsexOR-2) and five OR-coding genes have been identified (46, 47). Transcripts for all except MsexOR-3 were present in our data; MsexOR-2 and MsexOR-6 could also be extended to full-length. By using RACE-PCR, we were able to verify 47 OR genes as unigenes (Fig. 2, MsexOR-2 excluded), as well as one GR. Full-length OR-coding sequences seem to be present in 27 cases.
Fig. 2.
Aligned putative OR protein sequences of Manduca (red), Bombyx (blue), and Heliothis (green). The identified receptor candidates of Manduca are spread evenly inside the family. Two candidates belonging to the subgroup of male pheromone receptors (orange) have been identified. Additionally, female-specific ORs of Bombyx have been marked (blue). Two female-specific receptor groups also include receptor candidates of Manduca. Additional instances with clear homologs in all three species have been indicated by name. Coreceptors (MsexOR-2, BmorOR-2, and HR2) have been excluded.
Comparison with OR genes from Bombyx (27–30, 48) and Heliothis (refs. 25 and 49; Fig. 2) allowed several observations. In Lepidoptera, several sex-specific ORs have been identified, including a subgroup containing male-specific pheromone receptors (refs. 25, 28, 29, 50, and 51; marked orange in Fig. 2). Beside the reported MsexOR-1 and -4, we were not able to identify further members of this group. In addition to male-specific ORs, female-specific receptors with known function have been reported for Bombyx (ref. 52; marked in blue in Fig. 2). The comparison allowed us to ascertain the presence of putative homologs for BmOR-19 and BmOR-30. Interestingly, although BmOR-30 has exactly one homolog in Manduca (MsexOR-13), two homologs for the linalool-detecting BmOR-19 seem to be present (MsexOR-5 and -6).
We found several instances with conservation of OR-coding genes across the three analyzed species (Fig. 2, named but not labeled). In all three cases, one homolog per species exists, the coding gene seems comparatively conserved, and no unusual expression pattern or function has been reported.
Antennal Lobe.
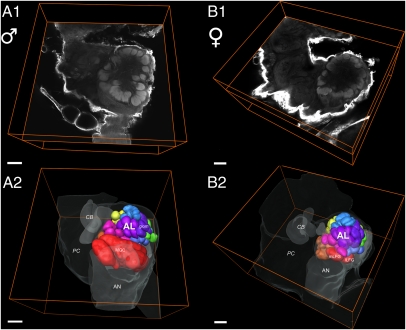
The number of glomeruli within the AL of most insects is correlated with the number of distinct functional populations of OSNs in the antenna and in the maxillary palp (12% in Drosophila; refs. 8, 20, and 21). As the number of distinct functional groups of neurons is associated with the total number of functional ORs, GRs (8, 20, 21), and IRs (7) on the antenna, we could estimate a total number of antennally expressed receptor genes based on the total number of glomeruli. For Manduca, this number has been determined as 63 ± 1 (53–55). We recreated the previous analysis by using a nonhistochemical approach based on confocal laser scanning microscopy followed by computer-assisted 3D reconstruction for both male and female animals (Fig. 3, Figs. S4 and S5, and Movies S1 and S2). A detailed analysis revealed the presence of a total of 70 ± 1 glomeruli in Manduca females and 68 glomeruli in males. By using landmarks, all glomeruli could be identified individually in both sexes (n = 3 each for males and females) and were designated according to Drosophila nomenclature (Movies S1 and S2; ref. 56). The male-specific macroglomerular complex (MGC; refs. 57 and 58) and the two female-specific glomeruli (mLFG and lLFG) as described by Rössler et al. (59) were easy to determine. Comparison of general AL architecture in terms of number of glomeruli and discounting MFG and LFGs revealed a small sexual dimorphism, with three non-LFG glomeruli (P5, VN3, VA4) present in females but not in males.
Fig. 3.
Example of the right hemisphere of the Manduca brain with focus on the AL and its innervation by antennal nerve fibers (AN). (A) Confocal micrographs extracting one single optical orthogonal slice from the 3D dataset of one male (A1) and one female AL (A2). (B) 3D reconstructions of the AL of both sexes depicting a ventral view of reconstructed glomeruli (B1: males, n = 68; B2: females, n = 70 ± 1) corresponding to optical slices in A. ALs display typical moth glomerular architecture: the striking sex-specific glomeruli, the male-specific MGC, and the medial large (mLFG) and lateral large female glomeruli (lLFG) are situated at the entrance of the antennal nerve and are illustrated in red. Brain outlines of adjacent neuropil areas serve as orientation guidelines. CB, central body; PC, protocerebrum; OL, optic lobe; AN, antennal nerve; glom, glomerulus. (Scale bars: 100 μm.)
Sex-Specific Gene Expression.
Based on the transcriptomic data from the present study, the larval midgut (60), and data present in National Center for Biotechnology Information (NCBI) databases, we constructed microarrays to scrutinize differential gene expression, using both male and female antennae. For analysis, differentially expressed genes were associated with GO terms (Figs. S6 and S7). In comparison with larval midgut expression, a somewhat small amount of gene objects were limited to male and female antennae (383 and 417, respectively; Fig. S6). In comparison, >8,000 gene objects present in the midgut were not detected in the antenna. However, the depleted antennal datasets were heavily enriched in terms associated with olfaction. Additionally, although the total number of transcripts appeared decreased, a surprisingly high percentage of terms signifying enzymes present in the data remained. Comparing male and female antennae, the most obvious difference was the higher complexity of the female antenna. In terms of number of transcripts, 729 were present in the female, whereas only 348 were found in the male. This result is further supported by the associated GO terms with the reduced datasets (Fig. S7). In female antennae the number of terms appearing for biological process and molecular function is approximately twice the number as for male antennae.
Discussion
We used next-generation sequencing to analyze the antennal transcriptome of the tobacco hornworm M. sexta. The sequence data (~22,000 contigs with an average size of 460 bases) allow an estimation of tissue complexity in terms of gene expression. By using the mean coding sequence size of 1.1–1.2 kb in the Bombyx genome as reference (32), we can estimate the total number of genes expressed to be ~7,000–8,000. Compared with the microarray data, the larval midgut exhibited >8,000 unique gene calls, whereas <1,000 were limited to the antenna (Fig. S6). Overall, the data indicate a low complexity of antennal gene expression.
Another intriguing result of our study is the large amount of transcripts of unknown function in the transcriptome. Although our knowledge regarding the function of genes in general is limited, especially in species that are not genetic models, a failure to associate GO terms for ~69% of the transcripts is remarkable and indicates high levels of unknown processes in this tissue. It seems possible that the nonannotated contigs are untranslated. However, comparison with the larval midgut data that has been generated using the same methods and does not exhibit the same percentage apparently indicates that this is not the case. Although a large portion of the nonannotated transcripts are common to both tissues, some are only present in the antennal data. Because of the specialized nature of the antenna, it is possible that these transcripts are involved in processes that are directly or indirectly connected to sensory perception.
Nonetheless, the annotated transcripts allow us to draw some conclusions. Surprisingly, we found an enrichment of terms connected to enzymatic activity. Although ODEs seem to be involved in removing active odor molecules from the sensillum lymph, possibly playing a crucial role in the retention of sensitivity, only a small number of enzyme families have been identified as containing ODE candidates (reviewed in ref. 9). For example, for the noctuid Spodoptera littoralis, 16 antennal esterases have been reported, with recent work indicating a role as ODEs (61). It is possible that a majority of ODEs from different enzyme families are still unidentified. Therefore, it seems likely that many of the transcripts coding for enzymes are indeed ODEs. Additionally, we noticed a large amount of regulatory transcripts in the dataset. This result might indicate epigenetic plasticity in the antenna, especially in light of studies demonstrating circadian changes in the pheromone sensitivity of Manduca (62–64). Indeed, postdevelopmental regulation of OR gene expression has been demonstrated in one other species (65).
As an indicator of our success in identifying antennally expressed receptors involved in olfaction as well as the completeness of our transcriptome, we used morphological data from the AL. We identified 54 receptor fragments from our transcriptomic dataset, compared with 73 unique glomeruli. However, we must consider that some neuron populations innervating glomeruli in Manduca may be derived from the maxillary palp (20, 21, 66, 67). With current knowledge on antennal physiology, we suspect the missing transcripts are expressed in very few cells, resulting in very low overall expression levels. A low representation of transcripts means a higher likelihood of either elimination in the normalization or being missed during random sequencing. However, it is possible to derive information on the identity of the missing receptors by comparison with other species. Assuming a similar organization in both Manduca and Bombyx, we missed 5 antennal IR transcripts out of 11 (19), with possibly functional homologs for at least one pseudogene in Bombyx (IR68a). Because IRs are expressed in OSNs associated with comparatively rare coeloconic sensilla (7), this result supports the hypothesis that our unsuccessful identification was due to low expression levels. Similarly, further comparison with two moth species with extensive reported OR gene sets (Bombyx and Heliothis) also indicates the identity of missed OR genes.
The best-described OR subgroup in Lepidoptera contains a number of male-specific receptors detecting active pheromone compounds in both Heliothis and Bombyx (29, 30, 68, 69). Published expression data for MsexOR-1 and MsexOR-4 indicate a role in the detection of the major pheromone component bombykal as well as the minor component E10,E12,Z14-16:Al (50, 51). However, a third pheromone component (E10,E12,E14-16:Al) is detected by a small, distinct neuron population (57, 70, 71). These data indicate that a third male-specific receptor eluded us. Additionally, the pheromone receptor group also contains non-sex-specific members not involved in pheromone communication (four in Bombyx) that seem to be expressed in very few cells and possibly detect plant odors (72). Again, we did not identify any homologs in Manduca. Similarly, we are lacking homologs for two female-specific receptors of Bombyx (BmOR-45 and -47). Because their function (52) is of high importance to Manduca (4), it seems apparent that their homologs are still elusive.
In total, we can predict the identity of 12 missing receptor genes, all of which seem to be expressed in low levels and with predicted functions. The presented data therefore will allow extensive analysis of the receptor population of a complex behaving moth.
Comparison with morphological data also allows us to draw additional conclusions from the microarray expression data. According to the AL data, five glomeruli are female-specific, and three are male-specific. These numbers indicate the presence of five female-specific and three male-specific OR genes. Although the identity of the male-specific receptor genes seems obvious (the three putative pheromone receptors, two of which are identified), the receptor correlates are less clear for the five female-specific glomeruli. Three receptors are expected to belong to the well-described LFG (73); obvious candidates are the two homologs of the Bombyx linalool-receptor BmorOR-19, MsexOR-5 and -6. Reisenman et al. (74) described two distinct glomeruli for the enantiomers of linalool. These data indicate that the detection of the two enantiomers is mediated by different neuron populations and therefore receptors, probably MsexOR-5 and -6. The expected third LFG-related receptor is probably MsexOR-13, the homolog of the female-specific BmOR-30.
Interestingly, the existence of data from lepidopteran species belonging to three different families (Bombycoidae, Noctuidae, and Sphyngidae) indicates that there are homologous receptors between the species. Given the evolutionary distance of the three families, and the generally positive selection of OR genes (75), these receptors might play a central role in lepidopteran olfaction. However, there is no reported function or ligand that readily presents itself as connected to these OR types, suggesting an unknown behavior or an unknown compound with a key role in established paradigms.
The data in this study present near-complete information on the molecular basis of a lepidopteran species besides the domesticated Bombyx mori. The chosen transcriptomic approach allowed an extensive analysis, which will be fundamental for future studies involving this complex model species as well as extensive cross-species comparison.
Materials and Methods
Animals.
Manduca (Lepidoptera, Sphingidae) moths were reared as described in Grosse-Wilde et al. (46). Detailed instructions are in SI Materials and Methods.
Extraction of Total RNA.
Antennae of adult animals were cut off close to the scapulum and transferred to an Eppendorf cup. Total RNA was extracted by using the TRIzol method. For a more detailed description, see SI Materials and Methods.
Expressed Sequence Tag Generation.
RNA extracted from four male and four female animals was unified and further purified by using the RNeasy MinElute Clean up Kit (Qiagen) following the manufacturer's protocol. To prevent overrepresentation of the most common transcripts, the reverse-transcribed mRNAs converted to double-stranded cDNAs (see below) were normalized by using the duplex-specific nuclease method (76). Normalized, full-length enriched cDNA libraries were generated by using a combination of the SMART cDNA library construction kit (BD Clontech) and the Trimmer Direct cDNA normalization kit (Evrogen) generally following the manufacturer's protocol but with several important modifications and enzyme replacements essentially as described (77). The resulting normalized ds-cDNAs were used as a template for NextGen sequencing on a Roche 454 FLX by using standard chemistry at the MPI for Molecular Genetics. Raw data have been deposited at the Short Read Archive (SRA), EMBL-EBI, www.ebi.ac.uk/ena/data/view/ERP000526 (accession no. ERP000526).
For additional description, see SI Materials and Methods.
RACE-PCR.
To extend fragments of candidate genes to full length, Marathon and SMART RACE-PCR kits (Clontech) were used, with gene-specific primers generated with eprimer3 according to recommended specifications and following the manufacturer's instructions.
Bioinformatics.
Multiple assemblies using either seqclean or the TGICL package (http://compbio.dfci.harvard.edu/tgi/), Seqman NGen (DNAStar), or CLC Genomics Workbench (CLCbio) were performed to facilitate comparison. In each case, reads and resulting contigs were screened for contaminants, low quality, and adaptor/linker sequences by using the respective options. GO Annotation was performed both by using Blast2GO (78, 79) and a Codequest Workstation W8 (Active Motif). For identification of ORs and IRs, custom databases for Blast and HMM profile searches were used. Candidate sequences for all mentioned gene families were analyzed further by using the Lasergene software suite (DNAStar). Predicted amino acid sequences were aligned by using MAFFT (80), followed by maximum-likelihood analysis with FastTree (81). Dendrograms were created by using MEGA4 (82) and colored in Adobe Illustrator (Adobe Systems). Identified olfactory genes are included in Dataset S1. Data used in the generation of Fig. 2 and S1–S3 are included as Dataset S2, S3, S4, and S5.
Microarrays and Data Analysis.
Sequence assembly of included Manduca antennal and gut ESTs (60) and all publicly available GenBank sequences was used with eArray (Agilent Technologies) for the design of 4× 44K microarrays based on 60-mer oligo probes.
For sex-specific antennal microarray hybridizations, RNA of three individuals of one sex was pooled per preparation, and five larvae each were dissected for gut tissue isolation with four biological replicates per sex (antennae) and tissue (gut), respectively. Double-purified total RNA was added to Agilent Technologies spike-in RNA and labeled by using QuickAmp amplification kit (Agilent Technologies) and the Kreatech ULS fluorescent labeling kit with cyanine 3-CTP dye following the manufacturer's instructions.
Amplified cRNA samples were used for microarray hybridization and scanned with the Agilent Microarray Scanner, and then data were extracted from TIFF images with Agilent Feature Extraction software (Version 9.1). Raw data output files were analyzed by using the GeneSpring GX11 and GeneSifter microarray analysis software programs. Data have been deposited at GEO, www.ncbi.nlm.nih.gov/geo (accession no. GSE27470).
More detailed descriptions are available in SI Materials and Methods.
Neuroanatomical Procedures.
Brains were dissected from the head capsule and fixed by using ice-cold 4% formaldehyde in PBS (pH 7.2). Tissues were permeabilized by dehydration and rehydration using a sequence of ethanol concentrations. Afterward brains were stained by using Lucifer yellow, dehydrated, and cleared with methyl salicylate. Image acquisition was performed by using a Zeiss LSM 510 (Carl Zeiss). Reconstruction was performed by using Amira 4.1.2 (Mercury Computer Systems). For a detailed description, see SI Materials and Methods.
Image Processing.
For video visualization, label surfaces were exported from AMIRA and visualized with ImageJ (Fiji). Fig. 3 was edited by using Adobe Photoshop CS4 (Adobe Systems) and compiled with Adobe Illustrator CS4 without any further modification on brightness or contrast.
Supplementary Material
Acknowledgments
We thank Sabine Kaltofen and Sylke Dietel for animal rearing; Richard Weniger for valuable support with AL reconstruction; Martin Niebergall and Steffi Gebauer-Jung for IT support; Bernd Timmermann (Max Planck Institute for Molecular Genetics, Berlin) for sequencing services; and Shannon Olsson for helpful comments on the manuscript. This work was supported by the Max Planck Society.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE27470) and the European Molecular Biology Laboratory-European Bioinformatics Institute database, www.ebi.ac.uk/ena/data/view/ERP000526 (accession no. ERP000526).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017963108/-/DCSupplemental.
References
- 1.Hildebrand JG. Analysis of chemical signals by nervous systems. Proc Natl Acad Sci USA. 1995;92:67–74. doi: 10.1073/pnas.92.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mustaparta H. Chemical information processing in the olfactory system of insects. Physiol Rev. 1990;70:199–245. doi: 10.1152/physrev.1990.70.1.199. [DOI] [PubMed] [Google Scholar]
- 3.Hansson BS, Carlsson MA, Kalinovà B. Olfactory activation patterns in the antennal lobe of the sphinx moth, Manduca sexta. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2003;189:301–308. doi: 10.1007/s00359-003-0403-5. [DOI] [PubMed] [Google Scholar]
- 4.Riffell JA, Lei H, Christensen TA, Hildebrand JG. Characterization and coding of behaviorally significant odor mixtures. Curr Biol. 2009;19:335–340. doi: 10.1016/j.cub.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JK, Strausfeld NJ. Structure, distribution and number of surface sensilla and their receptor cells on the olfactory appendage of the male moth Manduca sexta. J Neurocytol. 1990;19:519–538. doi: 10.1007/BF01257241. [DOI] [PubMed] [Google Scholar]
- 6.Angeli S, et al. Purification, structural characterization, cloning and immunocytochemical localization of chemoreception proteins from Schistocerca gregaria. Eur J Biochem. 1999;262:745–754. doi: 10.1046/j.1432-1327.1999.00438.x. [DOI] [PubMed] [Google Scholar]
- 7.Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Bruyne M, Baker TC. Odor detection in insects: volatile codes. J Chem Ecol. 2008;34:882–897. doi: 10.1007/s10886-008-9485-4. [DOI] [PubMed] [Google Scholar]
- 9.Vogt RG. Biochemical diversity of odor detection: OBPs, ODEs and SNMPs. In: Blomquist G, Vogt RG, editors. Insect Pheromone Biochemistry and Molecular Biology. San Diego: Academic Press; 2003. pp. 391–445. [Google Scholar]
- 10.Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- 11.Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robertson HM, Warr CG, Carlson JR. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci USA. 2003;100(Suppl 2):14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. 2007;445:86–90. doi: 10.1038/nature05466. [DOI] [PubMed] [Google Scholar]
- 14.Scott K, et al. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell. 2001;104:661–673. doi: 10.1016/s0092-8674(01)00263-x. [DOI] [PubMed] [Google Scholar]
- 15.Neuhaus EM, et al. Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster. Nat Neurosci. 2005;8:15–17. doi: 10.1038/nn1371. [DOI] [PubMed] [Google Scholar]
- 16.Sato K, et al. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- 17.Wicher D, et al. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- 18.Hallem EA, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Croset V, et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6:e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Couto A, Alenius M, Dickson BJ. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol. 2005;15:1535–1547. doi: 10.1016/j.cub.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 21.Fishilevich E, Vosshall LB. Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol. 2005;15:1548–1553. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- 22.Stocker RF, Singh RN, Schorderet M, Siddiqi O. Projection patterns of different types of antennal sensilla in the antennal glomeruli of Drosophila melanogaster. Cell Tissue Res. 1983;232:237–248. doi: 10.1007/BF00213783. [DOI] [PubMed] [Google Scholar]
- 23.Hansson BS, Ljungberg H, Hallberg E, Löfstedt C. Functional specialization of olfactory glomeruli in a moth. Science. 1992;256:1313–1315. doi: 10.1126/science.1598574. [DOI] [PubMed] [Google Scholar]
- 24.Engsontia P, et al. The red flour beetle's large nose: An expanded odorant receptor gene family in Tribolium castaneum. Insect Biochem Mol Biol. 2008;38:387–397. doi: 10.1016/j.ibmb.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 25.Krieger J, et al. Genes encoding candidate pheromone receptors in a moth (Heliothis virescens) Proc Natl Acad Sci USA. 2004;101:11845–11850. doi: 10.1073/pnas.0403052101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson HM, Wanner KW. The chemoreceptor superfamily in the honey bee, Apis mellifera: Expansion of the odorant, but not gustatory, receptor family. Genome Res. 2006;16:1395–1403. doi: 10.1101/gr.5057506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka K, et al. Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr Biol. 2009;19:881–890. doi: 10.1016/j.cub.2009.04.035. [DOI] [PubMed] [Google Scholar]
- 28.Krieger J, Grosse-Wilde E, Gohl T, Breer H. Candidate pheromone receptors of the silkmoth Bombyx mori. Eur J Neurosci. 2005;21:2167–2176. doi: 10.1111/j.1460-9568.2005.04058.x. [DOI] [PubMed] [Google Scholar]
- 29.Nakagawa T, Sakurai T, Nishioka T, Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science. 2005;307:1638–1642. doi: 10.1126/science.1106267. [DOI] [PubMed] [Google Scholar]
- 30.Sakurai T, et al. Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc Natl Acad Sci USA. 2004;101:16653–16658. doi: 10.1073/pnas.0407596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wanner KW, et al. A honey bee odorant receptor for the queen substance 9-oxo-2-decenoic acid. Proc Natl Acad Sci USA. 2007;104:14383–14388. doi: 10.1073/pnas.0705459104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.International Silkworm Genome Consortium. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol. 2008;38:1036–1045. doi: 10.1016/j.ibmb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Ashburner M, et al. The Gene Ontology Consortium. Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogt RG, Riddiford LM. Pheromone binding and inactivation by moth antennae. Nature. 1981;293:161–163. doi: 10.1038/293161a0. [DOI] [PubMed] [Google Scholar]
- 35.Leal WS, Nikonova L, Peng G. Disulfide structure of the pheromone binding protein from the silkworm moth, Bombyx mori. FEBS Lett. 1999;464:85–90. doi: 10.1016/s0014-5793(99)01683-x. [DOI] [PubMed] [Google Scholar]
- 36.Scaloni A, Monti M, Angeli S, Pelosi P. Structural analysis and disulfide-bridge pairing of two odorant-binding proteins from Bombyx mori. Biochem Biophys Res Commun. 1999;266:386–391. doi: 10.1006/bbrc.1999.1791. [DOI] [PubMed] [Google Scholar]
- 37.Györgyi TK, Roby-Shemkovitz AJ, Lerner MR. Characterization and cDNA cloning of the pheromone-binding protein from the tobacco hornworm, Manduca sexta: a tissue-specific developmentally regulated protein. Proc Natl Acad Sci USA. 1988;85:9851–9855. doi: 10.1073/pnas.85.24.9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson HM, et al. Diversity of odourant binding proteins revealed by an expressed sequence tag project on male Manduca sexta moth antennae. Insect Mol Biol. 1999;8:501–518. doi: 10.1046/j.1365-2583.1999.00146.x. [DOI] [PubMed] [Google Scholar]
- 39.Vogt RG, Rogers ME, Franco MD, Sun M. A comparative study of odorant binding protein genes: Differential expression of the PBP1-GOBP2 gene cluster in Manduca sexta (Lepidoptera) and the organization of OBP genes in Drosophila melanogaster (Diptera) J Exp Biol. 2002;205:719–744. doi: 10.1242/jeb.205.6.719. [DOI] [PubMed] [Google Scholar]
- 40.Vogt RG, Rybczynski R, Lerner MR. Molecular cloning and sequencing of general odorant-binding proteins GOBP1 and GOBP2 from the tobacco hawk moth Manduca sexta: Comparisons with other insect OBPs and their signal peptides. J Neurosci. 1991;11:2972–2984. doi: 10.1523/JNEUROSCI.11-10-02972.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gong DP, et al. Identification and expression pattern of the chemosensory protein gene family in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2007;37:266–277. doi: 10.1016/j.ibmb.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 42.Benton R, Vannice KS, Vosshall LB. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature. 2007;450:289–293. doi: 10.1038/nature06328. [DOI] [PubMed] [Google Scholar]
- 43.Forstner M, et al. Differential expression of SNMP-1 and SNMP-2 proteins in pheromone-sensitive hairs of moths. Chem Senses. 2008;33:291–299. doi: 10.1093/chemse/bjm087. [DOI] [PubMed] [Google Scholar]
- 44.Rogers ME, Krieger J, Vogt RG. Antennal SNMPs (sensory neuron membrane proteins) of Lepidoptera define a unique family of invertebrate CD36-like proteins. J Neurobiol. 2001;49:47–61. doi: 10.1002/neu.1065. [DOI] [PubMed] [Google Scholar]
- 45.Olivier V, Monsempes C, Francois MC, Poivet E, Jacquin-Joly E. Candidate chemosensory ionotropic receptors in a Lepidoptera. Insect Mol Biol. 2010;20:189–199. doi: 10.1111/j.1365-2583.2010.01057.x. [DOI] [PubMed] [Google Scholar]
- 46.Grosse-Wilde E, et al. Sex-specific odorant receptors of the tobacco hornworm Manduca sexta. Front Cell Neurosci. 2010;4:22. doi: 10.3389/fncel.2010.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patch HM, Velarde RA, Walden KK, Robertson HM. A candidate pheromone receptor and two odorant receptors of the hawkmoth Manduca sexta. Chem Senses. 2009;34:305–316. doi: 10.1093/chemse/bjp002. [DOI] [PubMed] [Google Scholar]
- 48.Wanner KW, et al. Female-biased expression of odourant receptor genes in the adult antennae of the silkworm, Bombyx mori. Insect Mol Biol. 2007;16:107–119. doi: 10.1111/j.1365-2583.2007.00708.x. [DOI] [PubMed] [Google Scholar]
- 49.Krieger J, et al. A divergent gene family encoding candidate olfactory receptors of the moth Heliothis virescens. Eur J Neurosci. 2002;16:619–628. doi: 10.1046/j.1460-9568.2002.02109.x. [DOI] [PubMed] [Google Scholar]
- 50.Miura N, Nakagawa T, Tatsuki S, Touhara K, Ishikawa Y. A male-specific odorant receptor conserved through the evolution of sex pheromones in Ostrinia moth species. Int J Biol Sci. 2009;5:319–330. doi: 10.7150/ijbs.5.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wanner KW, et al. Sex pheromone receptor specificity in the European corn borer moth, Ostrinia nubilalis. PLoS ONE. 2010;5:e8685. doi: 10.1371/journal.pone.0008685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson AR, et al. Molecular basis of female-specific odorant responses in Bombyx mori. Insect Biochem Mol Biol. 2009;39:189–197. doi: 10.1016/j.ibmb.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 53.Huetteroth W, Schachtner J. Standard three-dimensional glomeruli of the Manduca sexta antennal lobe: A tool to study both developmental and adult neuronal plasticity. Cell Tissue Res. 2005;319:513–524. doi: 10.1007/s00441-004-1016-1. [DOI] [PubMed] [Google Scholar]
- 54.Rospars JP, Hildebrand JG. Anatomical identification of glomeruli in the antennal lobes of the male sphinx moth Manduca sexta. Cell Tissue Res. 1992;270:205–227. doi: 10.1007/BF00328007. [DOI] [PubMed] [Google Scholar]
- 55.Rospars JP, Hildebrand JG. Sexually dimorphic and isomorphic glomeruli in the antennal lobes of the sphinx moth Manduca sexta. Chem Senses. 2000;25:119–129. doi: 10.1093/chemse/25.2.119. [DOI] [PubMed] [Google Scholar]
- 56.Laissue PP, et al. Three-dimensional reconstruction of the antennal lobe in Drosophila melanogaster. J Comp Neurol. 1999;405:543–552. [PubMed] [Google Scholar]
- 57.Hansson BS, Christensen TA, Hildebrand JG. Functionally distinct subdivisions of the macroglomerular complex in the antennal lobe of the male sphinx moth Manduca sexta. J Comp Neurol. 1991;312:264–278. doi: 10.1002/cne.903120209. [DOI] [PubMed] [Google Scholar]
- 58.Heinbockel T, Hildebrand JG. Antennal receptive fields of pheromone-responsive projection neurons in the antennal lobes of the male sphinx moth Manduca sexta. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1998;183:121–133. doi: 10.1007/s003590050240. [DOI] [PubMed] [Google Scholar]
- 59.Rössler W, Tolbert LP, Hildebrand JG. Early formation of sexually dimorphic glomeruli in the developing olfactory lobe of the brain of the moth Manduca sexta. J Comp Neurol. 1998;396:415–428. [PubMed] [Google Scholar]
- 60.Pauchet Y, et al. Pyrosequencing the Manduca sexta larval midgut transcriptome: Messages for digestion, detoxification and defence. Insect Mol Biol. 2010;19:61–75. doi: 10.1111/j.1365-2583.2009.00936.x. [DOI] [PubMed] [Google Scholar]
- 61.Durand N, et al. Characterization of an antennal carboxylesterase from the pest moth Spodoptera littoralis degrading a host plant odorant. PLoS ONE. 2010;5:e15026. doi: 10.1371/journal.pone.0015026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flecke C, Dolzer J, Krannich S, Stengl M. Perfusion with cGMP analogue adapts the action potential response of pheromone-sensitive sensilla trichoidea of the hawkmoth Manduca sexta in a daytime-dependent manner. J Exp Biol. 2006;209:3898–3912. doi: 10.1242/jeb.02432. [DOI] [PubMed] [Google Scholar]
- 63.Flecke C, Stengl M. Octopamine and tyramine modulate pheromone-sensitive olfactory sensilla of the hawkmoth Manduca sexta in a time-dependent manner. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2009;195:529–545. doi: 10.1007/s00359-009-0429-4. [DOI] [PubMed] [Google Scholar]
- 64.Schuckel J, Siwicki KK, Stengl M. Putative circadian pacemaker cells in the antenna of the hawkmoth Manduca sexta. Cell Tissue Res. 2007;330:271–278. doi: 10.1007/s00441-007-0471-x. [DOI] [PubMed] [Google Scholar]
- 65.Fox AN, Pitts RJ, Robertson HM, Carlson JR, Zwiebel LJ. Candidate odorant receptors from the malaria vector mosquito Anopheles gambiae and evidence of down-regulation in response to blood feeding. Proc Natl Acad Sci USA. 2001;98:14693–14697. doi: 10.1073/pnas.261432998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guerenstein PG, Christensen TA, Hildebrand JG. Sensory processing of ambient CO2 information in the brain of the moth Manduca sexta. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2004;190:707–725. doi: 10.1007/s00359-004-0529-0. [DOI] [PubMed] [Google Scholar]
- 67.Kent KS, Harrow ID, Quartararo P, Hildebrand JG. An accessory olfactory pathway in Lepidoptera: The labial pit organ and its central projections in Manduca sexta and certain other sphinx moths and silk moths. Cell Tissue Res. 1986;245:237–245. doi: 10.1007/BF00213927. [DOI] [PubMed] [Google Scholar]
- 68.Grosse-Wilde E, Gohl T, Bouché E, Breer H, Krieger J. Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur J Neurosci. 2007;25:2364–2373. doi: 10.1111/j.1460-9568.2007.05512.x. [DOI] [PubMed] [Google Scholar]
- 69.Grosse-Wilde E, Svatos A, Krieger J. A pheromone-binding protein mediates the bombykol-induced activation of a pheromone receptor in vitro. Chem Senses. 2006;31:547–555. doi: 10.1093/chemse/bjj059. [DOI] [PubMed] [Google Scholar]
- 70.Kaissling K-E, Hildebrand JG, Tumlinson JH. Pheromone receptor cells in the male moth Manduca sexta. Arch Insect Biochem Physiol. 1989;10:273–279. [Google Scholar]
- 71.Kalinová B, Hoskovec M, Liblikas I, Unelius CR, Hansson BS. Detection of sex pheromone components in Manduca sexta (L.) Chem Senses. 2001;26:1175–1186. doi: 10.1093/chemse/26.9.1175. [DOI] [PubMed] [Google Scholar]
- 72.Jordan MD, et al. Odorant receptors from the light brown apple moth (Epiphyas postvittana) recognize important volatile compounds produced by plants. Chem Senses. 2009;34:383–394. doi: 10.1093/chemse/bjp010. [DOI] [PubMed] [Google Scholar]
- 73.King JR, Christensen TA, Hildebrand JG. Response characteristics of an identified, sexually dimorphic olfactory glomerulus. J Neurosci. 2000;20:2391–2399. doi: 10.1523/JNEUROSCI.20-06-02391.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reisenman CE, Christensen TA, Francke W, Hildebrand JG. Enantioselectivity of projection neurons innervating identified olfactory glomeruli. J Neurosci. 2004;24:2602–2611. doi: 10.1523/JNEUROSCI.5192-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tunstall NE, Sirey T, Newcomb RD, Warr CG. Selective pressures on Drosophila chemosensory receptor genes. J Mol Evol. 2007;64:628–636. doi: 10.1007/s00239-006-0151-6. [DOI] [PubMed] [Google Scholar]
- 76.Zhulidov PA, et al. Simple cDNA normalization using kamchatka crab duplex-specific nuclease. Nucleic Acids Res. 2004;32:e37. doi: 10.1093/nar/gnh031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vogel H, Heidel AJ, Heckel DG, Groot AT. Transcriptome analysis of the sex pheromone gland of the noctuid moth Heliothis virescens. BMC Genomics. 2010;11:29. doi: 10.1186/1471-2164-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Conesa A, et al. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 79.Götz S, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Price MN, Dehal PS, Arkin AP. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.