Abstract
Ataxia with oculomotor apraxia 1 is caused by mutation in the APTX gene, which encodes the DNA strand-break repair protein aprataxin. Aprataxin exhibits homology to the histidine triad superfamily of nucleotide hydrolases and transferases and removes 5′-adenylate groups from DNA that arise from aborted ligation reactions. We report herein that aprataxin localizes to mitochondria in human cells and we identify an N-terminal amino acid sequence that targets certain isoforms of the protein to this intracellular compartment. We also show that transcripts encoding this unique N-terminal stretch are expressed in the human brain, with highest production in the cerebellum. Depletion of aprataxin in human SH-SY5Y neuroblastoma cells and primary skeletal muscle myoblasts results in mitochondrial dysfunction, which is revealed by reduced citrate synthase activity and mtDNA copy number. Moreover, mtDNA, not nuclear DNA, was found to have higher levels of background DNA damage on aprataxin knockdown, suggesting a direct role for the enzyme in mtDNA processing.
Ataxia with oculomotor apraxia 1 (AOA1) is an autosomal recessive genetic disorder caused by mutation in the APTX gene, which encodes the single strand-break DNA repair (SSBR) protein aprataxin (1, 2). Aprataxin is a member of the histidine triad (HIT) superfamily of nucleotide hydrolases and transferases, and it removes 5′-AMP groups that arise from aborted DNA ligation reactions (3–6). The clinical symptoms of AOA1 include global cerebellar atrophy characterized by loss of Purkinje cells, ocular motor apraxia, and motor and sensory neuropathy (7–13). The pathophysiology of AOA1 overlaps with both Friedreich's ataxia (FA) and Ataxia-Telangiectasia (A-T), rare disorders that present with similar neurological symptoms but different underlying genetic mutations (14). These and other autosomal recessive cerebellar ataxias can be broadly divided into two causative categories based on the function of the affected protein: ataxias induced by DNA repair or maintenance defects that include A-T, xeroderma pigmentosum (XP), and AOA1 and degenerative or metabolic ataxias that include FA (15–18).
Although AOA1 is associated with a DNA repair defect, AOA1 patients have fewer nonneurological symptoms than patients with other DNA repair defects. Most strikingly, AOA1 patients lack the cancer susceptibility associated with A-T, A-T–like disorder, and XP, and they do not manifest immunological deficiencies (or other peripheral symptoms) common in patients with A-T and A-T–like disorder. Cells from AOA1 patients are not profoundly hypersensitive to the genotoxic agents methyl methanesulphonate (MMS) or hydrogen peroxide (H2O2) (19, 20) or to compounds that generate DNA double strand breaks (DSBs) (19, 21). Furthermore, APTX knockout mice do not display a nuclear DNA repair defect, and neural cells from these mice have normal SSBR capacity (20). Consistent with this lack of repair deficiency, aprataxin is not recruited to nuclear DNA lesions in cells exposed to H2O2, ionizing radiation, mitomycin C, or MMS (22), and aprataxin-deficient human cells have no reported gross defect in genomic SSBR (19).
The lack of any definitive DNA repair defect in aprataxin-deficient cells is perplexing. Aprataxin has been reported to interact with and influence many proteins involved in base excision repair (BER) (19, 22–25). Of note, BER is active in both the nucleus and mitochondria, and recently, mitochondrial recruitment of repair cofactors, such as Cockayne syndrome B (CSB) and flap endonuclease 1 (FEN1), has confirmed that BER in the mitochondria has many, if not all, proteins and pathways active in nuclear BER (26–29). These facts led us to speculate that aprataxin might have a significant role in the maintenance of mtDNA. This hypothesis is also supported by similarities between AOA1 and diseases associated with mitochondrial dysfunction, such as FA (30). Like AOA1, FA patients are not susceptible to cancer but frequently present with peripheral neuropathy and progressive ataxia. FA and AOA1 patients are also reported to be deficient in coenzyme Q10, an essential component of the electron transport chain and potent antioxidant within the mitochondria (31–33).
Previous studies are equivocal regarding both the biological role and cellular localization of aprataxin. To further define the pathology of AOA1 and the biological role of aprataxin, we examined the subcellular localization of aprataxin in human cell lines and characterized mitochondrial function in aprataxin-deficient neural/muscular cells, cell types of potential clinical interest. Our results are consistent with the hypothesis that aprataxin plays an important role in the repair of mtDNA and that defects in aprataxin expression and/or function lead to mitochondrial dysfunction and oxidative stress.
Results
Mitochondrial Localization of Aprataxin in Human Neuroblastoma and Skeletal Muscle Cells.
The intracellular distribution of aprataxin was examined by immunofluorescence detection using an antibody against the N-terminal portion of the protein (see below). Aprataxin localized predominately to the nucleus of human SH-SY5Y neuroblastoma cells, with significant cytoplasmic staining also present (Fig. S1). We note that cytoplasmic aprataxin has been reported previously (34), although a functional role for aprataxin in the cytoplasm has not been described.
To determine whether cytoplasmic aprataxin localized to mitochondria, aprataxin antibody staining and the mitochondrial tracking dye Mitotracker Red (Molecular Probes) were covisualized by confocal laser scanning microscopy in SH-SY5Y cells and a second human neuroblastoma cell line, NT2. Aprataxin localization was also investigated in primary human skeletal muscle cells (HSMM), because patient reports suggest that muscle could be a target in AOA1 (31, 32). In all three cell types, aprataxin partially colocalized with Mitotracker Red (Fig. 1A, depicted in yellow). Pearson's correlation coefficient (r2) of the relative distribution of the two channels predicted a moderate colocalization (r2 = 0.179–0.359) when measuring the entire cell (Fig. 1B). However, negating the effect of nuclear aprataxin on the correlation coefficient by analyzing only mitochondrial regions of the cells revealed substantially higher levels of colocalization (r2 = 0.631–0.788). The colocalization observed is not an artifact of cross-channel noise or bleed from the red channel.
Fig. 1.
Aprataxin is detected in mitochondria. (A) Mitochondrial presence of aprataxin was investigated in neuroblastoma cells (SH-SY5Y and NT2) and primary human muscle myoblasts (HSMM) using antiaprataxin antibody (FITC) and Mitotracker Red counterstained with DAPI. Colocalization is a product of difference of mean (PDM) channel highlighting the positive correlation of voxels in the red and green channel, and merge denotes the merge of FITC, Mitotracker Red, and DAPI staining. Cells were viewed using Z-stack confocal microscopy. (Scale bars: NT2, 20 μm; HSMM, 10 μm; SH-SY5Y APTX, 20 μm.) (B) Pearson correlation coefficient (r2) between green (antibody stain) and red (Mitotracker) channels. The range of r2 values is presented for either the whole-image analysis or only a mitochondrial region of interest. Cells analyzed, antibody used, and sample size (n) are designated. For each whole-cell analysis, three mitochondrial colocalization comparisons were performed. (C) Colocalization of aprataxin with the mitochondrial transcription factor TFAM. Antibodies to aprataxin (FITC) and TFAM (CY5) were used, and visualization was performed as above. (Scale bar: 5 μm.)
To exclude the possibility that the aprataxin antibody interacted nonspecifically with Mitotracker Red, we also investigated whether aprataxin colocalized with transcription factor A mitochondrial (TFAM), a protein with a well-established role in mtDNA maintenance (Fig. 1C). The results determined that aprataxin also colocalizes with TFAM (r2 = 0.328–0.437 or 0.561–0.702).
N-Terminal Mitochondrial Targeting Sequence in Aprataxin.
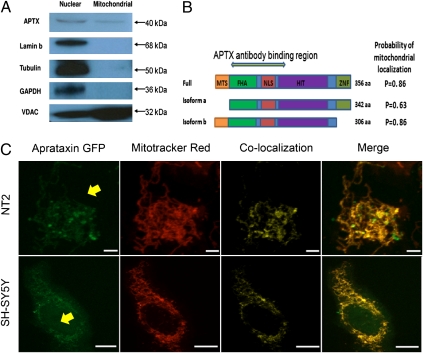
Mitochondrial localization of aprataxin was further investigated by performing Western blot analysis on highly purified nuclear and mitochondrial protein extracts from SH-SY5Y cells (35). The aprataxin antibody recognized a protein of ∼40 kDa in both the nuclear and mitochondrial fractions (Fig. 2A). To exclude the possibility of nuclear contamination of the mitochondrial fraction, the blot was probed for lamin b, cytoskeletal protein tubulin, housekeeping protein GAPDH, and voltage-dependent anion selective channel protein (VDAC), a mitochondrial marker. No nuclear or cytoplasmic contamination was detected in the mitochondrial extracts (Fig. 2A). Western blot results using a second independent antibody against aprataxin substantiated mitochondrial localization of the protein (Fig. S2).
Fig. 2.
Targeting of aprataxin to mitochondria. (A) Western blotting of purified extracts confirms mitochondrial localization of aprataxin. Membrane was probed with antiaprataxin antibody (ab31841), which detected an ∼40-kDa band in both nuclear and mitochondrial extracts. Lamin b and GAPDH/Tubulin antibodies were used to detect nuclear or cytoplasmic contamination of mitochondrial extracts, respectively. The mitochondrial protein VDAC was used to confirm mitochondrial enrichment of the mitochondrial fraction; 20 μg protein were loaded in each lane. Blot is representative of four separate experiments. (B) Full-length protein and two relevant isoforms of aprataxin. Isoform b has a distinct 14-aa N-terminal stretch that harbors the putative mitochondrial targeting sequence (MTS). The probability of mitochondrial localization was estimated using the mitochondrial prediction software MitoProt. FHA, fork head-associated; NLS, nuclear localization signal; HIT, histidine-triad; ZNF, zinc finger. Antigenic region for the aprataxin antibody (amino acids 1–177 of isoform a) is denoted. (C) C-terminal GFP tagged isoform b aprataxin (Aprataxin GFP) localizes to the mitochondria. The location of the fusion protein was compared with the Mitotracker Red mitochondrial marker. Additional details are in Fig. 1. The nucleus is indicated with a yellow arrow. (Scale bars: SH-SY5Y, 20 μm; NT2, 10 μm.)
There are multiple isoforms of aprataxin in the National Center for Biotechnology Information (NCBI) database that are distinguishable primarily by their N- or C-terminal sequence. Isoform a is considered to be the major isoform (22, 24, 36) (Fig. 2B). However, Caldecott (37) recently reported a full-length isoform (also known as isoform e) with a distinct N-terminal 14-aa sequence. The subcellular localization software MitoProt (38) predicted a higher relative potential of isoforms with the 14-aa N-terminal extension to localize to the mitochondria (Fig. 2B). Consistent with this observation, previous studies have shown that isoform a, which lacks this N-terminal extension, is largely nuclear (19, 24, 36).
To determine more directly whether the N terminus of aprataxin is involved in targeting the protein to mitochondria, isoform b (Fig. 2B) (one of the isoforms that has the putative mitochondrial localization N-terminal signal) was transiently expressed as a C-terminal GFP fusion protein in SH-SY5Y and NT2 cells. The cells were then exposed to Mitotracker Red to visualize mitochondrial distribution (Fig. 2C). GFP immunofluorescence was compared in cells with or without the aprataxin isoform b GFP fusion protein, allowing us to detect nonspecific overlap between the red mitochondrial channel and the green channel in control cells. Although the nucleus is not directly visible in the images, its position can be easily inferred (Fig. 2C, yellow arrow). Significantly, in both cell lines tested, the GFP-tagged fusion protein was almost exclusively localized to the mitochondria and surrounding cytoplasm (r2 = 0.457–0.626).
Expression of Aprataxin mRNA Encoding the Mitochondrial Targeting Sequence in Human Brain.
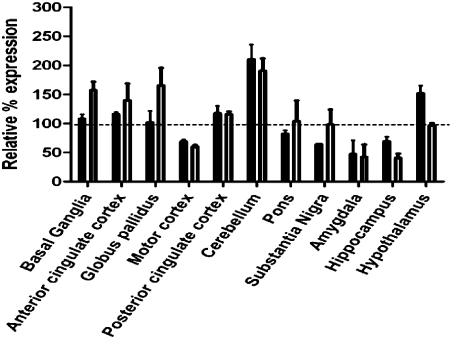
The NCBI database was queried with the 42-nt N-terminal mitochondrial targeting sequence (MTS), and the results suggest that transcripts possessing this sequence are expressed in many tissues, including human brain and muscle. RT-PCR was also performed using a primer that selectively amplifies transcripts harboring the MTS. Importantly, we confirmed using this method that aprataxin MTS-encoding transcripts are suppressed in aprataxin-depleted SH-SY5Y cells, confirming the specificity of the RT-PCR primers. RT-PCR for the aprataxin MTS-encoding transcripts and all aprataxin transcripts (total aprataxin) was then performed using cDNA from 12 regions of the brain from a single normal individual. The results showing total aprataxin (solid bars) and transcripts harboring the MTS (open bars) are presented in Fig. 3. Expression of total aprataxin transcript and aprataxin MTS-encoding transcript varied considerably among the different brain regions; however, both transcripts were most abundant in the cerebellum, the primary target for AOA1 disease progression.
Fig. 3.
Total APTX- and MTS-encoding mRNA levels in human brain regions. Total APTX- (solid bars) and MTS-encoding (unfilled bars) mRNA levels were measured in the indicated brain regions and cross-compared with all other brain regions. Graph represents relative mRNA expression. RT-PCR assay was done in triplicate. Error bars represent SEM.
Aprataxin Knockdown Causes Mitochondrial Dysfunction.
The effect of aprataxin knockdown on mitochondrial function was examined using a lentiviral-delivered shRNA system. The idea of targeting the MTS was determined to be problematic because of the complications of (i) developing an shRNA selective to the unique 14-aa stretch (or 42-nt sequence) and (ii) nonspecific effects against the full-length APTX transcript, which would likely result in reduced nuclear aprataxin protein as well. Thus, we initially trialed the SH-SY5Y neuroblastoma line using lentivirus expressing two different shRNA constructs, both of which target the HIT domain of aprataxin and thus, all APTX transcripts. Reduction of APTX mRNA and the encoded protein was verified by RT-PCR and Western blot analysis, respectively (Fig. S3). The SH-SY5Y aprataxin-depleted cells showed no acute growth defect and no gross changes in morphology, and they were not hypersensitive to menadione or MMS (Fig. S4).
To ascertain whether aprataxin knockdown resulted in mitochondrial dysfunction, cells were infected and grown for 14 d, and intracellular reactive oxygen species (ROS) were quantified using an ethidium-based probe. SH-SY5Y and HSMM aprataxin-depleted cells had a significantly higher level of ROS than control cells (Fig. 4A) as well as lower citrate synthase activity (a marker of intact mitochondria) (Fig. 4B) and reduced mtDNA copy number (assessed using a PCR-based strategy and GAPDH as an internal nuclear loading control) (Fig. 4B). We also examined the consequence of aprataxin-associated mitochondrial dysfunction by comparing the mitochondrial network in aprataxin-depleted HSMM cells with control cells (Fig. 4C). In the control cells, there is an extensive and well-organized network of mitochondria surrounding the nucleus, including regions of intense mitochondrial staining. In contrast, in aprataxin-deficient cells, the mitochondrial network was fragmented, and fluorescence was greatly diminished (contrast was enhanced for the aprataxin cells to allow visualization of the remaining mitochondrial network). These data (Fig. 4) provide strong evidence that aprataxin-depleted human cells are characterized by both mitochondrial dysfunction and a morphological change in mitochondrial organization.
Fig. 4.
Mitochondrial dysfunction in aprataxin-deficient cells. (A) Increased ROS levels were seen in SH-SY5Y cells (Upper) and HSMM (Lower). Gray lines and symbols represent aprataxin-deficient cells, and black lines and symbols represent scrambled control cells. Error bars are SEM (n = 3). (B) Reduced aprataxin causes mitochondrial dysfunction, regardless of differentiation status. Western blotting was used to determine residual aprataxin (APTX) level. Citrate synthase and mtDNA copy number were measured as described in Materials and Methods. Undiff., undifferentiated. SH-SY5Y cells were differentiated (Diff.) with retinoic acid and brain-derived growth factor. The broken line represents the value compared with the scrambled control cells (percent scrambled). All assays were conducted at least in triplicate. (C) Reduced levels of aprataxin causes loss of mitochondria and a breakdown in the organization of the mitochondrial network. The HSMM mitochondrial matrix was visualized using Mitotracker Red and counterstained using DAPI. Cells were viewed using Z-stack confocal microscopy. (D) Aprataxin-deficient cells have an increased amount of endogenous mtDNA damage. PCR amplification of a mitochondrial or nuclear DNA fragment was used to estimate the amount of endogenous DNA damage compared with scrambled cells. The long amplification products were normalized against the amount of input DNA. The assay was conducted in triplicate. (B and D) *P < 0.05; ***P < 0.01. Error bars are SEM.
Because AOA1 preferentially affects postmitotic tissue, mitochondrial dysfunction was evaluated in aprataxin-depleted SH-SY5Y cells that had been differentiated using a previously published protocol (39). As shown in Fig. 4B, differentiation of aprataxin-deficient neuroblastoma cells had no effect on aprataxin depletion-induced mitochondrial dysfunction, indicating that mitochondrial deficiency is independent of mitotic or differentiation status.
Finally, we explored whether mitochondrial defects are seen in two AOA1 patient-derived lymphoblast cell lines (L938/L939) compared with two control cell lines (C2ABR/C3ABR). AOA1 cells showed neither an increase in ROS production (Fig. S5A) nor a reduction in citrate synthase activity (Fig. S5B) relative to control cells. These findings are consistent with the fact that AOA1 patients have no immunological defects, and they indicate that expression, subcellular localization, and biological function of aprataxin may vary between different cell types. Of note, the Epstein–Barr transformation of the lymphoblastoid cell lines used in these experiments could have confounded the results by masking any impact of the aprataxin defect on mitochondrial function.
Reduced Intracellular Aprataxin Causes mtDNA Damage Accumulation.
Quantitative (Q)-PCR was used to measure mtDNA and nuclear DNA damage in SH-SY5Y cells with or without aprataxin knockdown. Q-PCR quantifies the relative amplification of input DNA, indirectly assessing the extent of blocking lesions present in the template DNA. The results show that aprataxin-deficient cells have reduced amplification of template DNA, and by calculation, they have ∼0.7 more steady-state lesions per 10 kb mtDNA than control cells (Fig. 4D). Conversely, knockdown cells displayed only 0.1 lesions more per 10 kb nuclear DNA than controls (Fig. 4D). These data indicate a significant increase in endogenous mtDNA damage, which may very well reflect unprocessed SSBs, but a negligible increase in endogenous nuclear DNA damage in aprataxin-depleted cells. Finally, we examined mtDNA integrity in control and aprataxin-deficient cells using a chloramphenicol (CAP) resistance assay, which records mutations in mitochondrial ribosomal RNA genes that are necessary to confer antibiotic tolerance (40). Aprataxin knockdown in SH-SY5Y cells resulted in increased mtDNA mutagenesis, likely reflective of the increased oxidative stress and associated DNA damage (Fig. S6).
Discussion
Previous research suggests that aprataxin localizes primarily to the nucleus in nonneuronal cell types (19, 22–24). The increased proportion of cytoplasmic staining in the neural-like SH-SY5Y cells observed herein supports findings from Asai et al. (34) that described that Purkinje cells had both nuclear and cytoplasmic aprataxin. Hence, although cytoplasmic aprataxin may be present at a low level in all cell types, it seems to be more predominant in neuron and neuron-like cell lines and tissues. We show herein that aprataxin is present in the mitochondria of human cells, we identify an N-terminal MTS, and we show that the mitochondrial localization of aprataxin has biological significance in relevant cell types. In particular, depletion of aprataxin was found to result in mitochondrial dysfunction and an accumulation of mtDNA damage.
Total APTX expression and expression of MTS-encoding transcripts were found to be highest in the cerebellum of a single individual. Although a limited survey, this observation is noteworthy, because AOA1 is characterized by early onset cerebellar ataxia, and severe cerebellar atrophy is prominent in AOA1 patient brains on autopsy (7, 9–11, 41). In addition, brain regions that have higher levels of transcripts harboring the MTS, such as the basal ganglia and its substructure (the globus pallidus), have also been implicated as a pathological target for AOA1 (42, 43). What regulates the cell type- and brain region-specific expression of the different aprataxin isoforms warrants further investigation.
The neurological specificity of AOA1 is likely related to the high metabolic demands of the target tissue and perhaps, other unique structural or physiologic variables. The high rate of metabolism in brain mitochondria would generate high levels of ROS, which could lead to increased mtDNA damage. In support, mtDNA damage accumulates in aprataxin-deficient neural cells under standard cell culture conditions, and it is feasible that such damage could cause mitochondrial dysfunction, as outlined in the mitochondrial theory of aging (44). Indeed, it is probable that the observed increase in ROS levels is directly related to the mitochondrial dysfunction. Consistent with the idea of oxidative stress in AOA1, elevated levels of oxidative DNA damage (8-oxoguanine) have been reported in patient fibroblasts and postmortem samples, although 8-oxoguanine lesions are not a direct substrate for aprataxin (23, 45).
Although AOA1 is primarily a neural disorder, there are reports of muscle dysfunction. In two studies (32, 33), the majority of AOA1 patients had below normal levels of CoQ10 (a potent antioxidant and component of the electron transport chain) in muscle. Histological analysis did not find ragged red fibers that typify clumps of diseased mitochondria, but the muscle did exhibit neurogenic atrophy. Interestingly, the decrease in CoQ10 had no functional impact on mitochondrial respiration. In our study, we observed that aprataxin-depleted primary human myoblast cells showed signs of mitochondrial dysfunction. This apparent discrepancy may involve the degree of aprataxin reduction or the amount of residual mitochondrial aprataxin activity. Regardless, muscle may be affected by aprataxin depletion or dysfunction but perhaps to a lesser extent than neuronal cells.
It is important to note that alternate proteins or pathways may compensate for the loss of aprataxin in the nuclear compartment. Such mechanisms could include long-patch BER and homologous recombination, which are particularly active in replicating cells (28, 46). With the assistance of nucleases, such as FEN1, 5′-AMP–modified DNA termini could be efficiently excised in the absence of aprataxin. The presence of back-up mechanisms in the nucleus could also explain the moderate sensitivity of aprataxin-deficient cells to DNA-damaging agents as well as the lack of genome instability and cancer predisposition in AOA1 patients.
The results presented herein support the hypothesis that the clinical disease and pathology associated with AOA1 may be caused by defective mtDNA maintenance and associated mitochondrial dysfunction. Notably, several autosomal recessive cerebellar ataxias have links to mitochondrial abnormalities, including a subset of DNA repair ataxias that are attributed to the breakdown of mtDNA maintenance processes. For example, infantile-onset spinocerebellar and mitochondrial recessive ataxias are the result of mutations in DNA helicase twinkle and DNA polymerase-γ, respectively (47, 48). Both of these disorders as well as other dominant ataxias associated with polymerase-γ mutations share more similarity to AOA1 than perhaps FA or A-T. AOA1 and the recessive cerebellar ataxias related to mtDNA breakdown share less common attributes as well, including mental retardation, early onset ataxia, and progressive atrophy of the cerebellum and cerebral cortex, with lesions present in the basal ganglia (47).
While preparing our article for submission, Das et al. (49) reported that tyrosyl-DNA phosphodiesterase 1 (TDP1), an enzyme defective in spinocerebellar ataxia with axonal neuropathy (SCAN1), also plays a role in mitochondria. This finding is of interest, because SCAN1 and AOA1 are clinically very similar, and we have confirmed experimentally by Western blot analysis that TDP1 is present in purified mitochondrial extracts. The knowledge that AOA1 and SCAN1 have mitochondrial involvement could change the clinical management of these disorders.
Materials and Methods
Cell Culture.
Human SH-SY5Y neuroblastoma cells, a subline of SK-N-SH (ATCC), and neuron-committed teratocarcinoma NT2 cells (Stratagene) were cultured in high-glucose DMEM media (Invitrogen). Both DMEM and RPMI 1640 were supplemented with 10% FBS (Sigma) and 1% penicillin-streptomycin (Invitrogen). Primary HSMMs (Lonza) were grown in skeletal muscle cell growth medium supplemented with the SkGM-2 bullet kit (Lonza). Control lymphoblastoid cell lines (C3ABR/C2ABR) and AOA1 patient-derived lines (L938/L939; gift from Martin F. Lavin) (22) are Epstein–Barr virus-transformed and were cultured in RPMI medium 1640 (Invitrogen).
Cell Viability Assay.
The cytotoxicity assay was conducted essentially as per ref. 50. Cells were exposed to MMS or menadione for 1 h at 37 °C and then allowed to recover in complete media for 24 h. Viability was determined using WST-1 dye (Roche), and absorbance was read at 450 nm using 600 nm as the reference wavelength.
Immunofluorescence.
Cells were fixed in 4% paraformaldehyde/PBS, blocked with 10% goat serum at room temperature for 60 min, and incubated with primary aprataxin antibody (ab31841; abcam) at 4 °C overnight. This antibody is predicted to target amino acids 1–177 of isoform a (19). Cells were washed with PBS and PBS-0.05% Tween 20 (Sigma), incubated for 60 min with Alexa fluor 488 goat anti-rabbit secondary antibody (Invitrogen), and mounted in prolong gold antifade reagent with DAPI (Invitrogen). For mitochondrial colocalization experiments, the cells were preincubated with Mitotracker Red CXMRos (Invitrogen) at 37 °C before fixation. For the TFAM experiments, a primary mouse monocolonal antibody was used (ab89818; abcam). Z-stacks of images were taken using a Nikon Plan Fluor 60×/0.50–1.25 oil objective mounted on a Nikon Eclipse TE2000 confocal microscope equipped with a Hamamatsu EM-CCD camera at 37 °C using prolong gold antifade mounting medium with DAPI and the Volocity-5 software (PerkinElmer). The image processing was limited to contrast enhancement.
C-Terminal GFP-Tagged Aprataxin.
A GFP-tagged ORF clone of the Homo sapiens aprataxin transcription variant 2 (NM_175069.1), which encodes isoform b, was purchased from OriGene. The plasmid used a generic pCMV6-AC-GFP backbone. The GFP plasmid was transfected into cells using the PolyJet transfection reagent (SigmaGen) as per the manufacturer's instructions; 3 d after transfection, live cells were stained with a Mitotracker Red dye, and colocalization was viewed using confocal microscopy as described above.
Isolation of Mitochondrial-Enriched Fractions and Immunoblot Analysis.
Mitochondrial-enriched protein fractions were isolated as described (35). The protein concentration was measured using a BCA kit (Pierce). Protein were separated on a 4–20% gradient gel (BioRad) and transferred to a PVDF membrane. Membranes were blocked with 4% BSA in PBS at room temperature. The membrane was then incubated overnight with primary antibody (ab31841; abcam or NB100-534; Novus) and incubated 1 h with the corresponding horseradish peroxidase-conjugated secondary antibody. For detection, ECL-plus chemiluminescent reagent was used (GE Healthcare).
Expression of APTX Transcript.
RT-PCR analysis of APTX and MTS-encoding transcripts was conducted on RNA from various regions of a normal human brain (Ambion). cDNA was made from total RNA using MultiScribe reverse transcriptase (Roche). A commercial TAQMAN probe (Applied Biosystems) for APTX was used to measure the major APTX transcripts; this probe bound to a conserved region in the HIT domain. A second probe was designed that recognized only transcripts harboring the MTS by targeting the unique 14-aa N-terminal region; this probe used the PrimeTime Std qPCR system (Integrated DNA Technologies). A probe for the housekeeping gene GAPDH was used as the internal loading control.
Lentivirus and Infection of Cells.
APTX-specific TRC shRNA-pLKO plasmid constructs (clone ID TRCN0000083640 and TRCN0000083642; Sigma) or a negative control scramble shRNA-pLKO.1 construct (Addgene) was cotransfected with the packaging plasmid pCMV-dr8.2 DVPR (Addgene) and envelope plasmid pCMV-VSV-G (Addgene) into human embryonic kidney 293T cells maintained in HyClone-characterized FBS (Thermo Fisher) using FuGENE6 transfection reagent (Roche). Media were collected 48 h after transduction, and virus titer was estimated using Lenti-X Go stix (Clontech). Infection of cells using lentivirus was conducted as described (39). Puromycin-resistant colonies were periodically checked by RT-PCR and/or standard Western blotting using aprataxin antibody (ab31841; abcam). Differentiation of SH-SY5Y cells was conducted as per ref. 39.
Detection of ROS.
The ROS marker dihydroethidium (DHE) (Invitrogen) was used to measure intracellular ROS, primarily superoxide. The working concentration of DHE varied with cell type, ranging between 2 and 6 μM. Fluorescence was measured at 37 °C using standard 520 nm excitation/610 nm emission at 5-min intervals for a minimum of 60 min using a FLUROstar Optima (BMG Labtech). The mitochondrial electron transport chain blocker antimycin A was used as a positive control to ensure that cellular dye uptake was not rate-limiting. Cell number was used to normalize the results. Statistical significance was calculated by comparing the linear regression of the two curves using GraphPad Prism 5 software (GraphPad).
Citrate Synthase Activity.
The citrate synthase assay is used as a quantitative marker for the content of intact mitochondria. The assay was adapted from ref. 51 with minor modifications. In brief, protein was extracted from aprataxin-deficient and control cells using RIPA buffer (50 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% Na, Deoxycholate, 0.5% Triton ×100 with protease inhibitor mixture) (Roche), and the amount of protein was quantified using bicinchoninic acid protein determination (Pierce). Protein samples were combined with incubation medium [0.1 mM Ellman's reagent (DTNB; Sigma), 0.25% Triton X-100, 0.5 mM oxalacetate, 0.31 mM acetyl CoA] and loaded into a 96-well plate on ice. Each sample was assayed for citrate synthase activity in triplicate with 50% protein input control to assure linearity. The kinetics of activity were measured over a 5-min period at 412 nm and 30 °C using a BioRad plate spectrophotometer. Commercial citrate synthase (Sigma) was used as the assay standard. Measurements were normalized against protein content. One sample t test was carried out on each group using the theoretical mean of 100%.
Measurement of DNA Damage and mtDNA Copy Number.
The assay was performed as described by ref. 52 with minor changes. DNA was extracted using the QIAamp DNA blood extraction kit (QIAGEN) and quantified using the Pico Green dsDNA Quantitation kit (Molecular Probes). The relative amount of initial mtDNA or nuclear DNA added to the Q-PCR was verified using a real-time PCR TAQMAN probe for the mitochondrial gene MTCOX-1 and GAPDH for genomic DNA (Applied Biosystems). This method proved more robust than the amplification of short-fragment mtDNA by Q-PCR used previously. Q-PCR primers used were identical to those previously published (52). We used the human mitochondrial primer sets 14,841 and 5,999 and the human nuclear primer sets 48,510 and 62,007 to amplify 8.9 and 13.5 kb products, respectively, that were verified by agarose gel electrophoresis. Amplification was conducted using an Eppendorf Mastercycler (Eppendorf). The amount of PCR product was quantitated using the Pico Green procedure and normalized against input mtDNA and genomic DNA. The quantitation of the mitochondrial copy number relative to a nuclear housekeeping gene was conducted in an identical manner to the mtDNA quantitation described above using the GAPDH TAQMAN probe (Applied Biosystems) as the internal genomic control. A one-sample t test was carried out on each group using the theoretical mean of 100%.
CAP Resistance Assay.
CAP resistance was used to measure mtDNA mutagenesis as described previously (53). In brief, for the selection assay, scrambled and aprataxin-deficient SH-SY5Y cells were seeded onto a six-well plate, and 24 h later, medium was supplemented with 300 μg/mL CAP. The culture was maintained for 10 d, and the number of colonies per well was counted. The relative survival rate was calculated by dividing the number of colonies in the presence of CAP by the number of colonies in the absence of CAP (i.e., plating control).
Supplementary Material
Acknowledgments
We thank Dr. Marie Rossi and Ms. Jingyan (Jane) Tian for assistance with some of the experimentation. This research was supported entirely by the Intramural Research Program of the National Institutes of Health, National Institute on Aging.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100084108/-/DCSupplemental.
References
- 1.Date H, et al. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat Genet. 2001;29:184–188. doi: 10.1038/ng1001-184. [DOI] [PubMed] [Google Scholar]
- 2.Moreira MC, et al. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat Genet. 2001;29:189–193. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- 3.Ahel I, et al. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 4.Brenner C. Hint, Fhit, and GalT: Function, structure, evolution, and mechanism of three branches of the histidine triad superfamily of nucleotide hydrolases and transferases. Biochemistry. 2002;41:9003–9014. doi: 10.1021/bi025942q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kijas AW, Harris JL, Harris JM, Lavin MF. Aprataxin forms a discrete branch in the HIT (histidine triad) superfamily of proteins with both DNA/RNA binding and nucleotide hydrolase activities. J Biol Chem. 2006;281:13939–13948. doi: 10.1074/jbc.M507946200. [DOI] [PubMed] [Google Scholar]
- 6.Rass U, Ahel I, West SC. Molecular mechanism of DNA deadenylation by the neurological disease protein aprataxin. J Biol Chem. 2008;283:33994–34001. doi: 10.1074/jbc.M807124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Arrigo S, et al. Ataxia with oculomotor apraxia type 1 (AOA1): Clinical and neuropsychological features in 2 new patients and differential diagnosis. J Child Neurol. 2008;23:895–900. doi: 10.1177/0883073808314959. [DOI] [PubMed] [Google Scholar]
- 8.Ferrarini M, et al. A novel mutation of aprataxin associated with ataxia ocular apraxia type 1: Phenotypical and genotypical characterization. J Neurol Sci. 2007;260:219–224. doi: 10.1016/j.jns.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 9.Le Ber I, et al. Cerebellar ataxia with oculomotor apraxia type 1: Clinical and genetic studies. Brain. 2003;126:2761–2772. doi: 10.1093/brain/awg283. [DOI] [PubMed] [Google Scholar]
- 10.Sugawara M, et al. Purkinje cell loss in the cerebellar flocculus in patients with ataxia with ocular motor apraxia type 1/early-onset ataxia with ocular motor apraxia and hypoalbuminemia. Eur Neurol. 2008;59:18–23. doi: 10.1159/000109256. [DOI] [PubMed] [Google Scholar]
- 11.Tranchant C, Fleury M, Moreira MC, Koenig M, Warter JM. Phenotypic variability of aprataxin gene mutations. Neurology. 2003;60:868–870. doi: 10.1212/01.wnl.0000048562.88536.a4. [DOI] [PubMed] [Google Scholar]
- 12.Yoon G, et al. Complete deletion of the aprataxin gene: Ataxia with oculomotor apraxia type 1 with severe phenotype and cognitive deficit. J Neurol Neurosurg Psychiatry. 2008;79:234–236. doi: 10.1136/jnnp.2007.127233. [DOI] [PubMed] [Google Scholar]
- 13.Onodera O. Spinocerebellar ataxia with ocular motor apraxia and DNA repair. Neuropathology. 2006;26:361–367. doi: 10.1111/j.1440-1789.2006.00741.x. [DOI] [PubMed] [Google Scholar]
- 14.Palau F, Espinós C. Autosomal recessive cerebellar ataxias. Orphanet J Rare Dis. 2006;1:47. doi: 10.1186/1750-1172-1-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richardson DR, et al. Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. Proc Natl Acad Sci USA. 2010;107:10775–10782. doi: 10.1073/pnas.0912925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 17.Stewart GS, et al. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 18.Dworaczek H, Xiao W. Xeroderma pigmentosum: A glimpse into nucleotide excision repair, genetic instability, and cancer. Crit Rev Oncog. 2007;13:159–177. doi: 10.1615/critrevoncog.v13.i2.20. [DOI] [PubMed] [Google Scholar]
- 19.Clements PM, et al. The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair (Amst) 2004;3:1493–1502. doi: 10.1016/j.dnarep.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 20.El-Khamisy SF, et al. Synergistic decrease of DNA single-strand break repair rates in mouse neural cells lacking both Tdp1 and aprataxin. DNA Repair (Amst) 2009;8:760–766. doi: 10.1016/j.dnarep.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becherel OJ, et al. CK2 phosphorylation-dependent interaction between aprataxin and MDC1 in the DNA damage response. Nucleic Acids Res. 2010;38:1489–1503. doi: 10.1093/nar/gkp1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gueven N, et al. Aprataxin, a novel protein that protects against genotoxic stress. Hum Mol Genet. 2004;13:1081–1093. doi: 10.1093/hmg/ddh122. [DOI] [PubMed] [Google Scholar]
- 23.Harris JL, et al. Aprataxin, poly-ADP ribose polymerase 1 (PARP-1) and apurinic endonuclease 1 (APE1) function together to protect the genome against oxidative damage. Hum Mol Genet. 2009;18:4102–4117. doi: 10.1093/hmg/ddp359. [DOI] [PubMed] [Google Scholar]
- 24.Hirano M, et al. DNA single-strand break repair is impaired in aprataxin-related ataxia. Ann Neurol. 2007;61:162–174. doi: 10.1002/ana.21078. [DOI] [PubMed] [Google Scholar]
- 25.Luo H, et al. A new XRCC1-containing complex and its role in cellular survival of methyl methanesulfonate treatment. Mol Cell Biol. 2004;24:8356–8365. doi: 10.1128/MCB.24.19.8356-8365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamenisch Y, et al. Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J Exp Med. 2010;207:379–390. doi: 10.1084/jem.20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aamann MD, et al. Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB J. 2010;24:2334–2346. doi: 10.1096/fj.09-147991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu P, et al. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol. 2008;28:4975–4987. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevnsner T, et al. Mitochondrial repair of 8-oxoguanine is deficient in Cockayne syndrome group B. Oncogene. 2002;21:8675–8682. doi: 10.1038/sj.onc.1205994. [DOI] [PubMed] [Google Scholar]
- 30.Stemmler TL, Lesuisse E, Pain D, Dancis A. Frataxin and mitochondrial FeS cluster biogenesis. J Biol Chem. 2010;285:26737–26743. doi: 10.1074/jbc.R110.118679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooper JM, Korlipara LV, Hart PE, Bradley JL, Schapira AH. Coenzyme Q10 and vitamin E deficiency in Friedreich's ataxia: Predictor of efficacy of vitamin E and coenzyme Q10 therapy. Eur J Neurol. 2008;15:1371–1379. doi: 10.1111/j.1468-1331.2008.02318.x. [DOI] [PubMed] [Google Scholar]
- 32.Le Ber I, et al. Muscle coenzyme Q10 deficiencies in ataxia with oculomotor apraxia 1. Neurology. 2007;68:295–297. doi: 10.1212/01.wnl.0000252366.10731.43. [DOI] [PubMed] [Google Scholar]
- 33.Quinzii CM, et al. Coenzyme Q deficiency and cerebellar ataxia associated with an aprataxin mutation. Neurology. 2005;64:539–541. doi: 10.1212/01.WNL.0000150588.75281.58. [DOI] [PubMed] [Google Scholar]
- 34.Asai H, et al. Protein kinase C gamma, a protein causative for dominant ataxia, negatively regulates nuclear import of recessive-ataxia-related aprataxin. Hum Mol Genet. 2009;18:3533–3543. doi: 10.1093/hmg/ddp298. [DOI] [PubMed] [Google Scholar]
- 35.Maynard S, de Souza-Pinto NC, Scheibye-Knudsen M, Bohr VA. Mitochondrial base excision repair assays. Methods. 2010;51:416–425. doi: 10.1016/j.ymeth.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi T, et al. Aprataxin, causative gene product for EAOH/AOA1, repairs DNA single-strand breaks with damaged 3′-phosphate and 3′-phosphoglycolate ends. Nucleic Acids Res. 2007;35:3797–3809. doi: 10.1093/nar/gkm158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 38.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 39.Kulkarni A, McNeill DR, Gleichmann M, Mattson MP, Wilson DM., 3rd XRCC1 protects against the lethality of induced oxidative DNA damage in nondividing neural cells. Nucleic Acids Res. 2008;36:5111–5121. doi: 10.1093/nar/gkn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wallace DC, Bunn CL, Eisenstadt JM. Cytoplasmic transfer of chloramphenicol resistance in human tissue culture cells. J Cell Biol. 1975;67:174–188. doi: 10.1083/jcb.67.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimazaki H, et al. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia: The aprataxin gene mutations. Neurology. 2002;59:590–595. doi: 10.1212/wnl.59.4.590. [DOI] [PubMed] [Google Scholar]
- 42.Salvatore E, et al. Nigrostriatal involvement in ataxia with oculomotor apraxia type 1. J Neurol. 2008;255:45–48. doi: 10.1007/s00415-007-0657-x. [DOI] [PubMed] [Google Scholar]
- 43.Sekijima Y, et al. Severe generalized dystonia as a presentation of a patient with aprataxin gene mutation. Mov Disord. 2003;18:1198–1200. doi: 10.1002/mds.10526. [DOI] [PubMed] [Google Scholar]
- 44.Harman D. Free radical theory of aging: An update. Increasing the functional life span. Ann N Y Acad Sci. 2006;1067:10–21. doi: 10.1196/annals.1354.003. [DOI] [PubMed] [Google Scholar]
- 45.Hirano M, Furiya Y, Kariya S, Nishiwaki T, Ueno S. Loss of function mechanism in aprataxin-related early-onset ataxia. Biochem Biophys Res Commun. 2004;322:380–386. doi: 10.1016/j.bbrc.2004.07.135. [DOI] [PubMed] [Google Scholar]
- 46.Liu P, Demple B. DNA repair in mammalian mitochondria: Much more than we thought? Environ Mol Mutagen. 2010;51:417–426. doi: 10.1002/em.20576. [DOI] [PubMed] [Google Scholar]
- 47.Hakonen AH, et al. Infantile-onset spinocerebellar ataxia and mitochondrial recessive ataxia syndrome are associated with neuronal complex I defect and mtDNA depletion. Hum Mol Genet. 2008;17:3822–3835. doi: 10.1093/hmg/ddn280. [DOI] [PubMed] [Google Scholar]
- 48.Finsterer J. Mitochondrial ataxias. Can J Neurol Sci. 2009;36:543–553. doi: 10.1017/s0317167100008027. [DOI] [PubMed] [Google Scholar]
- 49.Das BB, Dexheimer TS, Maddali K, Pommier Y. Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria. Proc Natl Acad Sci USA. 2010;107:19790–19795. doi: 10.1073/pnas.1009814107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Canugovi C, et al. The mitochondrial transcription factor A functions in mitochondrial base excision repair. DNA Repair (Amst) 2010;9:1080–1089. doi: 10.1016/j.dnarep.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Srere PA. In: Methods in Enzymology. Colowick SP, Kaplan NOP, editors. London: Academic; 1969. pp. 3–11. [Google Scholar]
- 52.Santos JH, Mandavilli BS, Van Houten B. In: Mitochondrial DNA: Methods and Protocols. Copeland WC, editor. Totowa, NJ: Humana Press; 2002. pp. 159–176. [Google Scholar]
- 53.de Souza-Pinto NC, et al. Novel DNA mismatch-repair activity involving YB-1 in human mitochondria. DNA Repair (Amst) 2009;8:704–719. doi: 10.1016/j.dnarep.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.