Abstract
Single-molecule FRET has been widely used for monitoring protein–nucleic acids interactions. Direct visualization of the interactions, however, often requires a site-specific labeling of the protein, which can be circuitous and inefficient. In addition, FRET is insensitive to distance changes in the 0–3-nm range. Here, we report a systematic calibration of a single molecule fluorescence assay termed protein induced fluorescence enhancement. This method circumvents protein labeling and displays a marked distance dependence below the 4-nm distance range. The enhancement of fluorescence is based on the photophysical phenomenon whereby the intensity of a fluorophore increases upon proximal binding of a protein. Our data reveals that the method can resolve as small as a single base pair distance at the extreme vicinity of the fluorophore, where the enhancement is maximized. We demonstrate the general applicability and distance sensitivity using (a) a finely spaced DNA ladder carrying a restriction site for BamHI, (b) RNA translocation by DExH enzyme RIG‐I, and (c) filament dynamics of RecA on single-stranded DNA. The high spatio-temporal resolution data and sensitivity to short distances combined with the ability to bypass protein labeling makes this assay an effective alternative or a complement to FRET.
Keywords: cis-trans isomerization, DNA–protein interaction, RNA–protein interaction, label free protein
Single-molecule Förster resonance energy transfer (FRET) has been a powerful tool in probing protein–nucleic acid interactions as demonstrated by numerous studies revealing unexpected dynamic movement and conformational changes of proteins that cannot be resolved by conventional ensemble techniques (1–4). Despite such advantages, FRET measurement is often limited to proteins that can be site-specifically labeled and the protein’s interaction with the corresponding DNA or RNA in a FRET-sensitive distance range of 3 to 8 nm (5). The conjugation of fluorescent dye to a protein involves mutagenesis (6) and/or chemical modifications (7), which may disrupt the structure and function of the protein. Furthermore, the labeling procedures are not straightforward and are often labor intensive yet with a low yield.
Recently we developed an alternative single molecule assay termed protein induced fluorescence enhancement (PIFE) whereby the emission of a fluorescent dye reports on its proximity to an interacting protein; i.e., the dye becomes brighter when a protein approaches its vicinity (8). This photophysical effect was originally employed in stop flow measurement (9) for monitoring directional movement of DNA motor proteins in ensemble (10–13), for following the dynamics of DNA and RNA polymerases on DNA (14, 15), and for detecting the motion of helicases on RNA or DNA at the single-molecule level (16, 17). This photophysical characteristic is exhibited by fluorophores such as Cy3, which undergoes cis-trans isomerization reaction. External factors such as protein reduce the rate at which the fluorophore isomerizes from the photo-active trans state to the photo-inactive cis state, resulting in an increase of quantum yield as well as in the fluorescence lifetime (18–20). For example, the RIG-I protein binding and unbinding to the double-stranded RNA (dsRNA) substrate bearing Cy3 or DY547 fluorophore was visualized as an abrupt increase and decrease in fluorescence intensity, respectively, whereas the ATP-driven translocation movement was seen as gradual fluctuations in the signal (16). In this report, we present a systematic characterization of the distance sensitivity of PIFE assay by using three protein systems, BamHI, RIG I, and RecA.
Results
BamHI Displays PIFE Sensitivity in 10 Base Pair Range.
We employed a restriction enzyme, BamHI, which binds to a unique recognition sequence (GGATCC) on double stranded DNA (dsDNA). We chose this system because it enables precise positioning of the protein at a known distance away from the fluorophore, and the rigidity of dsDNA allows for accurate distance estimation. We designed 40 base pair (bp) dsDNAs bearing the Cy3 fluorophore at one end and BamHI recognition sites at 1, 2, 3, 5, 7, 10, 12, and 15 bp away from the Cy3 and biotin at the other end (Fig. 1A). Each DNA construct was immobilized to a polymer-coated quartz surface via biotin neutravidin attachment. DNA intensities were measured to confirm that DNA alone does not display signal fluctuations. Over 95% of the single molecule traces reveal that the intensity is stable over a 2- to 3-min period without signal fluctuations. In addition, the overall intensity histogram obtained from DNA only data serves as a reference to which PIFE data is compared (Fig. S1A).
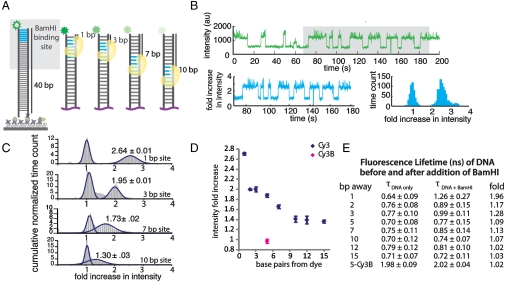
Fig. 1.
BamHI-induced PIFE effect. (A) BamHI recognition sequence (blue) was positioned at a known distance away from the Cy3 fluorophore (green) on a 40-bp double-stranded DNA. (B) A representative single-molecule trace obtained from 1-bp‐away site shows BamHI binding and dissociation as an abrupt increase and decrease in fluorescence signal, respectively (top). The raw intensity is normalized to the DNA only intensity (bottom left) and built into a histogram of fold increase for the single molecule (bottom right). (C) Cumulative histograms of intensity fold increase were built from over 200 molecules for 1-, 3-, 7-, and 10-bp‐away sites. (D) Summary of distance-dependent PIFE. Cy3B (pink) does not show PIFE effect. (E) Fluorescence lifetime of all DNA constructs before and after BamHI addition.
PIFE data is taken by flowing in the buffer (50 mM Tris-HCl pH 8, 10 mM CaCl2, 100 mM NaCl) containg BamHI (800 nM) to the DNA bound surface while maintaining the laser excitation level and the illumination field constant to minimize any external perturbation to the fluorescence signal. Calcium was used in place of magnesium to prevent BamHI-mediated cleavage. In this way, we were able to observe the site-specific labeling of BamHI without DNA cleavage. As a functional test, magnesium was added to trigger the cleavage at the end of the experiment. Upon addition of BamHI to DNA, Cy3 signals became brighter immediately (Fig. S1B). Fig. 1B, Upper, shows a representative single molecule trace taken from a DNA containing the 1-bp-away site, which shows a dynamic fluctuation in Cy3 signal, indicating a frequent binding and dissociation of BamHI. Such effect is more pronounced for 1- and 2-bp-away sites due to the unstable binding expected from the short flanking sequence on DNA (21). The intensities collected from a single molecule was normalized to the lower, i.e., unbound, intensity value and plotted as a histogram (Fig. S2) with the x axis in the unit of “fold increase in intensity” (Fig. 1B, Lower). The cumulative histogram from 200 individual molecules is shown in Fig. 1C. The intensity fold increase analyzed from all the sites displayed a distance dependence in the range of 1 bp to 10 bp with the maximal increase of 2.7-fold at the closest (1 bp) site. The maximal fold increase of fluorescent signal was independent of the laser intensity applied (Fig. S3). The large difference seen between 1 bp and 2 bp, which may be in part due to the asymmetric binding of BamHI to dsDNA helix (21), clearly indicates that PIFE can resolve a single bp distance, especially in the extreme vicinity of the fluorophore (Fig. 1D). Fluorescence lifetime measurements performed on the same set of DNA constructs also show a similar distance-dependent relationship (Fig. 1E) (19). Such changes were not observed with Cy3B, a nonisomerizable analog of Cy3, supporting the cis-trans isomerization as the basis for the PIFE effect (Fig. 1 D and E). Cy5, which also possesses the isomerizable linkage, showed 1.9-fold increase for the 1 bp site (Fig. S4). BamHI binding to the 35-bp-away site yielded about 1.3-fold increase, which is consistent with the residual enhancement obtained for the 12-bp- and 15-bp-away sites (Fig. 1D and Fig. S5A). As control experiments, we tested two nonspecific restriction enzymes, HindIII and XhoI, which rendered extremely low (< 5%) and transient binding occurrences, yielding broad distribution in histograms as expected from random contact with DNA (Fig. S5 B and C).
RIGh Translocation Data Provides a Continuous PIFE Detection within 12-bp Distance.
Next we sought to determine the distance dependence of PIFE using a motor protein, RIGh (truncation mutant of RIG I) (8). Triggered by 5′-triphosphate moiety, this antiviral protein translocates along a dsRNA and DNA∶RNA heteroduplex at an accelerated rate in an ATP dependent manner. The RIGh translocation data taken from 20, 30, and 40 bp labeled DNA∶RNA hybrid with Cy3 label were analyzed to determine the distance sensitivity of PIFE (Fig. 2A). We chose this system because (i) the PIFE effect of RIGh can be directly compared to the BamHI data because they both interact with a duplex nucleic acid, (ii) the translocation of RIGh yields a continuous readout of PIFE as the protein moves along the duplex axis, (iii) the highly repetitive cycles of translocation exhibited by RIGh enables monitoring multiple rounds of translocation signals from one RNA molecule, which reduces the level of heterogeneity in the data analyzed. The single molecule trace shows an abrupt increase followed by a gradual decrease, which is interpreted as RIGh binding the substrate adjacent to the Cy3 fluorophore and its translocation on the 40-bp duplex, respectively. The highest and the lowest intensity values collected from 80 traces indicate the maximal increase of 2.6-fold in intensity, which is similar to the case of BamHI (Fig. S6). This result indicates that the maximum increase of 2.6- to 2.7-fold intensity obtained from BamHI and RIGh may not be protein-dependent but more general.
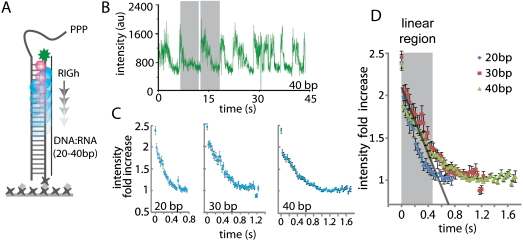
Fig. 2.
RIGh-induced PIFE effect. (A) Schematic of RIGh translocation on RNA∶DNA heteroduplex. (B) A single molecule trace of RIGh translocation. (C) Overlay of about 50 single-molecule traces from 20, 30, and 40 duplex. (D) Overlay of averaged translocation events from all three duplexes. The shaded region represents the PIFE-sensitive range at which translocation signals are most linear.
To obtain PIFE sensitive distance range, translocation events of similar periodicity were selected from 40 traces (Fig. 2B) and overlaid for 20, 30, and 40 bp (Fig. 2C). First, we calculated the binding occupancy of RIGh using the translocation dwell times of 20, 30, and 40 bp (Fig. S7). Assuming that RIGh protein translocates with a constant speed and through the entire length of the duplex of 20, 30, and 40 bps, we sought to calculate the length of duplex occupied by the protein. We performed dwell time analysis by taking peak-to-peak interval in PIFE traces obtained from the single-molecule experiments. (Fig. 2B) The average dwell times were plotted against the length of duplex and the x-intercept of 5 bp was interpreted as the binding occupancy. The resulting 5-bp binding occupancy indicates that we observe 15-, 25-, and 35-bp translocation of RIGh in single molecule PIFE traces. The overlay plot in Fig. 2D displays a linear decrease in the initial phase (gray bar) followed by a departure to a nonlinear pattern. To translate the time of translocation to the number of bp translocated, and hence the distance in nanometers, we performed linear regression analysis that revealed that the linear phase corresponds to approximately 12 bp, which is similar to the distance range observed with BamHI (Fig. S8). In addition, the sharp drop seen in the first two data points is likely due to the protein movement of 1 bp as seen in the BamHI case. Taken together, the PIFE assay allows detection over a 0- to 4-nm distance range, and the shape of PIFE along this distance may be protein-dependent. In addition, the level of fluorescence enhancement is likely to depend on the type of protein and the nature of protein–nucleic acid interaction. Therefore the distance dependence we provide based on the two protein systems should be considered a rough calibration
RecA Monomer Binding Is Visualized by PIFE Change on Single-Stranded DNA.
Next, we used RecA, which forms filament along ssDNA, to investigate the PIFE effect on ssDNA. We employed the experimental condition used by Joo et al. where single monomers of RecA binding on ssDNA was visualized by FRET (1). We used a partially duplexed DNA with a 3′ Cy3-labeled 13-nucleotide (nt) tail length to which up to four RecA molecules can bind (Fig. 3A).
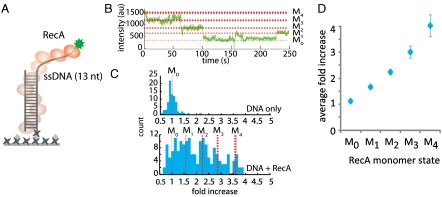
Fig. 3.
RecA mediated PIFE effect on ssDNA. (A) Schematic of RecA monomers forming a filament on single-strand DNA. (B) A representative single-molecule trace displaying RecA monomer binding and dissociation. PIFE levels correspond to M0 to M4 states. (C) Intensity histograms from the clustering analysis show a narrow single peak for DNA only (top) and appearance of distinct peaks corresponding to M0 to M4 states when RecA is added (bottom). (D) Plot of the average fold increase obtained from five independent measurements.
We optimized the illumination to obtain a narrow peak with well defined intensity for DNA-only condition (M0 in Fig. 3 B and C). The representative single-molecule trace shows multiple levels of intensities induced by RecA monomers binding and dissociation as expected from the low protein concentration applied (10–50 nM) (Fig. 3B). We selected 200 traces that exhibited intensity fluctuations and generated a histogram of the normalized intensity using a cluster analysis (Fig. S9). Five well-separated histogram peaks emerged, which represents DNA-only, one, two, three, and four monomer bound states (M0, M1, M2, M3, M4) (Fig. 3C). The normalized intensity data collected from five independent measurements yields highly reproducible PIFE changes of M0 to M4 distribution (Fig. 3D). Similarly, lifetime measurement showed multiple states corresponding to the status of RecA monomer binding (Fig. S10). Overall, the higher intensities observed in case of ssDNA (RecA) than dsDNA (BamHI, RIGh) can be partially attributed to the flexibility of ssDNA, which allows it to physically interact with the protein more efficiently, but the major contribution is likely due to the filament formation of RecA, which substantially raises the local viscosity around Cy3. Our result demonstrates that PIFE can be used to monitor ssDNA binding proteins with high sensitivity. Due to the flexibility of ssDNA, we cannot determine the distance sensitivity from this experiment. Therefore our RecA data points to the feasibility of using PIFE for single stranded DNA binding protein study rather than providing a calibration.
Discussion
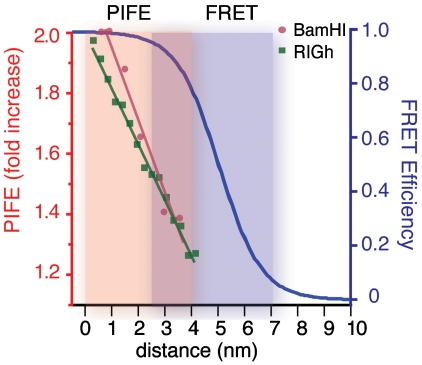
Biological interactions involve a wide range of distance changes between molecules. Cytoskeletal motors such as kinesin takes 8-nm steps across several micrometer-long microtubule axis (22), whereas DNA motors such as RNA polymerase and helicase move with a single-nucleotide step on DNA (23, 24). The PIFE method offers an efficient tool for exploring protein–protein or nucleic acid–protein contact by providing the signal within the experimentally useful range for this application. So far, PIFE effect was exhibited in dyes that possess cis-trans isomerizable carbon linkage such as Cy3, Cy5, and DY547 (16), whereas PIFE was not seen in Cy3B, which lacks this chemical structure. Based on these observations, we predict that the isomerizable fluorophores such as Dy548, Dy549, Alexa (25) series, and other Cyanine dyes will also have PIFE effect, whereas the nonisomerizable dyes such as Dy555 and Bodipy dyes will not (26).Here we present a systematic calibration of a unique single-molecule technique PIFE that displays a sharp distance sensitivity under 4-nm range, where FRET signal is insensitive (Fig. 4). Furthermore, the PIFE method expands the capacity of single-molecule fluorescence detection to an extensive degree, primarily because one can bypass the protein labeling. This makes PIFE a readily available tool for probing motions of nucleic acid motor proteins as an alternative or complement to FRET. Combined with FRET, PIFE is expected to reveal hidden dynamics in short distances such as several base pairs and nucleotides.
Fig. 4.
Distance sensitivity of PIFE vs. FRET. The circles (red) and squares (green) represent data obtained from the linear portion of the BamHI and RIGh experiments, respectively. The lines are their respective linear fit. The orange and purple shades indicate PIFE sensitivity (0–4 nm) and FRET sensitivity (2.5–7.5nm), respectively.
Materials and Methods
BamHI Substrates.
Oligonucleotides required to make the DNA duplex substrates were purchased from IDT DNA. The sequences used are listed with the BamHI recognition site colored in red (Table 1). The 35-bp site construct used for a control experiment had the following sequence: 5′ CGT ATG GAT CCA TAC GTA GCG TAG CGT AGC GTA GCG TAG G/Cy3 3′ and 5′ CCT ACG CTA CGC TAC GCT ACG CTA CGT ATG GAT CCA TAC G/3BiosG 3′. Cy3B 5-bp site DNA was achieved via labeling of amino modified oligonucleotides with Cy3B monofunctional N hydroxysuccinimidyl (NHS) ester, following the instructions of the supplier (GE Lifesciences). Briefly, 10 nmoles of amino modified oligonucleotides in 20 uL of 50 mM sodium tetraborate buffer pH 8.5 and 100 nmoles of Cy3B NHS ester dissolved in DMSO was added and incubated with rotation overnight at room temperature. The labeled oligonucleotides were purified by ethanol precipitation. The complementary DNA strands were annealed by heating at 95 °C for 2 min and slowly cooled to room temperature.
Table 1.
DNA sequence for BamHI experiment
| Recognition site 5′-Cy3 sequence | |
| 1 bp | /5Cy3/ TGG ATC CAT AGT AGC GTA GCG TAG CGT AGC GTA GCG TAG C |
| 2 bp | /5Cy3/ TAG GAT CCA TAG TAG CGT AGC GTA GCG TAG CGT AGC GTA G |
| 3 bp | /5Cy3/ TAT GGA TCC ATA CGT AGC GTA GCG TAG CGT AGC GTA GGC G |
| 5 bp | /5Cy3/ TGT ATG GAT CCA TAC GTA GCG TAG CGT AGC GTA GCG TAG G |
| 7 bp | /5Cy3/ AGC GTA TGG ATC CAT ACG TAG CGT AGC GTA GCG TAG CGT A |
| 10 bp | /5Cy3/ CGT ATA TAC GGG ATC CTA GCG TAG CGT AGC GTA GCG TAG G |
| 12 bp | /5Cy3/ AGC GTA GCG TAT GGA TCC ATA CGT AGC GTA GCG TAG CGT A |
| 15 bp | /5Cy3/ CGT ATA TAC GTA GCG GGA TCC TAG CGT AGC GTA GCG TAG G |
| Cy3B | /5Cy3B/CGT ATG GAT CCA TAC GTA GCG TAG CGT AGC GTA GCG TAG G |
| Recognition site 5′-biotin sequence |
|
| 1 bp | /5BiosG/ GCT ACG CTA CGC TAC GCT ACG CTA CGC TAC TAT GGA TCC A |
| 2 bp | /5BiosG/ CTA CGC TAC GCT ACG CTA CGC TAC GCT ACT ATG GAT CCT A |
| 3 bp | /5BiosG/ CGC CTA CGC TAC GCT ACG CTA CGC TAC GTA TGG ATC CAT A |
| 5 bp | /5BiosG/ CCT ACG CTA CGC TAC GCT ACG CTA CGT ATG GAT CCA TAC A |
| 7 bp | /5BiosG/ TAC GCT ACG CTA CGC TAC GCT ACG TAT GGA TCC ATA CGC T |
| 10 bp | /5BiosG/ CCT ACG CTA CGC TAC GCT ACG CTA GGA TCC CGT ATA TAC G |
| 12 bp | /5BiosG/ TAC GCT ACG CTA CGC TAC GTA TGG ATC CAT ACG CTA CGC T |
| 15 bp | /5BiosG/ CCT ACG CTA CGC TAC GCT AGG ATC CCG CTA CGT ATA TAC G |
| Cy3B | /5BiosG/ CCT ACG CTA CGC TAC GCT ACG CTA CGT ATG GAT CCA TAC G |
RIGh and RecA Reaction Condition and Substrates.
RIGh reaction conditions and constructs were described in previous work (8). RecA reaction conditions were conducted as described in Joo et al. (1). The DNA used in this assay was a partially duplexed DNA with18 mer duplex (5′ GCC TCG CTG CCG TCG CCA biotin 3′ and 5′ TGG CGA CGG CAG CGA GGC (T)13-3′) and 3′ poly T tail of 13 nucleotide.
Slide Preparation for Single-Molecule Experiment.
Both single-molecule PIFE and lifetime experiments were carried out on quartz slides. To minimize surface interactions with the protein, quartz slides and cover slips were coated with polyethylene glycol (PEG). Briefly, the slides and coverslips were cleaned and treated with methanol, acetone, KOH, burned, treated with aminosilane, and coated with a mixture of 3% m PEG and 97% biotin PEG (6).
Single-Molecule PIFE Measurement.
Prism type total internal reflection microscopy was used to acquire single molecule PIFE data. A 532-nm Nd∶YAG laser was guided through a prism to generate an evanescent field of illumination. A water-immersion objective was used to collect the signal and a 550-nm long pass filter was used to remove the scattered light. Cy3 signals were collected using a 630-nm dichroic mirror and sent to a CCD camera. Data were recorded, at a time resolution of 0.01 s, as a stream of imaging frames and analyzed with scripts written in IDL to give fluorescence intensity time trajectories of individual molecules (6).
BamHI Reaction Conditions.
Standard reaction buffer was 50 mM Tris-HCl pH 8.0, 10 mM CaCl2, 100 mM NaCl, with an oxygen scavenging system (1 mg/mL glucose oxidase, .4% (w/v) D glucose, 0.04 mg/mL catalase, and 1% v/v 2- mercaptoethanol). Measurements were performed at room temperature, 21 °C). BamHI (Invitrogen) was made to 800 nM in the reaction buffer and then added to a chamber that had 100-pM DNA immobilized on a polymer-coated quartz surface via biotin neutravidin linkage (5).
Single-Molecule Lifetime Measurements.
As described previously in the manuscript, single molecule lifetime measurements were taken with a home-built confocal pulsed laser with a piezo controlled electric stage (19). The laser beam was divided and split into two pathways, the first being sent to a photodiode and the second one reflected into a dichroic mirror and focused on the sample by an oil immersion objective. Emitted light was collected by the objective lens, focused on the pinhole, and then imaged onto the avalanche photodiode (APD). The signals from the APD and photodiode were sent to a time correlated single photon counting (TCSPC) device. The analysis of the lifetime was fitted in the fixed bin range with single exponential decay using a maximum likelihood estimator algorithm, as stated (19). The total number of photons collected in each histogram was 500. All measurements were taken at room temperature, 21 ± 1 °C.
Normalization Data Analysis for PIFE.
Single-molecule fluorescence time trajectories were viewed and analyzed using Matlab. Binding events were selected for BamHI protein system, while the minimum and maximum values from translocation events were selected for the RIGh translocation system. In both cases, the fluorescence intensities were binned and fitted to two Gaussian distributions, representing the intensities of the DNA-only region and that with protein bound. A fold increase was calculated with BamHI and each DNA construct to get the distance sensitive curve shown in Fig. 1D. For the RIGh data, further analysis was done to determine the PIFE sensitivity that was encoded in each translocation event. Assuming that RIGh translocates on dsRNA at a constant rate, dwell times of each event were taken and plotted for the 20-, 30-, and 40-bp duplex lengths. The dwell times were then fitted to a line, which x intercept represents the binding occupancy of RIGh (Fig. S7). The translocation traces from the 20-, 30-, and 40-bp duplex length were averaged. The least square regressions were taken of the intensity from the start of the translocation onset. A regression value close to 1 signifies that the values lie in a straight line. Thus, the translocation signals remain linear until 0.51 s, after which the r- value decreases dramatically (Fig. S8A). We used 0.51 s to calculate the linear portion for each duplex length to obtain that PIFE is sensitive until approximately 12 bp on the RIGh translocation system (Fig. S8B).
Clustering Data Analysis for PIFE.
Traces that showed the DNA-only intensity level and at least three additional states were selected (Fig. S9). A k-means clustering algorithm from Matlab was applied with four states. K- means clustering was implemented again, with five monomer states, to obtain fold increases on each monomer binding state. These distinct states remain consistent over different experimental conditions, included varying protein concentration from 10-nM RecA to 50-nM RecA.
Supplementary Material
Acknowledgments.
We thank T. Ha for his generous permission to use the TCSPC set up for fluorescence lifetime measurement; S. Doganay, Y. Ishitsuka, and X. Shi for technical assistance: and Y. Qiu and K. Ragunathan for helpful discussions. We appreciate the help of T. Ha, H. Koh, and D. Srinivasan on the manuscript. H.H. and S.M. were supported by National Institutes of Health Grant U19AI083025 (National Institute of Allergy and Infectious Diseases). H.K. was supported by the Korea Research Foundation Grant funded by the Korean government (KRF-2006-352-C00019).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017672108/-/DCSupplemental.
References
- 1.Joo C, et al. Real-time observation of RecA filament dynamics with single monomer resolution (Translated from eng) Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. (in eng) [DOI] [PubMed] [Google Scholar]
- 2.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA (Translated from eng) Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. (in eng) [DOI] [PubMed] [Google Scholar]
- 3.Abbondanzieri EA, et al. Dynamic binding orientations direct activity of HIV reverse transcriptase (Translated from eng) Nature. 2008;453:184–189. doi: 10.1038/nature06941. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woodside MT, et al. Direct measurement of the full, sequence-dependent folding landscape of a nucleic acid (Translated from eng) Science. 2006;314:1001–1004. doi: 10.1126/science.1133601. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ha T. Single-molecule fluorescence methods for the study of nucleic acids (Translated from eng) Curr Opin Struct Biol. 2001;11:287–292. doi: 10.1016/s0959-440x(00)00204-9. (in eng) [DOI] [PubMed] [Google Scholar]
- 6.Rasnik I, Myong S, Cheng W, Lohman TM, Ha T. DNA-binding orientation and domain conformation of the E. coli rep helicase monomer bound to a partial duplex junction: single-molecule studies of fluorescently labeled enzymes (Translated from eng) J Mol Biol. 2004;336:395–408. doi: 10.1016/j.jmb.2003.12.031. (in eng) [DOI] [PubMed] [Google Scholar]
- 7.Dorywalska M, et al. Site-specific labeling of the ribosome for single-molecule spectroscopy (Translated from eng) Nucleic Acids Res. 2005;33:182–189. doi: 10.1093/nar/gki151. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myong S, et al. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E. coli UvrD monomers along single-stranded DNA (Translated from eng) J Mol Biol. 2004;344(5):1287–1309. doi: 10.1016/j.jmb.2004.10.005. (in eng) [DOI] [PubMed] [Google Scholar]
- 10.Tomko EJ, Fischer CJ, Lohman TM. Ensemble methods for monitoring enzyme translocation along single stranded nucleic acids. Methods. 2010;51:269–276. doi: 10.1016/j.ymeth.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong CJ, Lohman TM. Kinetic control of Mg2+-dependent melting of duplex DNA ends by Escherichia coli RecBC (Translated from eng) J Mol Biol. 2008;378:761–777. doi: 10.1016/j.jmb.2008.03.023. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu CG, Bradford C, Lohman TM. Escherichia coli RecBC helicase has two translocase activities controlled by a single ATPase motor (Translated from eng) Nat Struct Mol Biol. 2010;17:1210–1217. doi: 10.1038/nsmb.1901. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niedziela-Majka A, Chesnik MA, Tomko EJ, Lohman TM. Bacillus stearothermophilus PcrA monomer is a single-stranded DNA translocase but not a processive helicase in vitro (Translated from eng) J Biol Chem. 2007;282:27076–27085. doi: 10.1074/jbc.M704399200. (in eng) [DOI] [PubMed] [Google Scholar]
- 14.Luo G, Wang M, Konigsberg WH, Xie XS. Single-molecule and ensemble fluorescence assays for a functionally important conformational change in T7 DNA polymerase. Proc Natl Acad Sci USA. 2007;104:12610–12615. doi: 10.1073/pnas.0700920104. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorokina M, Koh HR, Patel SS, Ha T. Fluorescent lifetime trajectories of a single fluorophore reveal reaction intermediates during transcription initiation (Translated from eng) J Am Chem Soc. 2009;131:9630–9631. doi: 10.1021/ja902861f. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myong S, et al. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA (Translated from eng) Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park J, et al. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps (Translated from eng) Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanborn M, Connolly B, Gurunathan K. Fluorescence properties and photophysics of the sulfoindocyanine Cy3 linked covalently to DNA. J Phys Chem B. 2007;11:11064–11074. doi: 10.1021/jp072912u. [DOI] [PubMed] [Google Scholar]
- 19.Sorokina M, Koh H, Patel S, Ha T. Fluorescent lifetime trajectories of a single fluorophore reveal reaction intermediates during transcription initiation. J Am Chem Soc. 2009;131:9630–9631. doi: 10.1021/ja902861f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aramendia PF, Negri RM, Roman ES. Temperature dependence of fluorescence and photoisomerization in symmetric carbocyanines. influence of medium viscosity and molecular structure. J Phys Chem-US. 1994;98:3165–3173. [Google Scholar]
- 21.Newman M, Strzelecka T, Dorner LF, Schildkraut I, Aggarwal AK. Structureof Bam HI endonuclease bound to DNA: Partial folding and unfolding on DNA binding (Translated from eng) Science. 1995;269:656–663. doi: 10.1126/science.7624794. (in eng) [DOI] [PubMed] [Google Scholar]
- 22.Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand (Translated from eng) Science. 2004;303:676–678. doi: 10.1126/science.1093753. (in eng) [DOI] [PubMed] [Google Scholar]
- 23.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase (Translated from eng) Nature. 2005;438:460–465. doi: 10.1038/nature04268. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase (Translated from eng) Science. 2007;317:513–516. doi: 10.1126/science.1144130. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dave R, Terry DS, Munro JB, Blanchard SC. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging (Translated from eng) Biophys J. 2009;96:2371–2381. doi: 10.1016/j.bpj.2008.11.061. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linck L, Resch-Genger U. Identification of efficient fluorophores for the direct labeling of DNA via rolling circle amplification (RCA) polymerase phi29 (Translated from eng) Eur J Med Chem. 2010;45:5561–5566. doi: 10.1016/j.ejmech.2010.09.005. (in eng) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.