Abstract
Protein-directed intracellular transport has not been observed in bacteria despite the existence of dynamic protein localization and a complex cytoskeleton. However, protein trafficking has clear potential uses for important cellular processes such as growth, development, chromosome segregation, and motility. Conflicting models have been proposed to explain Myxococcus xanthus motility on solid surfaces, some favoring secretion engines at the rear of cells and others evoking an unknown class of molecular motors distributed along the cell body. Through a combination of fluorescence imaging, force microscopy, and genetic manipulation, we show that membrane-bound cytoplasmic complexes consisting of motor and regulatory proteins are directionally transported down the axis of a cell at constant velocity. This intracellular motion is transmitted to the exterior of the cell and converted to traction forces on the substrate. Thus, this study demonstrates the existence of a conserved class of processive intracellular motors in bacteria and shows how these motors have been adapted to produce cell motility.
Keywords: murein cluster B, proton motive force
Because of its small size, the bacterial cell was long thought to be a disordered compartment where random collisions and diffusion drive enzymatic reactions and cellular processes (1). However, recent advances in light microscopy have shown that, akin to eukaryotic cells, bacteria are spatially organized by a complex cytoskeleton, potentially allowing directed sorting of proteins to specific subcellular sites (1). Despite the characterization of bacterial actins and tubulins, processive transport motors akin to myosins or kinesins have not been found. Because eukaryotic cell motility is driven, in part, by processive intracellular motors, studying how bacteria glide over solid surfaces may lead to the identification of similar types of motors.
Directed motility is a vital feature of the behavior of many organisms and often is essential for biofilm formation and virulence (2). Myxococcus xanthus, a rod-shaped, Gram-negative, soil-dwelling bacterium, uses a combination of gliding motility, termed “adventurous” (A), and pilus-driven twitching motility, termed “social” (S), to form organized multicellular structures (3). Directional control in M. xanthus is achieved by modulating the period of cellular reversals, wherein the leading and lagging poles exchange roles (3). Recent work has shown that a set of motility-regulatory proteins is localized at the two distinct poles in moving cells. Frizzy protein S (FrzS) and Adventurous gliding protein Z (AglZ) are found at the leading pole (4, 5), and RomR is located at the lagging pole (6). Every 6 min, on average, gliding direction is reversed, and the protein-localization pattern is switched. The frequency of these oscillations is regulated by the Frz chemosensory system that acts upstream of the Ras-like protein Mutual function for gliding protein A (MglA) to produce a dynamic and controlled cell polarity (7, 8).
Despite several decades of research, the physical mechanism driving gliding motility has remained difficult to define. Two general classes of models for force production in gliding bacteria have been proposed. The first class invokes the motion of substrate-bound motors on tracks inside the cell (5, 9, 10). The second class proposes that hydration of an extruded polyelectrolyte “slime” gel from the rear of the cell propels the cell forward (11). One key difference between these two models is the location of force generation at the cell surface: A distributed motor-based mechanism requires traction to be generated along the cell cylinder, whereas in the slime-extrusion model force is generated only at the rear of the cell (12).
We recently found indirect evidence for a distributed, motor-based mechanism of gliding motility by observing the subcellular localization of the gliding motility regulatory factor AglZ in moving M. xanthus cells (13). In gliding cells, cytoplasmic AglZ-YFP formed spatially periodic foci that remained fixed relative to the surface even as the cell moved by a distance of several microns. Based on this and other observations, we hypothesized that intracellular motors moving on cytoskeletal filaments in the cytoplasm transmit force through the cell wall to dynamic adhesion complexes attached to the substrate, causing the cell to move forward. The identification of such molecular motors is a critical step toward confirming this model of this cell locomotion.
Results and Discussion
Intracellular Transport Generates Traction Force During Cell Gliding.
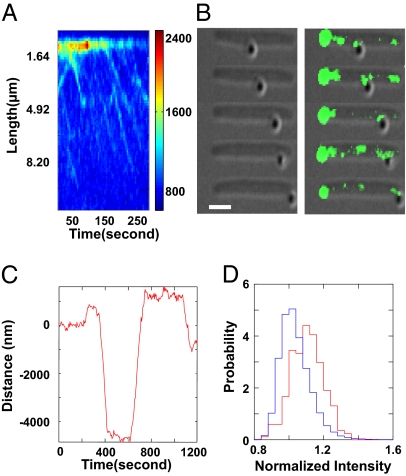
If AglZ-YFP is linked to intracellular motor-driven motion, this protein should exhibit unidirectional flow from the leading cell pole in the cell frame of reference. In cells immobilized on a chemically treated glass coverslip (SI Materials and Methods), AglZ-YFP does not distribute uniformly or form fixed foci. Instead, as expected, we observed the processive, unidirectional transport of AglZ clusters from the front of the cell, defined by the brightest pole, toward the back (Movie S1). This flow is observed most easily in kymographs of AglZ-YFP fluorescence from single cells (Fig. 1A). Moving clusters of AglZ appear as diagonal lines in the kymograph corresponding to unidirectional motion at a constant velocity. Line fits to trajectories from multiple cells yielded an average velocity of 6.0 ± 2.1 μm/min (91 trajectories from 35 cells). Almost all the trajectories, 94 ± 2%, were oriented away from the leading pole, the same relative motion between the cluster and cell pole as observed in gliding cells with fixed AglZ clusters (Fig. 1A). Multiple clusters could be found moving at the same time in a single cell, and, surprisingly, in a few instances we observed two clusters in the same cell moving in opposite directions at the same time (Fig. 1A). Further analysis of the ratio of forward- and reverse-moving foci in different genetic backgrounds may uncover details about how bidirectionality and control is achieved by motor complexes.
Fig. 1.
Intracellular transport drives extracellular membrane-bound motion. (A) Kymograph of AglZ-YFP fluorescence in an immobilized cell. Moving clusters of AglZ appear as diagonal lines in the kymograph corresponding to unidirectional motion at a constant velocity. (B) Colocalization of AglZ-YFP clusters with a gliding bead. A bead moves along an immobilized cell (DIC image, Left). An overlay of the DIC image and an AglZ-YFP fluorescence image (green) shows the colocalization of the gliding bead with a cluster of AglZ-YFP fluorescence (Right) (Scale bar, 2 μm.) (C) A position record of a bead moving on the side of an immobilized cell. (D) Histograms of AglZ-YFP fluorescence intensity in immobilized cells. The blue histogram is derived from the intensity of all pixels within a cell except those belonging to the bright leading pole. The red histogram is derived from a subset of these pixels, those that are within 1 pixel from the center of a moving bead. All the intensity values are normalized by the mean intensity of pixels within a cell, excluding the bright leading pole. The higher mean value represented by the peak of the red histogram is the result of a significant enhancement of AglZ-YFP fluorescence near moving beads.
We next probed whether traction forces are generated along the sides of cells, as predicted by the distributed motor-based model, using polystyrene beads bound to the outer surface of immobilized cells in a technique inspired by work on other gliding bacteria, particularly members of the bacteroides phylum (2, 9, 14). Similar to these previous observations, beads often were transported along the sides of the cell (Fig. 1B and Movie S2). Beads exhibited saltatory motion wherein motionless periods were interrupted by long runs (1.8 ± 1.2 μm) of unidirectional motion along the side of the cell (Fig. 1 B and C). During periods of fast motion, the bead velocity was 3.3 ± 1.8 μm/min. The speeds of AglZ-YFP clusters in immobilized cells, traveling beads on cell surfaces, and cells moving on agar (1.3 ± 1.8 μm/min) all have similar magnitudes, consistent with the notion that they reflect the activity of a common machinery. Subtle discrepancies in the exact magnitude of these speeds probably are caused by differences in the substrata and different applied loads experienced by the motility engine in each case. To examine the relative motion of multiple beads on a single cell, we artificially elongated cells by inhibiting the division-associated protein Penicillin-Binding Protein 3 (PBP3) with the drug cephalexin (15). Most of the beads on these cells, 93 ± 4%, moved in the same direction, away from the leading pole labeled with a bright AglZ focus, whereas 7% of the beads moved in the opposite direction, consistent with the behavior of AglZ-YFP cluster motion observed in immobilized cells. These results show that traction force is generated along the side of a cell and are consistent with the distributed motor-based model but are inconsistent with the slime-extrusion model of Wolgemuth et al. (11).
We performed several measurements that confirm that beads are powered by the cell-gliding machinery. First, we sought to connect the motion of the beads with the transport of AglZ along the axis of stationary cells by simultaneously measuring bead position and AglZ-YFP localization. Colocalizing AglZ-YFP and moving beads is challenging. Photobleaching of the AglZ-YFP foci occurs relatively quickly, whereas bead motion occurs sporadically, presumably because the unfunctionalized beads interact with the motor system only transiently. The difference in these time scales makes coincident measurement of AglZ-YFP fluorescence and bead velocity difficult. Nevertheless, whenever bead movement occurred rapidly after illumination, beads colocalized with AglZ-YFP (Fig. 1B). If AglZ-associated complexes of proteins drive extracellular motion, AglZ-YFP fluorescence intensity should be enhanced in the vicinity of moving beads. We compared the fluorescence intensity of regions of a cell that were within 82 nm (1 camera pixel) of the center of a moving bead with the overall fluorescence in the cell. Histograms of these two distributions show that AglZ-YFP fluorescence is enhanced near moving beads (Fig. 1D). Second, we added A22 to the medium and measured the effect on bead motion. A22 is a drug that has recently been shown to induce the depolymerization of the actin homolog MreB and reversibly to destabilize the AglZ-YFP complexes and inhibit gliding motility (16). After A22 treatment, processive bead motion was disrupted dramatically (Fig. S1C). Taken together, these data strongly suggest that gliding motility is driven by processive intracellular motors that interact with the MreB cytoskeleton.
Protein Transport and Motility Require the Proton Gradient.
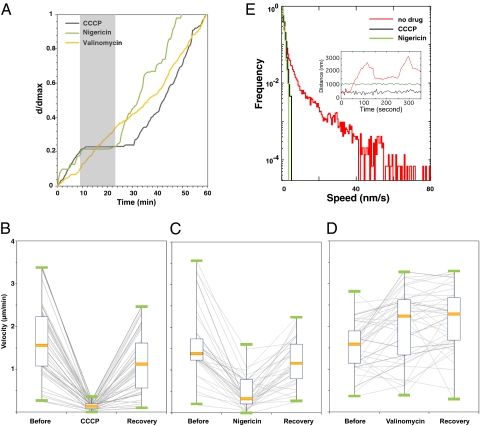
To guide the search for candidate motor genes, we sought to find the source of energy for gliding motility. ATP and the proton motive force (PMF) are common energy sources for molecular motors such as kinesin, myosin, and the bacterial flagellar motor. We used carbonyl cyanide-m-chlorophenylhydrazone (CCCP) to probe the dependence of gliding on the PMF. In our hands, CCCP at a concentration of 10 μM destroys the PMF in M. xanthus (Table S4) and rapidly abolishes cell movement (Fig. 2 A and B). This effect is reversible. When CCCP is washed out, cells regain their ability to glide. CCCP has the same reversible effect on bead movement using immobilized cells. To quantify the effect of drugs on bead motion, we calculated histograms of the speed of beads moving along immobilized cells (Materials and Methods). Upon the injection of CCCP, beads immediately stop moving (Fig. 2E). These results suggest that the PMF supplies the energy for gliding motors.
Fig. 2.
Gliding motility in M. xanthus is driven by the proton gradient. (A) Reversible effects of metabolic PMF-uncoupling drugs on single cell motility. The relative cumulated distances corresponding to the distance traveled by a cell at any given time over the maximum traveled distance by that cell at the end of the time lapse (d/dmax) are plotted over time. The gray rectangle indicates the time interval when the cells were in the presence of drugs. (B–D) Box plots of wild-type cell (n = 50) velocities before, during, and after treatment by CCCP (B), nigericin (C), and valinomycin (D). The solid orange bars represent the average velocity of the population during each condition. Each line represents a single cell before, during, and after treatment. (E) Histograms of bead gliding speeds. (Inset) Trajectories of gliding beads along nontreated (red), CCCP-treated (black), and nigericin-treated (green) cells. The absolute value of the first derivative of the trajectories is used to form speed histograms. Both CCCP and nigericin completely stop bead movement.
The PMF arises from gradients in both the chemical potential energy, in the form of a pH difference across the cell membrane, and electrical potential energy, caused by a voltage difference across the membrane. We used two drugs, nigericin and valinomycin, to uncover the relative roles that these two potential energies play in gliding motility. In M. xanthus cells, nigericin reduces the pH gradient without changing membrane potential, whereas valinomycin destroys the membrane potential with no change in the magnitude of the pH gradient (Table S4 and Fig S2D). When we added nigericin to gliding cells, motility was abolished, much as it was in the presence of CCCP. Cell and bead motion were stopped, and AglZ-YFP foci disappeared (Fig. 2 A, C, and E and Figs. S1A and S2A). As with CCCP, the effect of nigericin was reversible. In contrast, valinomycin has no effect on either cell or bead movement (Fig. 2 A and D). From these results, we conclude that gliding and bead motion are energized directly by the proton gradient. We further confirmed that CCCP and nigericin treatment did not affect the intracellular ATP pools during the short timescales relevant for gliding motility experiments (Fig S2C and Materials and Methods). Additionally, nigericin treatment had no significant effect on twitching motility, which uses the hydrolysis of ATP as an energy source, showing that this drug does not affect motility systems that are powered by ATP (Fig. S2B).
A Proton Channel Powers Force Generation.
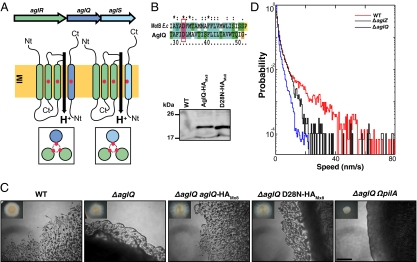
Taken together, the above data show that gliding motility in M. xanthus is driven by a proton gradient, suggesting that the mechanism underlying bacterial gliding and swimming may be linked to a common form of molecular motor, a proton channel. Bacterial motors that make use of a proton gradient are widespread and power flagellar rotation [Motility proteins AB (MotAB)], ATP synthesis (F1FO), and macromolecular transport across the cell envelope [Tolerant proteins QR (TolQR), Excretion of an inhibitor of Colicin B proteins BD (ExbBD)]. We searched the Myxococcus genome for homologs of MotAB and TolQR/ExbBD (Fig S3A) and found one particular locus, aglRQS (MXAN6862-60), which fulfilled all the expected criteria for a gliding motor candidate. Transposon insertions in aglR (MXAN6862) and aglS (MXAN6860) have been described as specifically inactivating gliding and not twitching motility (17). Sequence analysis indicates that AglR is a TolQ/ExbB/MotA homolog, whereas AglQ (previously MXAN6861) and AglS are TolR/ExbD/MotB homologs (Fig. 3A and Fig. S3 B–D). AglR, AglQ, and AglS all contain the key residues in the predicted lumen of the channel that enable proton conduction, and theoretically both AglRQ and AglRS can form functioning channels (Fig. 3A and Fig. S3 B–D) (18).
Fig. 3.
The aglRQS locus encodes a proton-conducting channel essential for gliding motility. (A) Proton-conducting channels encoded by the AglRQS proteins. The proposed membrane topologies of the proteins are inspired by work with Escherichia coli TolQR. Proton-conducting residues are systematically conserved in AglR, -Q, and -S (red dots). (Insets) Top view of the potential heterotrimeric channels. (B) Conserved aspartate in the AglQ and E. coli MotB channel-forming transmembrane helix. The D28N substitution does not affect the stability of a functional AglQ-HA protein. (C) The channel function of AglQ is strictly required for gliding motility. Single-cell gliding motility of aglQ mutants cannot be detected at the edges of colonies on hard (1.5%) agar, but motility on soft (0.5%) agar, which detects only S-motility, is intact. As expected a ΔaglQ ΩpilA is nonmotile on both substrata. (Scale bar, 1 mm.) (D) Histograms of bead gliding speeds on wild-type (red), ΔaglZ (black), and ΔaglQ (blue) cells. Compared with wild-type cells, gliding motility is perturbed in ΔaglZ cells. However, speeds faster than 30 nm/s are still observed. In contrast, gliding motility in ΔaglQ cells is disrupted more severely, with a narrower speed histogram and no events showing speeds faster than 30 nm/s.
Other TolQR homolog pairs are found in the Myxococcus genome but are less likely to be the true gliding motor (Fig S3A). Nan et al. (10) identified AglX and AglV as necessary for gliding motility. However, these genes are found in a putative operon upstream from TolA, TolB, and Peptidoglycan-associated lipoprotein (Pal) homologs and thus probably are involved in global maintenance of the cell envelope (19). Therefore we favor the idea that deletion of these proteins inhibits gliding through pleiotropic effects on the cell exterior, e.g., by the maintenance of external adhesion structures within the motor complex. AglRQS homologs also are found in a third cluster on the Myxococcus genome (MXAN3005-3003; Fig S3A). However, in-frame deletion of the MXAN3004 gene did not abolish gliding motility (Fig S4), suggesting that these genes are cryptic or perform a distinct, nongliding function.
To test the role of the AglRQS system directly, we characterized motility in strains containing in-frame deletions in aglR, aglQ, and aglS. All three deletions eliminate gliding motility but do not affect pilus-based twitching motility when assayed by colony morphology or single-cell analysis (Fig. 3C and Fig. S4). Cells containing a double deletion of aglR, -Q, or -S in combination with pilA do not exhibit any motility in either assay (Fig. 3C and Fig. S4). All agl deletions were fully complemented when aglR, -Q, and -S were expressed ectopically, showing that all three agl genes are essential for gliding motility (Fig. 3C and Fig. S4). A point mutation in a conserved residue of AglQ (D28N), predicted to abolish H+ binding within the lumen of the proton channel (20), does not affect protein stability but completely abolishes cell gliding, indicating that proton transport is required for gliding motility (Fig. 3 B and C). Finally, to test whether AglR, -Q, and -S form a complex, we searched for proteins that may associate with AglQ in vivo by conducting immunoprecipitation experiments using HA-tag fusion constructs of the wild-type and D28N mutant forms of AglQ as bait. Matching the spectra obtained from liquid chromatography-tandem mass spectrometry (LC-MS/MS) of the eluted trypsin-digested peptides against the Myxococcus sp. proteome resulted in the unambiguous identification of AglR and AglS in these samples but not in control samples derived from cells lacking the fusion proteins (Fig. S5). Thus, we expect that both AglR and AglS associate physically with AglQ-containing motility complexes.
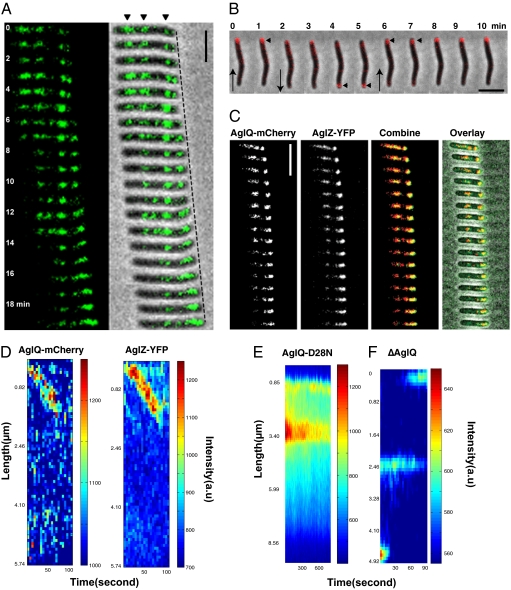
The AglRQS motor proteins exhibit the same intracellular localization dynamics as AglZ. To examine the localization of this motor system, we constructed a C-terminal fluorescent fusion of AglQ to mCherry (Fig. S6A). Cells expressing this fusion move with reduced velocity, 0.1 ± 0.1 μm/min compared with 1.3 ± 1.8 μm/min in wild-type cells, demonstrating that the fusion is partially defective but still proficient for motility. As does AglZ-YFP, this construct forms a bright polar spot at the leading pole and fixed periodic adhesion complexes that colocalize with AglZ-YFP in gliding cells (Fig. 4 A and C). The bright spot switches to the new leading pole when cells reverse direction (Fig. 4B). In stationary cells, AglQ-mcherry traffics away from the head in dynamic foci that colocalize with AglZ-YFP (Fig. 4D). Transport also occurred at a reduced speed compared with the speed measured for AglZ-YFP spots in a wild-type background (1.0 ± 0.5 μm/min vs. 6.0 ± 2.1 μm/min in wild type). Consistent with this interpretation, the velocity of bead trafficking also was reduced in these cells (0.8 ± 0.4 μm/min). The observation that this construct exhibits reduced velocity for both gliding and cytoplasmic transport lends further support to the conclusion that intracellular motion is connected to cell motility. In cells treated with nigericin, AglQ-mCherry foci dispersed rapidly and condensed at the cell pole, similar to the pattern of localization seen in nigericin-treated AglZ-YFP cells (Fig. S1B). Finally, an AglQ (D28N)-mCherry fusion also assembled bright fluorescent clusters that remained stationary because of the lack of channel activity (Fig. 4E and Fig. S6B), confirming that the mutant protein assembles paralyzed motor complexes. Thus, cluster formation and/or maintenance requires an intact pH gradient, whereas cluster motion requires an active AglRQS complex.
Fig. 4.
AglQ is a component of the gliding motility engine. (A) AglQ-mCherry localizes to fixed internal clusters in moving cells (black arrowheads). (B) AglQ-mCherry oscillates from pole to pole when moving cells reverse. (Scale bar, 2 μm.) (C) AglQ-mCherry colocalizes with AglZ-YFP in moving cells. (D) Kymographs of AglQ-mCherry (Left) and AglZ-YFP (Right) fluorescence in an immobilized cell. AglQ-mCherry and AglZ-YFP colocalize and are transported together along the cell from head to tail. (E) Kymograph of AglQ(D28N)-mCherry fluorescence in an immobilized cell. (F) Kymograph of AglZ-YFP fluorescence in an immobilized ΔaglQ cell. The AglZ-YFP clusters retain a fixed position at all times, but directional pole-to-pole oscillation still is observed.
To examine further the role of this proton channel in gliding motility, we examined the dynamics of AglZ-YFP in an aglQ-deletion strain. On agar, these cells contain a single bright polar spot and several observable periodic foci across the length of the cell. However, the periodic foci are not dynamic and remain fixed, slowly losing fluorescence intensity via photobleaching (Fig. S7). In addition, kymographs do not show any evidence of transport of AglZ in these cells (Fig. 4F). That AglZ-YFP still forms midcell clusters in an aglQ mutant suggests that the AglRQS system is required for the movement but not for the connection of cytoplasmic proteins such as AglZ and MreB to the substrate. Most interestingly, although the periodic foci remained fixed, the bright polar spot did exhibit reversal dynamics (Fig. 4F and Fig. S7). After an average of 6.4 ± 1.4 min (n = 20 cells), the bright polar spot switched poles even though cells and the periodic foci remained stationary. This observation strongly suggests that the periodic relocation of polarly localized proteins from one pole to the other, which generates directional reversal, does not require cell motion or a functioning gliding apparatus.
The distributed motor-based model of gliding motility supported by the data presented here requires the global coordination of a number of individual moving proteins to produce directional force and gliding. It is highly likely, therefore, that gliding motility mutants might exhibit a complex set of phenotypes relating to defects in directionality, coordination, and/or core motor function. The role that specific genes play in these different functions can be found by using a combination of the motility assays described here. Moving-bead experiments provide information on the motion of single motor complexes, whereas cell gliding presumably requires a sufficient level of coordination between multiple motor units to produce motion. For example, many previously studied gliding motility genes, such as aglZ, can be thought of as purely regulatory, because their disruption can be rescued by deletion of genes upstream in the control pathway, such as frzCD (21). Consistent with this concept, the motion of beads bound to the side of aglZ-deleted cells is severely perturbed but not completely abolished. In multiple instances in time, ΔaglZ cells powered the motion of beads with speeds faster than 30 nm/s, as seen in speed histograms (Fig. 3D). However, gliding motility in a ΔaglQ background is not restored by a second-site frz mutation, and beads on ΔaglQ cells show a much more dramatic reduction in the level of movement (Fig. 3D).
To transmit mechanical forces from the cytoplasmic membrane, where the AglRQS motors lie, to the cell exterior, a mechanical linkage must be established between the inner and outer membranes. In a recent publication, Nan et al. (10) reported observations of MreB-dependant, PMF-driven rotation of the periplasmic protein Adventurous gliding motility protein U (AgmU). The authors also used theoretical modeling to show that PMF-driven motor proteins running along helical tracks might produce gliding motility through a viscous interaction with the substrate that is mediated by AgmU. Our data strongly indicate that the AglRQS complex is the gliding-associated, PMF-driven motor, although a number of important avenues remain to be explored experimentally: How is traction force transduced from AglRQS to the substrate, and does it link directly through AgmU? What are the molecular components of the full transducing complex, and how is force transmitted through the structural cell wall? Are viscous or elastic contributions dominant in the interaction with the substrate?
Here, we show that a widely conserved class of bacterial motors, which includes both the flagellar motor and the gliding motor, can drive intracellular protein transport in bacteria and suggest that gliding motility emerged through the recruitment of these motors. This type of motor-based locomotion is likely to be quite widespread, because externally bound beads also are propelled along the sides of members of the Bacteroidetes phylum, although in those systems the molecular engine remains to be characterized (2). In addition, the existence of intracellular trafficking in bacteria opens up the exciting possibility that transport might be widely used to localize proteins for many other bacterial processes.
Materials and Methods
Bacterial Strains, Plasmids, and Growth.
Plasmids were introduced into M. xanthus by electroporation. Mutants and transformants were obtained by homologous recombination. Detailed construction schemes of the strains and plasmids and the sequences of all primers are shown in Tables S1–S3.
Measurement of the Effect of Drugs on Cell Gliding.
Drug-injection experiments with gliding cells were performed as previously described (22) on A+S− cells (ΔpilA) to ascertain that the drugs specifically affected gliding motility. Briefly, the injection experiments were conducted in a custom diffusion chamber where cells were immobilized on a thin layer of TPM agar and chemicals reached the cells by diffusion through the agar (22). Injections were performed by a coupled computerized injector system at a flow rate of 10 μL/s. Typically, CCCP (10 μM), valinomycin (40 μM), and nigericin (100 μM) were injected in TPM medium [10 mM Tris (pH 7.6), 8 mM MgSO4, 100 mM KH2PO4) containing 10 mM glucose. When effects were detected, reversibility was checked after the diffusion chamber was flushed with TPM-glucose.
Measurement of Membrane Potential, Intracellular ATP Level, and pH.
The effect of CCCP, valinomycin, and nigericin on membrane potential was measured with the standard lipophilic cation tetraphenylphosphonium (TPP+) as described previously (23). Details about the procedure are given in SI Materials and Methods.
Intracellular ATP levels were measured at different times after drug addition [100 μM nigericin, 10 μM CCCP, or 50 mM arsenate (47.5 mM sodium arsenate; 2.5 mM potassium arsenate, pH 8.0)] to 106 exponentially growing cells with a standard luminescence assay using luciferase ATP-dependent light emission and the ATP Bioluminescence Assay Kit HS II as described by the manufacturer (Roche Applied Bioscience). Bioluminescence expressed in arbitrary units was measured with an Infinite M200 microplate reader from Tecan.
The effects of metabolic poisons on intracellular pH were measured with the dye BCECF-AM (Molecular Probes), a standard pH fluorescent reporter probe (SI Materials and Methods).
Coimmunoprecipitation of the AglRQS Complex.
Procedures for the preparation of solubilized AglRQS complex, coimmunoprecipitation, and mass spectrometry analysis are provided in SI Materials and Methods.
Western Blotting.
Western blotting was performed as described previously (13) with 1/1,000 dilutions of anti-HA (Roche) or anti-mCherry (kind gift from V. Géli, Université d'Aix-Marseille, Marseille, France) antisera.
Imaging of Cell Gliding.
Time-lapse experiments of gliding motility were performed over TPM agar using an automated and inverted TE2000-E-PFS epifluorescence microscope (Nikon). Details can be found in SI Materials and Methods.
Optical Trap.
Our optical trap is built on a modified Nikon TE2000 inverted microscope with both differential interference contrast (DIC) and epifluorescence modules. A Nd:YVO4 laser (1,064 nm; Spectra Physics) is used to generate the trapping potential. For position detection, the scattering of an 855-nm diode laser (Bluesky Research) is detected by a position-sensitive detector (Newfocus). The trap and sample are steered using a closed-loop piezo-driven tip-tilt mirror and stage, respectively (Mad City Labs).
Surface Coating for Cell Immobilization.
A fluid tunnel slide was formed with a microscope slide and a clean glass coverslip separated by two layers of double-sided tape, and 20 μL agarose DMSO (Sigma Aldrich) solution (0.75% wt/vol) was injected into the fluid tunnel. Ten minutes later the tunnel was washed with 400 μL distilled water, and 20 μL of the overnight cell culture was injected into the tunnel. After 30 min, floating cells were flushed out with 400 μL TPM solution containing 10 mM glucose. For drug treatments, the corresponding drug solution was injected into the tunnel during an experiment.
Bead Preparation.
For all bead experiments, we used polystyrene beads 0.5 μm in diameter (Bangs Labs) diluted in TPM solution (0.01% wt/vol). To study the motion of beads on a cell surface, freely floating beads were trapped in solution and then were stuck gently on the top of immobilized cells.
Kymograph Analysis and Bead Tracking.
For kymograph analysis and bead tracking, images were taken every 10 s using the modified Nikon TE2000 inverted microscope with a 100×/1.49 oil immersion objective lens (Nikon) and a CCD camera (Andor Technology). A laser-based, 3D feedback method was used to overcome drift of the microscope focus during time-lapse imaging by monitoring the forward scattered light pattern of the 855-nm detection laser sent through a coverslip-bound polystyrene bead 0.5 μm in diameter (Bangs Labs). The output of the position-sensitive detector (PSD) was held constant by adjusting the position of the 3D closed-loop piezo-driven stage (Mad City Labs) using a modified proportional–integral–derivative (PID) algorithm. Custom software written in Matlab was used to construct the kymograph and to track bead motion (SI Materials and Methods).
Construction of the Speed and Fluorescence Intensity Histograms.
The time-dependent position of a bead was smoothed with a second-order Savitzky–Golay filter with a fixed window size of 25 s and differentiated to obtain the instantaneous velocity. The absolute value of the instantaneous velocity was used to construct the speed histogram. For fluorescence intensity histograms, fluorescence intensity was normalized by the average intensity from each cell. The area under the curve was normalized to 1 to create a normalized histogram.
All errors are SDs unless otherwise specified. For measured fractions, f, the SD is calculated using the binomial distribution 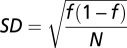 .
.
Supplementary Material
Acknowledgments
We thank Vincent Géli for the gift of the α-mCherry antiserum and Zhaomin Yang for pBJΔpilA. We also thank Thierry Doan, Romé Voulhoux, and Gérard Michel for advice on detergents and membrane preparations; Adrien Ducret for help with pH measurements; Adrien Ducret, Yi Deng, and Gordon Berman provided help with data processing and valuable discussions, and Adèle Sanràn-Cune for critical reading of the manuscript. We acknowledge the assistance of David H. Perlman in the Princeton Mass Spectrometry Center, which is supported by National Institutes of Health Grant P50GM071508. This work was funded by a joint Human Frontier Science Program Young Investigators Award (RGY0075/2008) (to T.M. and J.W.S.). M.W. is supported by a Ministere de l'Education Nationale de la Recherche et de Technologie thesis fellowship.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 7283.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101101108/-/DCSupplemental.
References
- 1.Mignot T, Shaevitz JW. Active and passive mechanisms of intracellular transport and localization in bacteria. Curr Opin Microbiol. 2008;11:580–585. doi: 10.1016/j.mib.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Jarrell KF, McBride MJ. The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol. 2008;6:466–476. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- 3.Mignot T, Kirby JR. Genetic circuitry controlling motility behaviors of Myxococcus xanthus. Bioessays. 2008;30:733–743. doi: 10.1002/bies.20790. [DOI] [PubMed] [Google Scholar]
- 4.Mignot T, Merlie JP, Jr, Zusman DR. Regulated pole-to-pole oscillations of a bacterial gliding motility protein. Science. 2005;310:855–857. doi: 10.1126/science.1119052. [DOI] [PubMed] [Google Scholar]
- 5.Mignot T. The elusive engine in Myxococcus xanthus gliding motility. Cell Mol Life Sci. 2007;64:2733–2745. doi: 10.1007/s00018-007-7176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leonardy S, Freymark G, Hebener S, Ellehauge E, Søgaard-Andersen L. Coupling of protein localization and cell movements by a dynamically localized response regulator in Myxococcus xanthus. EMBO J. 2007;26:4433–4444. doi: 10.1038/sj.emboj.7601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Franco M, Ducret A, Mignot T. A bacterial Ras-like small GTP-binding protein and its cognate GAP establish a dynamic spatial polarity axis to control directed motility. PLoS Biol. 2010;8:e1000430. doi: 10.1371/journal.pbio.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leonardy S, et al. Regulation of dynamic polarity switching in bacteria by a Ras-like G-protein and its cognate GAP. EMBO J. 2010;29:2276–2289. doi: 10.1038/emboj.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lapidus IR, Berg HC. Gliding motility of Cytophaga sp. strain U67. J Bacteriol. 1982;151:384–398. doi: 10.1128/jb.151.1.384-398.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nan B, et al. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc Natl Acad Sci USA. 2011;108:2498–2503. doi: 10.1073/pnas.1018556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolgemuth C, Hoiczyk E, Kaiser D, Oster G. How myxobacteria glide. Curr Biol. 2002;12:369–377. doi: 10.1016/s0960-9822(02)00716-9. [DOI] [PubMed] [Google Scholar]
- 12.Kaiser D. Are there lateral as well as polar engines for A-motile gliding in myxobacteria? J Bacteriol. 2009;191:5336–5341. doi: 10.1128/JB.00486-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mignot T, Shaevitz JW, Hartzell PL, Zusman DR. Evidence that focal adhesion complexes power bacterial gliding motility. Science. 2007;315:853–856. doi: 10.1126/science.1137223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pate JL, Chang L-YE. Evidence that gliding motility in procaryotic cells is driven by rotary assemblies in the cell envelopes. Curr Microbiol. 1979;2:59–64. [Google Scholar]
- 15.Pogliano J, Pogliano K, Weiss DS, Losick R, Beckwith J. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc Natl Acad Sci USA. 1997;94:559–564. doi: 10.1073/pnas.94.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mauriello EM, et al. Bacterial motility complexes require the actin-like protein, MreB and the Ras homologue, MglA. EMBO J. 2010;29:315–326. doi: 10.1038/emboj.2009.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Youderian P, Burke N, White DJ, Hartzell PL. Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner. Mol Microbiol. 2003;49:555–570. doi: 10.1046/j.1365-2958.2003.03582.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhou J, et al. Function of protonatable residues in the flagellar motor of Escherichia coli: A critical role for Asp 32 of MotB. J Bacteriol. 1998;180:2729–2735. doi: 10.1128/jb.180.10.2729-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nan BY, Mauriello EMF, Sun IH, Wong A, Zusman DR. A multi-protein complex from Myxococcus xanthus required for bacterial gliding motility. Mol Microbiol. 2010;76:1539–1554. doi: 10.1111/j.1365-2958.2010.07184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goemaere EL, Cascales E, Lloubès R. Mutational analyses define helix organization and key residues of a bacterial membrane energy-transducing complex. J Mol Biol. 2007;366:1424–1436. doi: 10.1016/j.jmb.2006.12.020. [DOI] [PubMed] [Google Scholar]
- 21.Mauriello EM, Nan B, Zusman DR. AglZ regulates adventurous (A-) motility in Myxococcus xanthus through its interaction with the cytoplasmic receptor, FrzCD. Mol Microbiol. 2009;72:964–977. doi: 10.1111/j.1365-2958.2009.06697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ducret A, et al. A microscope automated fluidic system to study bacterial processes in real time. PLoS ONE. 2009;4:e7282. doi: 10.1371/journal.pone.0007282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cascales E, Christie PJ. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc Natl Acad Sci USA. 2004;101:17228–17233. doi: 10.1073/pnas.0405843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.