Abstract
Prohead RNA (pRNA) is an essential component in the assembly and operation of the powerful bacteriophage ϕ29 DNA packaging motor. The pRNA forms a multimeric ring via intermolecular base-pairing interactions between protomers that serves to guide the assembly of the ring ATPase that drives DNA packaging. Here we report the quaternary structure of this rare multimeric RNA at 3.5 Å resolution, crystallized as tetrameric rings. Strong quaternary interactions and the inherent flexibility helped rationalize how free pRNA is able to adopt multiple oligomerization states in solution. These characteristics also allowed excellent fitting of the crystallographic pRNA protomers into previous prohead/pRNA cryo-EM reconstructions, supporting the presence of a pentameric, but not hexameric, pRNA ring in the context of the DNA packaging motor. The pentameric pRNA ring anchors itself directly to the phage prohead by interacting specifically with the fivefold symmetric capsid structures that surround the head-tail connector portal. From these contacts, five RNA superhelices project from the pRNA ring, where they serve as scaffolds for binding and assembly of the ring ATPase, and possibly mediate communication between motor components. Construction of structure-based designer pRNAs with little sequence similarity to the wild-type pRNA were shown to fully support the packaging of ϕ29 DNA.
Keywords: bacteriophage phi29, oligomeric RNA, molecular motor, gp16
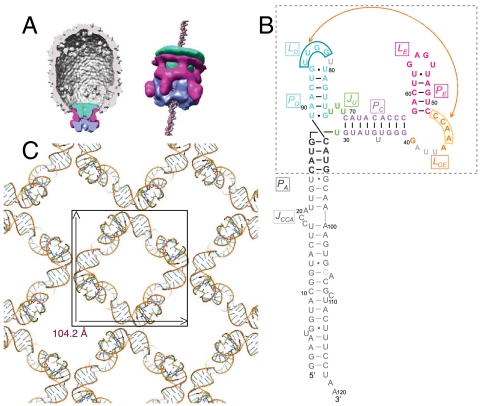
During the assembly of the Bacillus subtilis bacteriophage ϕ29 and other double-stranded DNA phages, an ATP-driven ring motor plays a crucial role in packaging viral DNA to near crystalline density inside preformed protein shells (proheads) (1). In ϕ29, the packaging motor is comprised of three ring structures (Fig. 1A). The head-tail connector, a dodecameric ring of gp10, is embedded in the portal vertex of the head and provides a channel for entry and exit of the genome (2). An oligomeric ring of ϕ29 encoded prohead RNA (pRNA) encircles the protruding end of the connector, displaying five “spokes” that project away from the head (2, 3). Subunits of the viral packaging ATPase gp16, a member of the FtsK/HerA ring motor family (4), form a ring that contacts the five spokes of pRNA, completing the packaging motor (3, 5, 6). Single-molecule laser tweezers studies showed that the ϕ29 DNA packaging motor is one of the strongest molecular motors known, generating approximately 65 pN force (compared to approximately 3 pN for muscle myosin) (7). The requirement of a RNA-ring structure in the assembly and function of the ϕ29 DNA packaging motor contrasts with equivalent motors in other dsDNA phages such as T4, SPP1, lambda, and P22 where protein–protein contacts anchor ring ATPases to the phage proheads (1). Cryoelectron microscopy (cryo-EM) 3D reconstructions (2, 3, 8) show that pRNA is strategically positioned in the ϕ29 packaging motor to link the capsid, connector and ATPase components of the motor (Fig. 1A).
Fig. 1.
The pRNA is an essential component of the bacteriophage ϕ29 DNA packaging motor. (A) Model of the ϕ29 DNA packaging motor derived from cryo-EM 3D reconstructions (3). The head-tail connector (green), pRNA (magenta), and gp16 ring-ATPase (blue) are positioned at the portal vertex of the prohead. (B) Sequence and secondary-structure model of the 120b pRNA Domain I. Nucleotides included in the crystal structure are colored: gray, PA; green, three-way junction; violet, PC; orange, LCE; magenta, PE and LE; and cyan, PD and LD. The crystallized pRNA oligomerization domain is in the boxed area. In light gray are nucleotides deleted in the final crystallization construct (pRNAmin). The 4-bp PA sequence was changed to a 5′-GGCG helix (12). Residue numbers refer to the wild-type sequence. (C) The I422 crystal lattice of pRNAmin viewed along the fourfold symmetry axis, revealing the tetrameric pRNA ring. The 3D lattice is formed via PA–PA and PE–PE crystal stacking above and below.
Besides being an essential component of the motor, the ϕ29 pRNA multimer is a rare example of a self-assembling RNA that functions at the quaternary structure level, extending the evolvement of unique functions for RNA. Whereas full-length pRNA is a 174b transcript, a 120b form lacking the 54-nt 3′-Domain II is fully competent for DNA packaging and phage assembly in vitro (9, 10). Phylogenetic analysis (11), combined with mutagenesis and biochemistry (9, 10), established a secondary-structure model for the 120b pRNA in which three major helices are organized around a U-rich 3-way junction (Fig. 1B). The prohead binding and oligomerization capacity of the 120b form resides in a 71b fragment (Fig. 1B), with the remaining 39-nt PA helix providing the binding site for the gp16 ATPase (3, 12). Early studies demonstrated that pRNA oligomerization is mediated by an intermolecular pseudoknot formed by base-pairing between an internal loop LCE in one pRNA and the LD loop of another pRNA (13, 14).
The oligomerization state of pRNA on the prohead has been controversial. Free pRNA oligomerizes in solution, and dimers and hexamers have been reported (13–16). Early studies concluded that pRNA forms a hexameric ring in the ϕ29 packaging motor (13, 14, 16), whereas cryo-EM reconstructions of pRNA bound to the ϕ29 prohead showed a pentameric RNA ring contacting the protruding dodecameric connector (Fig. 1A) (2, 3, 8). Cryo-EM analyses also demonstrated that the PA helices extend out from the pRNA ring, forming the five spokes that contact the fivefold-symmetric ring ATPase (3). Structural models of the pRNA ring as either a hexamer (13, 17) or a pentamer (2) were proposed. These models, which differ significantly, were based on computational methods integrating the phylogenetic, mutagenesis, and biochemical observations (13, 17), and cryo-EM reconstructions (2).
In this study, we report the 3.5 Å oligomeric pRNA crystal structure lacking only the ATPase binding domain. The pRNA protomers were found to assemble in a head-to-tail fashion into a tetrameric ring in the crystal lattice. The structure revealed the intermolecular pseudoknot formation in great detail, and demonstrated a few “design principles” that favor inter- rather than intramolecular interactions in pRNA. Limited flexibility observed in each pRNA protomer provided insight into how it is able to assemble into different oligomeric ring structures in solution. The overall shape of the pRNA protomer in the crystal structure agreed extremely well with its corresponding cryo-EM density, and several EM docking approaches converged to suggest that pRNA assembles into a pentameric ring in the DNA packaging motor. EM docking combined with chemical probing and mutagenesis provided unique insight into pRNA function. In particular, we identified an essential set of specific interactions that anchor the pentameric pRNA ring to the fivefold symmetric prohead shell, providing an updated model for how the ϕ29 packaging motor assembles and functions. Lastly, we demonstrate our understanding of the pRNA structure and function by designing a pRNA molecule that retains the same secondary structure, but conserved sequence at only a few key locations, that is fully active in in vitro ϕ29 DNA packaging.
Results
Arrangement of the pRNA Ring in the Crystal Lattice.
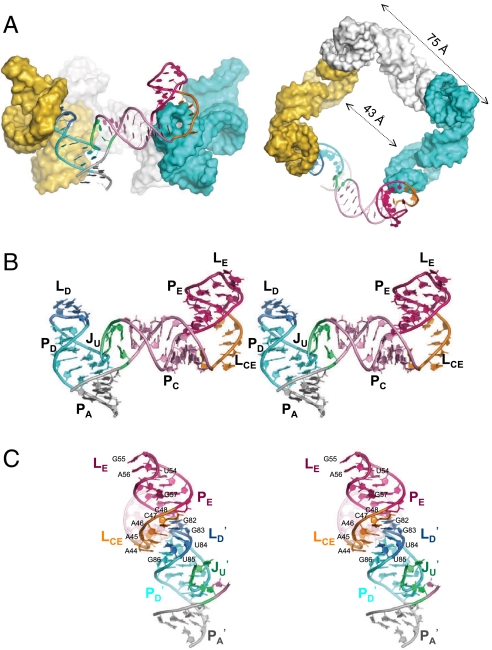
The crystal structure of the ϕ29 pRNA prohead-binding domain (Fig. 1B) was solved at a resolution of 3.5 Å. The starting crystallization construct was the 71b form (bases 25–95) that retains full prohead binding and oligomerization properties, but contains a shortened PA helix (12). To improve crystal diffraction, the U35 bulge in the PC helix, A41U42U43 of the LCE loop, and U81 in the LD loop were removed (Fig. 1B); these changes were based on previous mutagenesis studies (12, 15). When these modifications were introduced into the packaging-competent 120b form of pRNA, DNA packaging activity was retained (Fig. S1A). This construct crystallized as a symmetrical tetrameric ring occupying only 10% of the volume in the I422 crystal lattice (Fig. 1C). The electron density map allowed unambiguous tracing of the entire RNA molecule (Fig. S1B). The ring is not flat; its four edges undulate up and down, and the tilted legs protrude out as spokes at the four vertices (Fig. 2A). The narrowest inner diameter of the tetrameric ring is 43 Å, the outer diameter is 75 Å from edge to edge, and the ring is 104 Å across. As discussed below, intermolecular interactions are mediated by base pairing between the LCE and LD loops of adjacent pRNA protomers.
Fig. 2.
Crystal structure of the tetrameric pRNA ring. (A) Side and top-down views of the tetrameric pRNA ring assembled via protomer head-to-tail base-pairing interactions. (B) Stereo view of the pRNA protomer, which adopts an extended conformation resembling the letter “τ” rotated counterclockwise. (C) Stereo view of the intermolecular interface. The LCE–LD interaction mediates the continuous base stacking from PE of one pRNA protomer to PA of its neighbor, creating an RNA superhelix. All coloring schemes are as in panel B.
Two Flexible Joints and Lack of Tertiary Interactions in the pRNA Protomer.
The individual pRNA protomer in the tetramer ring lacks strong tertiary interactions and adopts a rather extended conformation (Fig. 2B). Its three major helical elements are organized around the U-rich three-way junction (JU); helices PA and PD stack coaxially, whereas the PC helix branches out at an approximately 70° angle. At the distal end of the PC helix, the LCE loop kinks the direction of the PE helix sharply by approximately 110°. At the end of PE, the 6-membered LE loop adopts an unexpectedly compact conformation, where U53 and U58 form a non-Watson–Crick pair to buttress the U54G55A56 triloop, leaving G57 as a base flip-out. This LE loop conformation is similar to an independently determined NMR structure of the E-loop hairpin (rmsd of all atoms alignment is approximately 2 Å) (18). The crystal structure also revealed two flexible joints in the JU bulge and the LCE loop flanking PC. At JU, the U29 and the tri-U linker (U72–74) possess relatively high flexibility, as evidenced by their significantly higher temperature B factors and weaker electron densities, and by their nonstandard local geometry, including a U-turn motif at U74 (Fig. S1B). Previously, it was shown that deletion of the tri-U linker in JU abolished DNA packaging function (12). However, introducing a strand scission at this site using a circularly permuted pRNA restored activity in the absence of the bulge (19). Thus, it appears that maintaining a degree of flexibility at the U-rich three-way junction is critical for pRNA function. At the LCE loop, the unpaired G40 also has weak corresponding electron densities, even though this loop was shortened by removing A41U42U43 for crystallization (Fig. S1B). Thus flexibility at both ends of PC are likely important for pRNA function.
Intermolecular Base Pairing Mediates pRNA Oligomerization.
In contrast to a lack of tertiary interactions within each pRNA protomer, the tetrameric ring is formed by strong interactions between protomers. The crystal structure confirmed that pRNA ring formation is mediated by LCE–LD loop interactions, forming the so-called “intermolecular pseudoknot” (Fig. 2C). Clearly, all four potential Watson–Crick base pairings (A45A46C47C48 in LCE to G82G83U84U85 in LD of the adjacent protomer) are involved, in contrast to previous mutagenesis studies that showed that the two G/C base-pairs are sufficient for DNA packaging in vitro (13, 15, 16). In fact, our structure revealed a fifth pseudoknot interaction as a base triple between A44 in LCE and G86•U80 in LD. Here the G86•U80 wobble pair below LD likely plays a supporting role in intermolecular pseudoknot formation by widening the surrounding minor groove to enable the unusual coplanar juxtaposition of four bases in the LD loop. This explains previous results where replacing the G•U wobble with a G-C pair significantly impairs pRNA multimerization in solution (20). The nonessential U81 deleted from LD in the crystallization construct would likely adopt a U-turn motif to facilitate the strand reversal between U80 and G82.
Unexpectedly, the intermolecular base pairing was also found to be an integral component of RNA superhelices that transect the ring as tilted spokes. These superhelices arise from the continuous stacking of multiple helical elements from two adjacent pRNAs: PE, LCE of one pRNA aligning with LD, PD, and PA of the adjacent pRNA (Fig. 2 A and C). These superhelices dominate the structure of oligomeric pRNA, providing unique insight into its assembly and function (see below).
A Pentameric pRNA Model Through cryo-EM Docking.
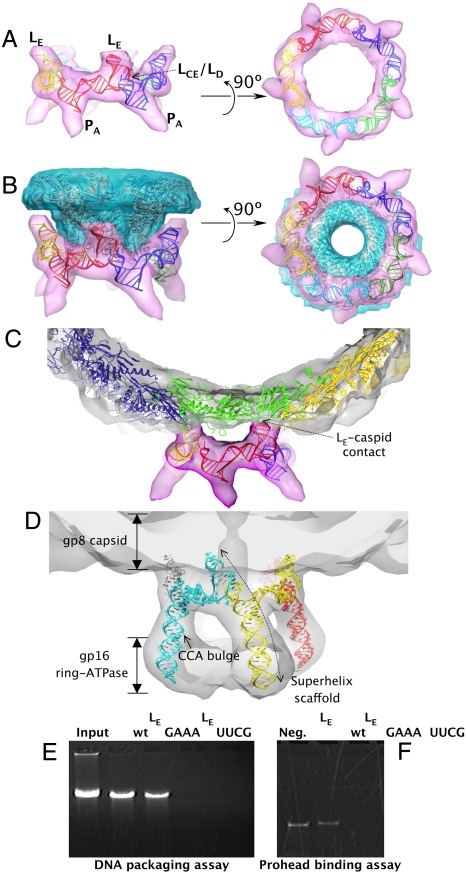
To understand the architecture and function of pRNA as part of the ϕ29 prohead, the pRNA crystal structure was fit into a 12.7 Å resolution cryo-EM 3D reconstruction of the prohead/pRNA complex (21) using the automated molecular-docking procedure in the Chimera program (Fig. 3) (22). The pRNA protomer fits very well into the RNA envelope (Fig. S2A), as the LCE and LD loops are positioned to interact with their respective neighboring pRNAs and the truncated PA helix aligned well with the spoke density known to be the PA helix, the site of the ATPase gp16 binding. Further, the orientation of the LE loop of pRNA correlated very well with previously unassigned cryo-EM density that contacts the phage shell. Overall, the excellent alignment of the crystal structure with the known structural features of pRNA supports the notion that the crystal structure reflects its native conformation in the packaging motor.
Fig. 3.
Docking of the pRNA crystal structure into the ϕ29 prohead cryo-EM envelopes. (A) Side and bottom views of the pentameric pRNA ring structure flexibly docked into its cryo-EM envelope (21). (B) Side and bottom views of the pentameric pRNA ring and the dodecameric connector structures (2) fit into their corresponding cryo-EM envelopes. Note the gap between the pRNA and the connector at the LE region. (C) Side view of the pRNA ring and a pseudoatomic model of the gp8 capsid protein fit into the cryo-EM envelope. Note the pRNA-gp8 contacts in the LE region. The connector structure was omitted for clarity. (D) Side view of the modeled 120b pRNA pentameric ring superimposed with the cryo-EM envelopes of the pRNA and the pRNA-bound gp16 ATPase (3), contoured at 1.7σ. The ATPase density is weakened at this contour level. Note the inferred pRNA–gp16 interactions at the lower portion of PA. (E, F) Effect of LE loop substitutions on in vitro DNA packaging and prohead binding. (E) Proheads reconstituted with 120b pRNA having either the LE-to-GAAA or -UUCG tetraloop substitution were tested for in vitro DNA packaging. The packaged DNA protected from DNase digestion was extracted from the phage head and analyzed on an agarose gel. The Input lane shows the quantity of DNA added to a packaging reaction. The negative control omits ATP from the packaging reaction. (F) For prohead binding, proheads were incubated with pRNAs with LE loop substitutions, purified, and RNA content analyzed on denaturing urea-PAGE.
Next, both rigid-body and flexible docking approaches were used to dock crystallographic pRNA protomers sequentially into the EM densities of pRNA, producing pentameric pRNA models that fit the EM envelope quite well (correlation coefficients over 0.8 by Situs program (23); Fig. S2 A and B; see SI Discussion for details). The flexible docking procedure that takes into account the inherent flexibility in JU produced a pentameric pRNA model that satisfied all geometric constraints and agreed extremely well with the cryo-EM densities (correlation coefficient of 0.84; Fig. 3A).
Considering recent reports of a pRNA hexamer on the prohead using single particle analysis (24, 25), a symmetric hexameric pRNA ring was generated in silico using a similar flexible docking approach (Fig. S3A). Notably, the inner pore of this hexameric pRNA model is approximately 95 Å (Fig. S3A), significantly larger than the approximately 82 Å pentameric pRNA ring generated by cryo-EM docking (Fig. 3A and Fig. S3C). Whereas a pentameric pRNA ring makes close contacts with the lower rim of the connector portal (Fig. 3B), a large gap is present between a hexameric ring and the connector portal, suggesting that a hexameric pRNA ring would fit poorly on the prohead (Fig. S3). In addition, a hexameric pRNA ring would presumably have six, rather than five, projecting spokes in cryo-EM reconstruction (8). Enforcing sixfold symmetry during reconstruction (8, 26) yielded a ring, but without the spokes known to be comprised of PA (3) (approximately 45% of the 120b pRNA mass) due to the imposition of sixfold averaging.
pRNA-Prohead Contacts Critical for pRNA Symmetry.
To further examine pRNA-prohead contacts relevant to pRNA assembly and motor function, the crystal structure of the dodecameric gp10 connector (2) and a pseudoatomic model of the capsid protein gp8 (21) were also docked into the EM envelope. Excellent shape complementarity was found at the pRNA–connector interface (Fig. 3B), with contact confined to the ring portion of pRNA (including PC, LCE–LD, and the 3-way junction) and the protruding narrow end of the connector. This interface is consistent with a directed hydroxyl radical probing/mapping study that showed increased cleavage sensitivity in this region of the pRNA when a probe was tethered to the narrow end of the connector in proheads (27). The symmetry mismatch between a pentameric pRNA and the dodecameric connector dictates that the contacts to each pRNA protomer are nonidentical, and therefore more likely to be nonspecific electrostatic interactions rather than base-specific interactions. The positively charged RKR residues at the N-terminus of the connector are likely candidates for such interactions, as deletion of these residues leads to loss of pRNA binding (27).
In contrast, a set of symmetric interactions are found between the LE loop of each pRNA protomer and the gp8 capsid subunits that abut the upper rim of the connector (Fig. 3C). Earlier cryo-EM studies (3, 21) reported a contact, and our new structural model of pRNA defines this interaction. gp8 subunits abutting the connector adopt a conformation distinct from subunits in the rest of the prohead in that the BIG2 surface domains are rotated 180° away from the connector (3, 21). These five unique gp8 surfaces at the capsid portal vertex appear to form specific, symmetric contacts with a minor groove-facing dinucleotide stack (G55 and A56) in the LE loop of the five pRNA protomers. The contacts anchor the pRNA ring to the ϕ29 prohead.
To provide a pseudoatomic model of a fully functional pRNA oligomer in the packaging motor, the truncated PA helix of the crystal structure was extended using an ideal A-form RNA to generate a 120b pRNA model. This RNA was flexibly docked into the 18 Å prohead/pRNA/ATPase motor cryo-EM reconstruction (3) (Fig. 3D). The structure of the pRNA in the motor is dominated by five relatively rigid superhelical scaffolds that serve to connect the head to the ATPase. The central positioning of the intermolecular interaction in the superhelix supports the hypothesis that the intermolecular base pairing is retained in the active packaging motor. Approximately 80% of the bases in 120b pRNA would comprise these superhelices, suggesting that pRNA in the motor may now best be described as five superhelices, joined by flexible linkers, that span the entire packaging motor assemblage (Fig. 3D). Although not included in the 120b pRNA model, the essential C18C19A20 bulge in the PA helix is expected to point toward the gp16 moiety, consistent with the protection of C18C19 in pRNA footprinting studies with gp16 (5).
Mutagenesis and Chemical Probing Confirm the Importance of the pRNA–gp8 Interaction.
The symmetric interaction between the five LE loops and five gp8 subunits surrounding the connector suggests a role in pRNA recruitment and motor assembly. Because previous single base substitutions in LE did not yield a definitive phenotype (10), the entire loop was substituted to verify the functional importance of the putative pRNA-capsid (gp8) contact. The wild-type LE sequence (UGAGUU) was replaced with either a GAAA or UUCG tetraloop in the 120b form of pRNA to test the effect of LE substitution on in vitro DNA packaging. Both tetraloops are known to reproducibly adopt stable tertiary structures (28, 29), and NMR data suggested that the UUCG substitution does not change pRNA folding (20). The GAAA tetraloop resembles LE in that both contain a dinucleotide purine stack facing the minor groove that can potentially interact in a similar fashion with gp8 (Fig. S4A). In contrast, the UUCG tetraloop conformation is quite distinct from the wild-type LE conformation, and thus it was expected to be defective for prohead binding (Fig. S4B). Confirming these predictions, proheads reconstituted with GAAA-pRNA retained near wild-type prohead-binding capacity and in vitro DNA packaging (Fig. 3 E and F). In contrast, the prohead binding and packaging activities with UUCG-pRNA were undetectable, likely due to the inability to establish functional contacts with gp8. The importance of the LE loop structure is supported by its high sequence conservation among ϕ29-relative pRNAs (11) and systematic evolution of ligands by exponential enrichment studies of the prohead binding domain (10).
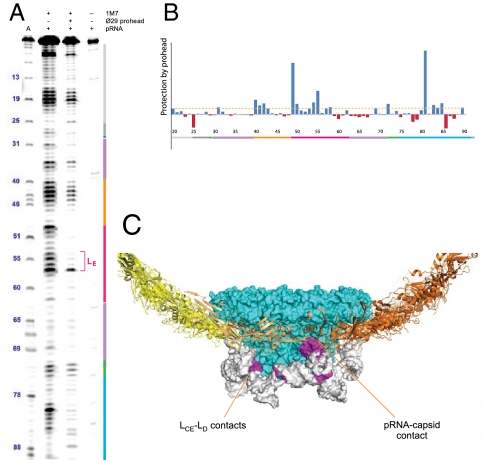
SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) was used to probe the conformation of the 120b pRNA and its molecular contacts with the ϕ29 prohead. SHAPE probes the RNA conformation and the protein-binding footprint based on the accessibility of each ribose 2′ hydroxyl to acylation reagents such as 1M7 (30). The reactivity of the free pRNA (Fig. 4A) agreed well with the secondary structure revealed by our crystal structure. In the prohead-bound pRNA sample, several regions of pRNA became less reactive, and these residues correlated well with the predicted gp8 and gp10 contacts on the prohead (Fig. 4 B and C). The pRNA residues in proximity of the connector, including those in LD, LCE, and the three-way junction, became significantly protected, presumably due to burial or loss of flexibility upon pRNA ring formation around the connector portal. In addition, significant protection was found for LE loop residues (Fig. 4A) that are in contact with the gp8 capsid in our cryo-EM docking results. Thus, SHAPE analysis and the mutagenesis results support a direct contact between pRNA and the surrounding capsid gp8 subunits on the ϕ29 prohead.
Fig. 4.
SHAPE chemical probing and footprinting analysis of pRNA conformation and its interactions with the ϕ29 prohead. (A) SHAPE reactivity of the free and prohead-bound pRNA. The residue numbers and color-coded secondary structures are marked beside the SHAPE lanes. The adenosine sequencing ladder (lane 1) migrates 1-nt faster than the SHAPE lanes. Background due to reverse transcriptase pausing is shown in lane 4. (B) Quantitative analysis of the pRNA SHAPE reactivity profile to reveal “footprints” due to pRNA-prohead interactions. Positive values indicate protection due to prohead binding. (C) Protected regions of pRNA (in magenta) were mapped onto the ϕ29 prohead-pRNA structure model.
Designer pRNAs Fully Reconstitute Wild-Type pRNA Function.
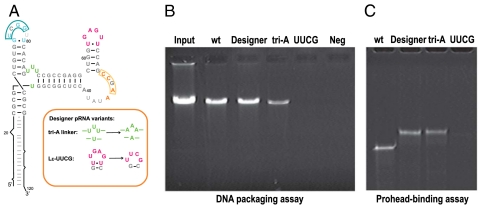
To test our understanding of the pRNA structure-function relationship in prohead binding, we designed functionally competent pRNA molecules based on the accumulated genetic, biochemical and structural data. A120-nt designer pRNA was generated that preserved secondary structure throughout most of the RNA molecule, but that retained only the specific sequences of the LE loop, the U-rich 3-way junction needed for flexibility, and a paired LCE–LD interface (Fig. 5A); the ATPase binding PA helix, not included in the crystallization construct, had the wild-type sequence. This designer pRNA retained near wild-type levels of prohead binding and DNA packaging activities (Fig. 5 B and C). In contrast, a designer pRNA construct lacking the LE-prohead contact (UUCG–LE) had no detectable activities (Fig. 5 B and C). To determine if the U-rich 3-way junction plays additional roles besides maintaining flexibility, U29 and U72–74 were replaced with adenosines. This construct still retained approximately 25% of the wild-type DNA packaging activity (Fig. 5 B and C), consistent with this region playing a structural role rather than a sequence-dependent role. Because pRNA residues that contact the connector were varied, whereas the shell contacts were preserved, these experiments further verify the sequence-independent nature of the pRNA–connector interaction and the functional significance of the specific LE–gp8 interactions in directing the correct assembly of the ϕ29 DNA packaging motor.
Fig. 5.
Activity of designer pRNAs. (A) A designer pRNA or variants where the Ju flexible bulge was replaced with adenines or with the LE-to-UUCG substitution were generated. These RNAs were then assayed for in vitro DNA packaging activity (B) and prohead binding (C).
Discussion
Principles of RNA Oligomerization Derived from the pRNA Structure.
Determination of the oligomeric pRNA crystal structure provides a direct observation of a self-assembled RNA quaternary structure larger than a homodimer. Whereas homodimeric RNA structures can be assembled from two similar or identical modules through kissing loops, higher-order structure formation requires the involvement of two distinct interacting modules, such as the LCE and LD sequences in the ϕ29 pRNA. Here the limited flexibility within each pRNA protomer dictates the range of oligomeric ring structures that can be assembled from the same building block. This principle has been applied to tectonic RNA design studies. For example, introducing two different interaction modules into distal ends of a right-angle-, three-way junction-, or tRNA-motif led to oligomeric RNA ring formation (31).
Interestingly, base-pairing modules in a three-way junction architecture are frequently found to mediate intramolecular interactions in the RNA world. However, the same framework in pRNA is tailored for inter- rather than intramolecular interaction. First, the continuous base stacking of LCE/LD below PE projects the two arms PC and PD away from each other at roughly a right angle (Fig. 2). Second, the mismatch in length between the PC and PD arms serves as a deterrent to alignment for intramolecular interaction. Last, the limited flexibility at the three-way junction cannot accommodate a connection between PC and PD that would be required for an intramolecular interaction. These design principles utilized in a naturally occurring oligomeric RNA could inform and assist future RNA tectonics design.
Assembly of pRNA.
Anchoring of the pRNA to the prohead is the final step in prohead assembly (9, 10) and involves multiple events and components. Previously it was shown that the intermolecular pseudoknot (13–16) and the interaction of pRNA with the RKR residues of the gp10 connector N-terminus (27) were required for prohead binding. Here we demonstrate the additional requirement for a functional LE loop-shell contact. All three contacts are required, as mutation of any one disrupts prohead binding. A possible scenario for this multistep process is that the positively charged N-termini of the connector subunits recruit the pRNA to the vicinity of the head, while the LE-shell contacts and intermolecular base-pairing then confer the final stable conformation of the bound pRNA ring.
As for pRNA symmetry, the excellent fit of the pRNA crystal structure into the cryo-EM density and the identification of the specific LE-shell contact, verified by mutagenesis and SHAPE analysis, rationalizes the observed pentameric symmetry seen in cryo-EM (see also SI Text). Further, the positioning of the intermolecular pseudoknot that links pRNAs as a central part of the superhelices that form the five spokes of the ring (Fig. 3) make it unlikely that a sixth pRNA protomer may be in a unique position in the ring that is not detected in cryo-EM; if so, it would not be connected by the base-pairing seen at the LCE–LD junction (Fig. 2C). Taken together, these conclusions provide strong support for the pentameric, rather than hexameric, symmetry of pRNA on the prohead.
Function of pRNA.
pRNA clearly has a demonstrated role in motor assembly. The multiple interactions that occur in the pRNA prohead-binding domain, visualized here, serve to anchor pRNA directly to the head, with the spokes portion of the pRNA superhelices then providing the scaffold for the assembly of the ring ATPase. This function of pRNA as a scaffold is reminiscent of the 7S RNA in the eukaryotic signal recognition particle (SRP), which provides a scaffold to chaperone the assembly of SRP proteins. The activity of the packaging ATPase gp16 is pRNA-dependent, as the ATPase activity of gp16 alone is weak and only reaches full activity when bound to pRNA on the prohead (10). A putative ATP-binding function for pRNA has been reported (32), although our structural and mutagenesis data suggested that it is unlikely to be a native function of pRNA (see SI Text and Fig. S5).
Finally, pRNA may play a role in communication between the ATPase and the connector and capsid, given its position as the central connecting hub within the motor. Recent single-molecule laser tweezers studies show that the ϕ29 packaging motor is highly coordinated, with at least four gp16 ATPases participating during each mechanochemical cycle (33). In addition, phage packaging motors have been shown to respond to the amount of DNA packaged, because the motor slows during packaging as pressure builds within the head (7). Both phenomena require communication and coordination in the motor and, in ϕ29, the pRNA is likely involved in both processes given its central position. Given this, the intermolecular pseudoknot may be crucial as it appears to provide dual function: It is the structural linkage between pRNA protomers that provides connectivity within the RNA ring, and it is a central component of the RNA superhelices that provides a structural basis for communication between all of the protein components.
The dsDNA bacteriophages are thought to share a common packaging mechanism (reviewed in ref. 1). As pRNA is unique to ϕ29-like phages, RNA-mediated ATPase assembly and communication would likely be carried out by protein subdomains in these other motor assemblages. Indeed, whereas the ϕ29 ATPase is notably smaller than other dsDNA packaging ATPases (the large subunit of the terminase complex), the gp16–pRNA complex is typically considered functionally equivalent (1). Cryo-EM (34) and genetic evidence (1) for interaction of the packaging ATPases of other phages with the connector has been reported, and in the case of T4 a pentameric symmetry of the ATPase on the head has been established (34). Whether there is also a head shell interaction in ATPase assembly that confers symmetry, as seen in pRNA, remains to be determined. Many dsDNA phages also have a “head-full” mechanism that is believed to report the amount of DNA packaged to the translocating ATPase-terminase complex (1) and “cross-talk” between the connector and ATPase has been demonstrated in SPP1 (35). In ϕ29, pRNA would be the mediator of cross-talk because there is no direct contact between the ATPase and the prohead proteins. Whereas the ATPases in the ϕ29 motor have been shown to be highly coordinated, with a potential role assigned to pRNA, in T4 the ATPase subunits are proposed to have a mechanism similar to a monomeric helicase (34, 36). If this holds true for other phage systems, pRNA may have a unique role in contributing to motor coordination. Because the potential role of pRNA in communication can be structurally segmented from the other catalytic and mechanical components involved in packaging, study of pRNA provides an unique opportunity to dissect principles of motor communication that may be common to other phage packaging systems.
Experimental Procedures
RNA Crystallization, Phasing, and Structure Refinement.
The pRNA crystal structure was determined by the single isomorphous replacement anomalous scattering method from a tantalum bromide cluster derivative (Table S1). RNA construct design, crystal optimization, phasing, and refinement are described in detail in SI Experimental Procedures.
Docking of the pRNA into Cryo-EM Envelopes.
The detailed EM docking procedure is described in SI Experimental Procedures.
In Vitro Prohead Binding and DNA Packaging Assay.
In vitro prohead binding and DNA packaging assays were carried out as described (37) with minor modifications (see SI Experimental Procedures).
SHAPE Probing Analysis.
SHAPE analysis was carried out as described (30) with minor modifications (see SI Experimental Procedures).
Supplementary Material
Acknowledgments.
We thank the beam line staff at Advanced Photon Source ID24 and Macromolecular Diffraction at Cornell High Energy Synchrotron Source for assistance in data collection, Marc Morais for sharing EM maps, Kevin Weeks for sharing SHAPE reagents, and Rockney Atz and Marc Morais for helpful discussions and comments on the manuscript. Work in the Grimes lab was supported in part by Public Health Service Grants GM-059604 and DE-003606 from the National Institutes of Health, and the Ke lab by GM-059604 and GM-086766.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates for the pRNA structure have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3R4F).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016690108/-/DCSupplemental.
References
- 1.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 2.Simpson AA, et al. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morais MC, et al. Defining molecular and domain boundaries in the bacteriophage phi29 DNA packaging motor. Structure. 2008;16:1267–1274. doi: 10.1016/j.str.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burroughs AM, Iyer LM, Aravind L. Comparative genomics and evolutionary trajectories of viral ATP dependent DNA-packaging systems. Genome Dyn. 2007;3:48–65. doi: 10.1159/000107603. [DOI] [PubMed] [Google Scholar]
- 5.Koti JS, et al. DNA packaging motor assembly intermediate of bacteriophage phi29. J Mol Biol. 2008;381:1114–1132. doi: 10.1016/j.jmb.2008.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee TJ, Guo P. Interaction of gp16 with pRNA and DNA for genome packaging by the motor of bacterial virus phi29. J Mol Biol. 2006;356:589–599. doi: 10.1016/j.jmb.2005.10.045. [DOI] [PubMed] [Google Scholar]
- 7.Smith DE, et al. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 8.Morais MC, et al. Cryoelectron-microscopy image reconstruction of symmetry mismatches in bacteriophage phi29. J Struct Biol. 2001;135:38–46. doi: 10.1006/jsbi.2001.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo P. Structure and function of phi29 hexameric RNA that drives the viral DNA packaging motor: Review. Prog Nucleic Acid Res Mol Biol. 2002;72:415–472. doi: 10.1016/s0079-6603(02)72076-x. [DOI] [PubMed] [Google Scholar]
- 10.Grimes S, Jardine PJ, Anderson D. Bacteriophage phi 29 DNA packaging. Adv Virus Res. 2002;58:255–294. doi: 10.1016/s0065-3527(02)58007-6. [DOI] [PubMed] [Google Scholar]
- 11.Bailey S, et al. Phylogenetic analysis and secondary structure of the Bacillus subtilis bacteriophage RNA required for DNA packaging. J Biol Chem. 1990;265:22365–22370. [PubMed] [Google Scholar]
- 12.Reid RJ, Zhang F, Benson S, Anderson D. Probing the structure of bacteriophage phi 29 prohead RNA with specific mutations. J Biol Chem. 1994;269:18656–18661. [PubMed] [Google Scholar]
- 13.Zhang F, et al. Function of hexameric RNA in packaging of bacteriophage phi 29 DNA in vitro. Mol Cell. 1998;2:141–147. doi: 10.1016/s1097-2765(00)80123-9. [DOI] [PubMed] [Google Scholar]
- 14.Guo P, Zhang C, Chen C, Garver K, Trottier M. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 15.Chen C, Zhang C, Guo P. Sequence requirement for hand-in-hand interaction in formation of RNA dimers and hexamers to gear phi29 DNA translocation motor. RNA. 1999;5:805–818. doi: 10.1017/s1355838299990350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen C, Sheng S, Shao Z, Guo P. A dimer as a building block in assembling RNA. A hexamer that gears bacterial virus phi29 DNA-translocating machinery. J Biol Chem. 2000;275:17510–17516. doi: 10.1074/jbc.M909662199. [DOI] [PubMed] [Google Scholar]
- 17.Hoeprich S, Guo P. Computer modeling of three-dimensional structure of DNA-packaging RNA (pRNA) monomer, dimer, and hexamer of Phi29 DNA packaging motor. J Biol Chem. 2002;277:20794–20803. doi: 10.1074/jbc.M112061200. [DOI] [PubMed] [Google Scholar]
- 18.Harris S, Schroeder SJ. Nuclear magnetic resonance structure of the prohead RNA E-loop hairpin. Biochemistry. 2010;49:5989–5997. doi: 10.1021/bi100393r. [DOI] [PubMed] [Google Scholar]
- 19.Zhang C, Tellinghuisen T, Guo P. Use of circular permutation to assess six bulges and four loops of DNA-packaging pRNA of bacteriophage phi29. RNA. 1997;3:315–323. [PMC free article] [PubMed] [Google Scholar]
- 20.Kitamura A, Jardine PJ, Anderson DL, Grimes S, Matsuo H. Analysis of intermolecular base pair formation of prohead RNA of the phage phi29 DNA packaging motor using NMR spectroscopy. Nucleic Acids Res. 2008;36:839–848. doi: 10.1093/nar/gkm874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morais MC, et al. Conservation of the capsid structure in tailed dsDNA bacteriophages: The pseudoatomic structure of phi29. Mol Cell. 2005;18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 23.Wriggers W, Milligan RA, McCammon JA. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J Struct Biol. 1999;125:185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- 24.Shu D, Zhang H, Jin J, Guo P. Counting of six pRNAs of phi29 DNA-packaging motor with customized single-molecule dual-view system. EMBO J. 2007;26:527–537. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao F, Zhang H, Guo P. Novel mechanism of hexamer ring assembly in protein/RNA interactions revealed by single molecule imaging. Nucleic Acids Res. 2008;36:6620–6632. doi: 10.1093/nar/gkn669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ibarra B, et al. Topology of the components of the DNA packaging machinery in the phage phi29 prohead. J Mol Biol. 2000;298:807–815. doi: 10.1006/jmbi.2000.3712. [DOI] [PubMed] [Google Scholar]
- 27.Atz R, Ma S, Gao J, Anderson DL, Grimes S. Alanine scanning and Fe-BABE probing of the bacteriophage ϕ29 prohead RNA-connector interaction. J Mol Biol. 2007;369:239–248. doi: 10.1016/j.jmb.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ennifar E, et al. The crystal structure of UUCG tetraloop. J Mol Biol. 2000;304:35–42. doi: 10.1006/jmbi.2000.4204. [DOI] [PubMed] [Google Scholar]
- 29.Cate JH, et al. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 30.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 31.Severcan I, Geary C, Verzemnieks E, Chworos A, Jaeger L. Square-shaped RNA particles from different RNA folds. Nano Lett. 2009;9:1270–1277. doi: 10.1021/nl900261h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shu D, Guo P. A viral RNA that binds ATP and contains a motif similar to an ATP-binding aptamer from SELEX. J Biol Chem. 2003;278:7119–7125. doi: 10.1074/jbc.M209895200. [DOI] [PubMed] [Google Scholar]
- 33.Moffitt JR, et al. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun S, et al. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 35.Oliveira L, Henriques AO, Tavares P. Modulation of the viral ATPase activity by the portal protein correlates with DNA packaging efficiency. J Biol Chem. 2006;281:21914–21923. doi: 10.1074/jbc.M603314200. [DOI] [PubMed] [Google Scholar]
- 36.Sun S, Kondabagil K, Gentz PM, Rossmann MG, Rao VB. The structure of the ATPase that powers DNA packaging into bacteriophage T4 procapsids. Mol Cell. 2007;25:943–949. doi: 10.1016/j.molcel.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 37.Zhao W, Morais MC, Anderson DL, Jardine PJ, Grimes S. Role of the CCA bulge of prohead RNA of bacteriophage o29 in DNA packaging. J Mol Biol. 2008;383:520–528. doi: 10.1016/j.jmb.2008.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.