Abstract
The Soybean Genomics and Microarray Database (SGMD) attempts to provide an integrated view of the interaction of soybean with the soybean cyst nematode and contains genomic, EST and microarray data with embedded analytical tools allowing correlation of soybean ESTs with their gene expression profiles. SGMD provides analytical tools to mine the microarray data quickly by integrating many analysis methods within the database itself. The expression profiles of genes at time intervals during the first 8 days of nematode invasion is searchable by gene name or GenBank accession number. Recent developments include the addition of a searchable database for soybean cyst nematode ESTs and photographs of the invasion process at time points examined using microarrays. SGMD is completely accessible from the web at: http://psi081.ba.ars.usda.gov/SGMD/default.htm.
INTRODUCTION
Soybean (Glycine max L. Merr.) is the most important grain legume crop grown in the United States. The estimated value of soybean is 10.6 billion dollars. There are over 60 million acres planted with an average production of 32.4 bushels (1.14 cubic meter) per acre. The soybean cyst nematode (SCN), Heterodera glycines Ichinoe, is the major pest of soybean, causing an estimated $1 billion in damages throughout the USA per annum. This is more than the soybean loss from all other pests combined (1). The soybean genome is not fully sequenced yet, and little is known about the functions of many soybean genes, especially related to disease and pest resistance. The Soybean Genomics and Microarray Database (SGMD; http://psi081.ba.ars.usda.gov/SGMD/default.htm) is a public database that links soybean ESTs with gene expression data and provides embedded analytical tools for data mining. SGMD was established in 1999 to serve as a sequence and microarray database for the soybean community with its primary focus to store and analyze EST and microarray data generated from experiments involving the interaction of soybean with the SCN. The database currently stores over 50 million rows of microarray data (from USDA labs and collaborators) and almost 20 000 ESTs. Many of the ESTs are printed on microarray slides, allowing the correlation of expression levels with function. More recently, SGMD has broadened its scope to include public soybean ESTs and SCN ESTs. SGMD also serves as a means of novel gene discovery through EST analysis (2). Numerous applications to conduct statistical analysis on DNA microarray data have been integrated into the SGMD interface for rapid data analysis. Analytical tools include analysis of variance (ANOVA) and t-tests, which have been integrated using SQL procedures, thereby eliminating the need for third-party software.
DATABASE COMPONENTS
SGMD is a relational database built on SQLServer2000 and is housed at the Beltsville Agricultural Research Center in Beltsville, MD, USA. The web interface to SGMD is divided into three categories: microarray experiments, the sequence database and collaborations.
Microarray experiments
SGMD’s schema conforms to the minimal information about a microarray experiment (MIAME) guidelines set forth by the Microarray and Gene Expression Databases (MGED) group (3,4). MIAME aims to outline the minimum information required to unambiguously interpret microarray data and to subsequently allow independent verification of these data at a later stage if required. Data for a number of microarray experiments are accessible, among them a time series experiment examining soybean gene expression profiles during SCN invasion from 6 h to 8 days.
A number of web-based applications have been built, among them ANOVA and t-test applications that perform these tests to determine reproducibility and significance of measurements, respectively (Fig. 1). This is done on-the-fly from any web browser, but it works best using Internet Explorer. Other web-based tools allow the user to perform binary queries to identify commonly induced genes between two or more experiments. Cluster analysis data are also stored and accessible from the website (Fig. 2).
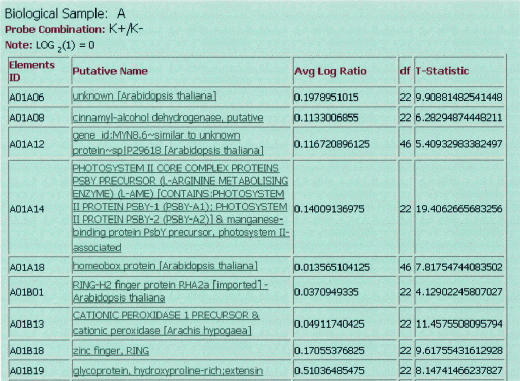
Figure 1.
Snap shot of a query result showing the genes that were found to be statistically induced by a t-test. Clicking on the name of the gene will return a PubMed query listing relevant citations on that gene.
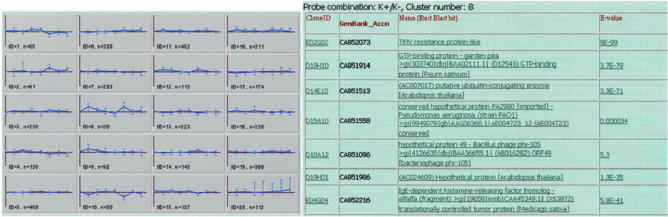
Figure 2.
K-means clustering of a time series data. Cluster profiles are on the left. Clicking on any profile (rectangle) will return the list of genes in that cluster (right). From there researchers can get sequence data, do PubMed searches and view Blast results for any gene by clicking on the appropriate links.
Sequence database
The sequence database provides access to EST sequences stored at SGMD. These include soybean ESTs sequenced at the USDA, Clemson University and Genome Solutions, Inc (GSI). All relevant information about every EST is stored, including the cloning vector and bacterial host strain, insert size, dbEST ID and GenBank accession number, type of clone (cDNA or genomic) and Blast results, which include E-value, score and identity percentage (5) (Fig. 3). Perl scripts were written to extract this information from the Blast results and are available through the authors. The database also contains results of EST analysis and contig assembly for specific GenBank root libraries.
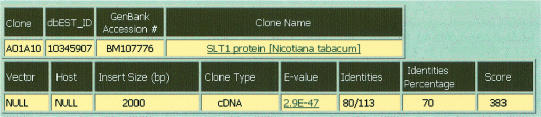
Figure 3.
Result of a query showing the gene name and all relevant information on that gene. Clicking on the name results in a PubMed search and clicking on the E-value displays the most recent Blast search result.
Since the soybean genome is not fully sequenced yet, EST sequences such as the ones stored at SGMD will play an important role in gene identification and discovery, as they have in other organisms (6–8).
Collaborations
SGMD stores a number of microarray experiments that investigate issues other than the soybean–SCN interaction. Among them are microarray experiments designed to detect genes important to the retention of the flat worm Ascaris suum in the intestine of swine (9) and to study the expression of genes in chicken upon infection by coccidiosis, an important disease of poultry (10).
DATABASE ACCESS
SGMD is accessible at http://psi081.ba.ars.usda.gov/SGMD/Default.htm. Raw data from microarrays can be downloaded and the manager of the database can be contacted by email at nalkhar3@gmu.edu.
FUTURE PERSPECTIVES
Work is underway to include pathway information, time series data analysis applications, and pictures documenting cytological events correlating with microarray profiling time points. Also, work is underway to implement MAGE-OM (Microarray Gene Expression–Object Model) and MAGE-ML (Microarray Gene Expression–Markup language), the data exchange model and the data exchange format, respectively (11), as set forth by MGED.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Rana Khan, Imed Chouikha, Peggy MacDonald, Hunter Beard and Kris Pilitt for their technical expertise in generating the microarray and sequence data. This work is supported by the United Soybean Board (USB) and the CSREES-National Research Initiative (NRI) competitive grants program (grant no. 99-35302-8189). N.A. is supported by a pre-doctoral fellowship from the Beltsville Agricultural Research Center (BARC).
REFERENCES
- 1.Wrather J.A. (2001) Soybean disease loss estimates for the United States 1996–2000. http://aes.missouri.edu/delta/research/soyloss.stm. [DOI] [PubMed]
- 2.Alkharouf N., Khan,R. and Matthews,B.F. (2003) Analysis of expressed sequence tags in roots of resistant soybean plants infected by the soybean cyst nematode. Genome, in press. [DOI] [PubMed] [Google Scholar]
- 3.Brazma A., Hingamp,P., Quackenbush,J., Sherlock,G., Spellman,P., Stoeckert,C., Aach,J., Ansorge,W., Ball,C.A., Causton,H.C. et al. (2001) Minimum Information About a Microarray Experiment (MIAME)—towards standards for microarray data. Nature Genet., 29, 365–371. [DOI] [PubMed] [Google Scholar]
- 4.Stoeckert C.J., Causton,H.C. and Ball,C.A. (2002) Microarray databases: standards and ontologies. Nature Genet., 32 (Suppl.), 469–473. [DOI] [PubMed] [Google Scholar]
- 5.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim S., Ahn,K.P. and Lee,Y.H. (2001) Analysis of genes expressed during rice–Magnaporthe grisea interactions. Mol. Plant–Microbe Interact., 14, 1340–1346. [DOI] [PubMed] [Google Scholar]
- 7.Kruger W.M., Pritsch,C., Shao,S. and Muehlbauer,G. (2002) Functional and comparative bioinformatics analysis of expressed genes from wheat spikes infected with Fusarium graminearum. Mol. Plant–Microbe Interact., 15, 445–455. [DOI] [PubMed] [Google Scholar]
- 8.Ewing R.M., Kahla,A.B., Poirot,O., Lopez,F., Audic,S. and Claverie,J.M. (1999) Large-scale statistical analyses of rice ESTs reveal correlated patterns of gene expression. Genome Res., 9, 950–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morimoto M., Zarlenga,D., Beard,H., Alkharouf,N., Matthews,B.F. and Urban,J.F.,Jr (2003) Ascaris suum: cDNA microarray analysis of 4th stage larvae (L4) during self-cure from the intestine. Exp. Parasitol., 104, 113–121. [DOI] [PubMed] [Google Scholar]
- 10.Min W., Lillehoj,H.S., Kim,S., Zhu,J.J., Beard,H., Alkharouf,N. and Matthews,B.F. (2003) Host gene expression profiling in chicken–Eimeria interactions using cDNA microarrays. Appl. Microbiol. Biotechnol., 62, 392–399. [DOI] [PubMed] [Google Scholar]
- 11.Spellman P.T., Miller,M., Stewart,J., Troup,C., Sarkans,U., Chervitz,S., Bernhart,D., Sherlock,G., Ball,C., Lepage,M. et al. (2002) Design and implementation of microarray gene expression markup language (MAGE-ML). Genome Biol., 3, RESEARCH0046–0046.9. [DOI] [PMC free article] [PubMed] [Google Scholar]