Abstract
Growing evidence suggests that sensory neuron synapses not merely pass, but actively encode sensory information and convey it to the central nervous system. The chemosensory preferences of Caenorhabditis elegans, as manifested in the direction of chemotaxis, are reversibly regulated by prior experience at the level of sensory neurons; the attractive drive is promoted by diacylglycerol (DAG) signaling, whereas the counteracting repulsive drive requires PtdIns(3,4,5)P3 signaling. Here we report that the two opposing drives require a class IIA phosphatidylinositol transfer protein (PITP), PITP-1, which localizes to the sensory neuron synapses. In pitp-1 mutants, attraction behavior to salt is reduced, whereas conditioned repulsion from salt is eliminated: the mutants inflexibly show weak attraction behavior to salt, irrespective of prior experience. To generate flexible behavioral outputs, attraction and repulsion, PITP-1 acts in the gustatory neuron ASER and likely regulates neurotransmission from ASER, as pitp-1 mutations do not affect the ASER Ca2+ response to sensory stimulus. Furthermore, full attraction to salt is restored in pitp-1 mutants by expression of the phosphatidylinositol transfer domain alone, and also by mutations of a DGK gene that cause accumulation of DAG, suggesting that PITP-1 serves for DAG production via phosphatidylinositol transport and, hence, regulates synaptic transmission. In addition to gustatory behavior, olfactory behaviors and osmotic avoidance are also regulated by PITP-1 in the sensory neurons that detect each sensory stimulus. Thus, PITP-1–dependent phosphatidylinositol transport is essential for sensory neuron synapses to couple sensory inputs to effective behavioral responses.
Keywords: lipid transport, RdgB, PITPNM, PI3K pathway
The sensory information detected by the peripheral neurons is represented as a topographic neural map in the mammalian brain (1). In contrast, in the relatively simple nervous system of invertebrates devoid of extensive neural maps, much of the information processing occurs at the level of local sensory circuits, or in extreme cases, sensory neurons themselves (2–5). The fewer connections between sensory and motor neurons demand the processing at the sensory neuron level be more dominant. In the neural circuit of Aplysia gill-withdrawal reflex, for example, mechanosensory neurons innervating the siphon make direct connections with gill motorneurons. Presynaptic facilitation is caused at the synapses by aversive conditioning that induces short- and long-term memory (3).
Consisting of only 302 neurons, the nervous system of the nematode Caenorhabditis elegans is even more compact (6). Despite being quite compact, C. elegans displays a rich repertoire of sensory behaviors, and moreover, each behavior is plastic and changes with experience (7). A complete reversal in behavior is observed when worms are starved in the presence of NaCl, which is normally an attractive taste. Animals previously soaked in NaCl-containing buffer without food show avoidance behavior against NaCl instead of attraction (8). The switch of behavior between attraction and avoidance is reversible and is largely regulated at the level of the gustatory neuron ASER, wherein the switch requires two phospholipid signaling pathways: a diacylglycerol (DAG)/PKC pathway and a PI3K pathway (9, 10). High or low DAG/PKC signals seem to be associated with attraction or avoidance behaviors, respectively. Genetic or pharmacological activation of the DAG signaling pathway eliminates the plasticity of chemotaxis and causes constitutive attraction behavior to salt. Similarly, the plasticity is eliminated by mutations of the PI3K pathway, which likely transduces starvation signals. Genetic analysis suggests that the DAG/PKC pathway acts downstream of the PI3K pathway although the precise mechanism of the interaction is not clear. Another known regulator of lipid signals involved in the sensory response is TTX-7/IMPase (myo-inositol monophosphatase) (11). Loss of TTX-7 leads to thermotaxis defects because of inositol depletion, as well as defects in salt chemotaxis.
Despite the accumulating evidence for roles of phosphatidylinositol (PtdIns)-derived signals in the regulation of chemotaxis, little is known about where these signals arise and act within sensory neurons. The DAG signaling likely affects the synaptic output of sensory neurons by analogy to its role in neuromuscular junction (12, 13). It also interacts with the guanylyl cyclase GCY-28, which is localized to axons rather than dendritic sensory endings, in olfactory plasticity (4). In general, the localized generation of inositol lipids can be based on the local delivery of PtdIns, the precursor for all phosphoinositides. Phosphatidylinositol transfer proteins (PITPs) contain a hydrophobic cavity that can sequester PtdIns or phosphatidylcholine and transport the lipids to various membrane compartments from the endoplasmic reticulum, where these phospholipids are synthesized. Here we describe an essential role of PITP-1, the sole C. elegans ortholog of the conserved class IIA PITPs, in the regulation of salt chemotaxis. PITP-1 functions for attraction behavior to salt apparently by regulating neurotransmission from ASER, via synaptic transport of PtdIns, which is required for DAG production, and also functions for plasticity of salt chemotaxis possibly through production of PIP3. Our results suggest that phospholipids at sensory neuron synapses are important regulatory elements in the sensory circuits mediating salt chemotaxis.
Results
Reduced Attraction to NaCl and Loss of Chemotaxis Plasticity in pe1209 Mutants.
The single gustatory neuron ASER senses a number of chemicals and usually mediates attraction behavior to the sensory cues (14). NaCl is sensed mainly by ASER and normally induces attraction behavior, but elicits avoidance behavior after worms are exposed to NaCl under starvation conditions (designated as NaCl conditioning) (8). In these behaviors, ASER not only mediates sensation of salt in both attraction and avoidance, but also appears to be the site of plasticity (9). To explore the regulatory mechanism of the plasticity, we performed a genetic screen for mutants defective in the plasticity by insertional mutagenesis using the Drosophila transposon Mos1 (15) and isolated a mutant, pe1209. The pe1209 animals showed attraction behavior to salt even after NaCl conditioning (Fig. 1A). In addition, we observed reduction of attraction to salt in mock-conditioned pe1209 mutants compared with wild-type animals (Fig. 1A). Thus, the pe1209 animals showed a unique combination of two phenotypes, reduced attraction to salt, and eliminated plasticity after exposure to salt.
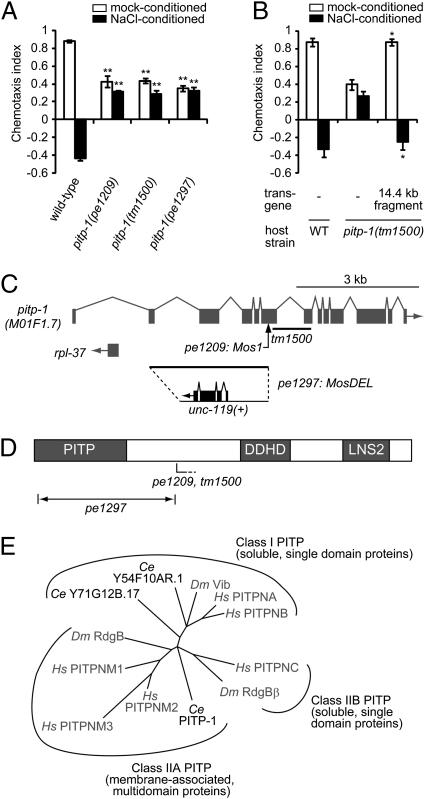
Fig. 1.
pitp-1 encodes a class IIA PITP. (A and B) Attraction behaviors to NaCl and chemotaxis plasticity in wild-type and pitp-1 mutant animals. Error bars represent SEM. (A) pitp-1 mutants show two phenotypes: decreased attraction to salt (mock-conditioned) and eliminated plasticity of chemotaxis (NaCl-conditioned). **P < 0.001 compared with wild-type, Dunnet test. (B) The two phenotypes of pitp-1(tm1500) mutants were fully rescued by introduction of a PCR-amplified 14.4-kb genomic fragment. *P < 0.01, t test. (C) Genomic organization of the pitp-1 locus. The Mos1 insertion in pe1209 is indicated by an arrow, and the tm1500 deletion is indicated by a line. In pe1297, the 2,951-bp region from second exon to the Mos1 insertion site in pe1209 was replaced by a 2.2-kb Caenorhabditis Briggsae unc-119(+) genomic DNA. (D) Schematic of the domain structures of PITP-1. The approximate positions of the mutations are indicated. PITP, DDHD domain (DDHD), and LNS2 domain (LNS2) are indicated. (E) Phylogenic analysis of full-length sequences of PITP orthologs. C. elegans PITPs are two class I PITPs (Y54F10AR.1 and Y71G12B.17) and a class IIA PITP (PITP-1). Other species are abbreviated as follows: Hs, Homo sapiens; Dm, Drosophila melanogaster. The dendrogram was generated with ClustalW and NJplot.
pe1209 Is an Allele of the Class IIA PITP, pitp-1.
By inverse PCR on genomic DNA from pe1209 animals and sequencing (15), we identified a Mos1 insertion into a previously uncharacterized gene M01F1.7, which encodes an ortholog of class IIA PITPs conserved among vertebrates and invertebrates (Fig. 1C) (16). We hereafter refer to this gene as pitp-1. A deletion mutation in the pitp-1 gene, pitp-1(tm1500), showed the two defects similar to those of pitp-1(pe1209) (Fig. 1A). The defects of attraction and plasticity in pitp-1(tm1500) were rescued by introduction of a PCR fragment corresponding to the genomic region of pitp-1 (Fig. 1B).
The PITPs consist of small soluble proteins containing a single PITP domain (class I and class IIB PITPs) and larger membrane-associated proteins (class IIA PITPs) (Fig. 1E) (16). Class IIA PITPs are multidomain proteins with an N-terminal PITP domain, a central DDHD domain implicated in metal binding, and a C-terminal LNS2 domain of unknown function (Fig. 1D) (16). Previous studies in Drosophila have shown that mutations in the class IIA PITP, retinal degeneration B (rdgB), cause light-dependent photoreceptor degeneration (17, 18). In the Drosophila photoreceptors, RdgB is thought to transfer PtdIns to the rhabdomere, the membrane-rich organelle specialized for phototransduction, where PtdIns(4,5)P2 is hydrolyzed by a light-activated phospholipase C (19, 20).
Because pe1209 and tm1500 mutations are predicted to cause premature stop codons caused by a transposon insertion and a frame shift, the two mutations probably cause truncated gene products containing the N-terminal PITP domain and might represent weak alleles. To obtain a null allele of pitp-1, we used MosDEL, a recently developed method to generate a targeted deletion in the C. elegans genome (21). The inserted Mos1 transposon in pitp-1(pe1209) unc-119(ed3) was remobilized to cause a double-strand break, resulting in deletion of the adjacent 3-kb sequence and simultaneous insertion of a wild-type copy of the unc-119 gene (Fig. 1C). Based on the nearly complete elimination of the PITP domain, the generated pitp-1(pe1297) is a candidate null allele. The pitp-1(pe1297) mutants showed behavioral phenotypes similar to the other two alleles, suggesting that they are also loss-of-function alleles (Fig. 1A). We mainly used pitp-1(tm1500) in the rest of this study.
Spatial and Temporal Requirement of PITP-1 in the Regulation of Salt Chemotaxis.
To determine the site of action of PITP-1, we examined the expression pattern of a Ppitp-1::venus transcriptional reporter. We observed weak fluorescence in many, but not all, neurons throughout the nervous system (Fig. 2 A and B). Coexpression with mCherry driven by various promoters confirmed that pitp-1 is expressed in sensory neurons ASE and AWC (Fig. 2 C–E and Fig. S1 A–C). We did not detect expression in ASH by using this reporter (Fig. S1 D–F). The widespread but specific expression pattern in the nervous tissue is analogous to the expression pattern of RdgB in the Drosophila brain (20).
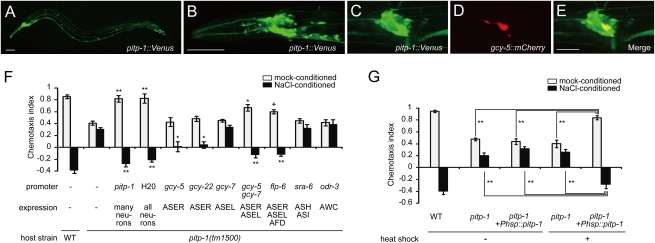
Fig. 2.
Spatial and temporal sites of PITP-1 action. (A and B) A transcriptional reporter of pitp-1, pitp-1::venus, is expressed in neurons. Whole body (A) and the head region (B). (Scale bars, 50 μm.) (C–E) Expression of pitp-1 in ASER. pitp-1::venus (C), ASER marker gcy-5::mCherry (D), and merged image (E). (Scale bar, 20 μm.) (F) Cell-specific rescue of pitp-1 mutants by cDNA expression. **P < 0.001, *P < 0.01, +P < 0.05, Dunnett test. (G) Defects of pitp-1(tm1500) animals were rescued by the temporal expression of pitp-1 by a heat-shock promoter at the adult stage. Error bars represent SEM. **P < 0.001, Tukey test.
Consistent with the neuronal expression pattern, the two behavioral defects of attraction and plasticity in pitp-1 mutants were fully restored by the pitp-1 cDNA expressed under the pan-neuronal H20 promoter (Fig. 2F). We next performed cell-specific rescue experiments of pitp-1 mutants, using neuron type-specific promoters (Fig. 2F). The attraction defect was partially rescued by expression of pitp-1 in both ASER and ASEL, but not by expression in either one alone. In contrast, the plasticity defect was partially rescued by expression in ASER alone. Whereas expression of pitp-1 solely in ASEL had no rescue activity, expression in both ASEL and ASER rescued the plasticity better than expression in ASER alone. We tested whether pitp-1 functions also in downstream interneurons AIA, AIB, AIY, and AIZ (Fig. S2). Expression of pitp-1 in the interneurons failed to rescue the defects of pitp-1 mutants (Fig. S2A), but seemed to enhance the rescue activity when combined with expression of pitp-1 in ASER (Fig. S2B). These results suggest that pitp-1 acts mainly in ASER, additionally in ASEL, and possibly in downstream interneurons.
To investigate whether PITP-1 functions in the mature nervous system, we performed rescue experiments by transient gene expression. Transient expression of pitp-1 cDNA at the adult stage using the heat-shock promoter fully rescued the behavioral defects of pitp-1 mutants (Fig. 2G). This result indicates that PITP-1 acts in the mature neural circuit. In Drosophila, the light-induced retinal degeneration results from mutations in rdgB (17, 18). In contrast, the overall morphology of the ASER neuron was normal, and there was no loss of ASE neurons in pitp-1 mutants (Fig. S3 A and B). These results, in conjunction with the acute rescue of the behavioral phenotypes, suggest that the neural circuit mediating salt chemotaxis does not undergo degeneration in pitp-1 mutants.
PITP-1 Is Required for Olfaction and Osmotic Avoidance.
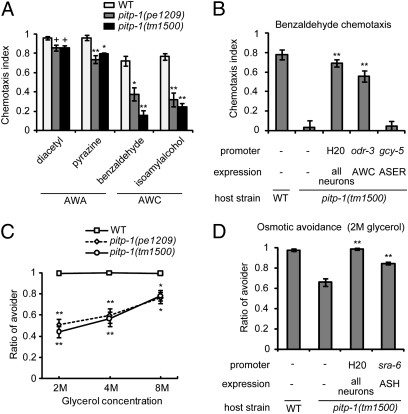
To examine functions of PITP-1 in other sensory modalities, pitp-1 mutants were subjected to behavioral assays for olfaction and osmosensation. The AWA and AWC olfactory neurons mediate attraction behaviors to various odorants (22). The pitp-1 mutants were defective for chemotaxis to AWC-sensed odorants (benzaldehyde and isoamyl alcohol) (Fig. 3A). They also showed minor defects in chemotaxis to AWA-sensed odorants (diacetyl and pyrazine) (Fig. 3A). The defect in benzaldehyde chemotaxis was rescued by expressing pitp-1 cDNA in AWC, but not in ASER (Fig. 3B). The pitp-1 mutants were also defective in osmotic avoidance, which is mediated by the ASH polymodal sensory neurons (Fig. 3C) (23, 24). The osmotic avoidance defect was partially rescued when pitp-1 cDNA was expressed in ASH (Fig. 3D). Thus, PITP-1 is essential for a variety of sensory behaviors. In contrast, locomotion and egg-laying appeared grossly normal in pitp-1 mutants, suggesting that abnormalities are largely confined to sensory functions.
Fig. 3.
pitp-1 mutants exhibit defects in olfaction and osmotic avoidance. (A) Chemotaxis to volatile attractants. (B) Chemotaxis to benzaldehyde was rescued by pitp-1 cDNA expression in AWC, but not in ASER. (C) Osmotic avoidance against high concentrations of glycerol. (D) Rescue of defective osmotic avoidance by cell-specific expression of pitp-1 in ASH. Error bars represent SEM. **P < 0.001, *P < 0.01, +P < 0.05, Dunnett test.
PITP-1 Localizes to the Presynaptic Regions.
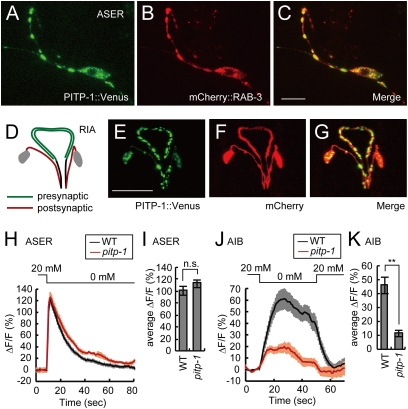
To determine the subcellular localization of PITP-1, a functional PITP-1::Venus fusion protein was specifically expressed in ASER (Fig. 4 A–C and Fig. S4O). PITP-1::Venus was mainly localized in a punctate pattern along the axon and in the cell body, and colocalized with the synaptic vesicle marker mCherry::RAB-3 in the axon, indicating that PITP-1::Venus is localized to presynaptic sites. A few inconsistent puncta were often observed along the dendrites (Fig. S4F), but this might be mislocalization because of overexpression of the transgene. In addition to the presynaptic localizations, we observed PITP-1::Venus puncta consistently at the ciliary transition zone (Fig. S4 A–E). UNC-104 (KIF1A) is essential for trafficking synaptic vesicles from the cell body to the presynaptic sites in C. elegans (25). In unc-104 mutants, PITP-1::Venus localization decreased in the axon and increased in the dendrite and the cell body (Fig. S4 I–K). We additionally examined PITP-1::Venus localization in the RIA interneurons, which have a neurite clearly divided into pre- and postsynaptic regions (Fig. 4D) (6). PITP-1::Venus was enriched in the distal presynaptic regions of the RIA neurites (Fig. 4 E–G). In summary, PITP-1::Venus is mainly localized to the presynaptic sites, where its activity for PtdIns transport may be required.
Fig. 4.
PITP-1 localizes to presynaptic regions and is required for interneuron response. (A–C) Subcellular localization of PITP-1 in the ASER neuron. ASER cell body and axon is shown. PITP-1::Venus translational fusion (A), mCherry::RAB-3 (B), and merged image (C). (Scale bar, 10 μm.) (D) Schematic diagram of the RIA interneurons. RIA neurite is polarized; the proximal region is postsynaptic (red) and the distal region is presynaptic (green). (E–G) Presynaptic localization of PITP-1::Venus in RIA neurons. The glr-3 promoter was used to induce expression in RIA. PITP-1::Venus translational fusion (E), mCherry (F), and merged image (G). (Scale bar, 20 μm.) (H and J) Average Ca2+ responses in the ASER sensory neurons (H) and the AIB interneurons (J) of wild-type (n = 19 for ASER, 20 for AIB) and pitp-1(tm1500) mutants (n = 21 for ASER and AIB) expressing G-CaMP in each neuron type to changes in NaCl concentration shown at the top of each response curve. The shaded area around each trace represents SEM. (I and K) Average ASER (I) and AIB (K) fluorescence change in the 9.6-s and 40-s window after NaCl downstep, respectively, in wild-type and pitp-1(tm1500) animals. Error bars represent SEM. **P < 0.001, n.s. not significant, t test.
pitp-1 Mutations Reduce the Ca2+ Response in AIB Interneurons Without Affecting the ASER Response.
RdgB is essential for sensory transduction of light in Drosophila photoreceptors, where phototransduction is mediated by hydrolysis of PtdIns(4,5)P2 by a phospholipase C (19). Although the sensory transduction in ASER is mediated by guanylyl cyclases and cGMP-gated channels (26, 27) and does not apparently require phospholipids, the reduced chemoattraction of pitp-1 mutants and the localization of PITP-1 at the base of cilium suggested a possible role for PITP-1 in sensory transduction. To investigate this possibility, we asked whether sensory transduction is perturbed in pitp-1 mutants by Ca2+ imaging of ASER. To monitor Ca2+ responses in ASER, we used G-CaMP, a genetically encoded Ca2+ sensor that increases fluorescence intensity in response to intracellular Ca2+ elevation (28, 29). As previously reported (30), upstep and downstep of external NaCl concentration lead to reduction and elevation of Ca2+ levels, respectively, in the wild-type ASER neuron (Fig. 4H and Fig. S5A). Unlike the case in Drosophila photoreceptors, pitp-1 showed similar ASER responses to wild-type across a range of NaCl concentrations examined (Fig. 4 H and I and Fig. S5 A and C). Ca2+ responses of ASER in pitp-1 mutants were also similar to wild-type animals after NaCl conditioning (Fig. S5 B and C). In addition, we observed that the ASH response to high osmolarity glycerol was mostly normal in pitp-1 mutants (Fig. S5D). We further examined the cilia morphology of ASER and olfactory neurons AWC, but found no difference between wild-type animals and pitp-1 mutants (Fig. S3 C–L). These results suggest that PITP-1 is not required for primary sensory transduction in ASE and that the defects of attraction and plasticity are likely caused at downstream processes, such as neurotransmission. To investigate this possibility, we monitored NaCl downstep-evoked Ca2+ responses in AIB interneurons, which receive direct synaptic connections from ASER (6). Wild-type AIB neurons were activated by NaCl downstep and maintained the elevated Ca2+ levels for more than half a minute (Fig. 4J). In contrast, pitp-1 mutants showed a reduced increase in AIB Ca2+ levels, consistent with the hypothesis that synaptic transmission from ASER is compromised in pitp-1 mutants (Fig. 4 J and K).
Presynaptic Function of PITP-1.
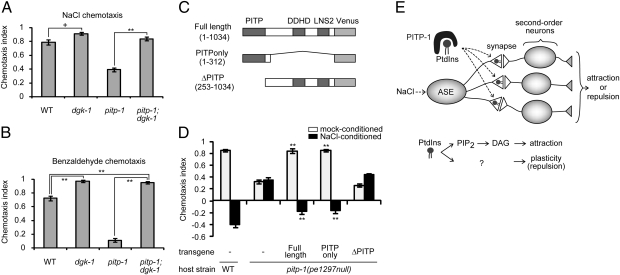
We next investigated the role of PITP-1 in presynaptic sites. The localizations of PITP-1 to presynaptic regions suggested a possible role in transport of PtdIns to synaptic membranes (Fig. 4 A–C). Based on this assumption, loss of PITP-1 function may lead to decreased availability of PtdIns. In presynapses, PtdIns(4,5)P2 is hydrolyzed to produce DAG (31), which facilitates cholinergic neurotransmission at the C. elegans neuromuscular junction (32, 33). In ASER and AWC, DAG signaling promotes attraction to salt and odors respectively, possibly by enhancing synaptic release from the sensory neurons (10, 12, 34). Thus, the reduction of attraction to salt and odors in pitp-1 mutants can be caused in part by a reduction of synaptic DAG signaling. To test this possibility, we examined whether a mutation of dgk-1 suppresses the pitp-1 chemotaxis phenotypes: dgk-1 encodes a DAG kinase (DGK) that reduces DAG levels by phosphorylating DAG to phophatidic acid (33, 35). Loss-of-function mutations in dgk-1 lead to enhanced neurotransmission at the neuromuscular junction through elevation of DAG levels (33, 35). A loss-of-function mutation in dgk-1 nearly completely suppressed pitp-1 defects of attraction to both salt and benzaldehyde (Fig. 5 A and B), suggesting that synaptic DAG signaling acts downstream of or in parallel to PITP-1 to promote attraction behaviors. These results suggest a role for PITP-1 in PtdIns transport to synaptic membranes for DAG production (Fig. 5E).
Fig. 5.
pitp-1 and DAG signaling interact downstream of sensory transduction.(A and B) Effect of dgk-1(sy428) mutation on chemotaxis to salt (A) and benzaldehyde (B), under pitp-1(tm1500) mutant background. Error bars represent SEM. **P < 0.001, +P < 0.05, Tukey test. (C and D) Structure-function analysis of PITP-1. Schematic representation of pitp-1 deletion constructs tagged with Venus (C) and the results of rescue experiments (D). The pitp-1 promoter was used to express the deletion constructs of pitp-1. Error bars represent SEM. **P < 0.001, Dunnett test. (E) Model for PITP-1 function in ASER. PITP-1 transports PtdIns to the presynaptic membrane and promotes salt attraction via DAG production and plasticity of salt chemotaxis through unknown signals derived from PtdIns.
In contrast to the reduced attraction phenotypes, the pitp-1 defect in plasticity of salt chemotaxis was not suppressed by dgk-1 mutations (Fig. S6A). Similarly, the pitp-1;dgk-1 double-mutants showed a strong defect in olfactory adaptation to benzaldehyde (Fig. S6B), where preexposure to benzaldehyde elicits aversive responses to the odor in wild-type animals (34, 36). These observations imply the requirements for other PtdIns derivatives, such as PtdIns(4,5)P2, PtdIns(3,4,5)P3, or Ins(1,4,5)P3, in the plasticity of chemotaxis.
PITP Domain Is Sufficient for the Regulation of Salt Chemotaxis.
PITP-1 is a multidomain protein with a PITP domain at the N terminus and other less well-defined domains. Using the putative null allele, pitp-1(pe1297), we found that the N-terminal fragment containing the PITP domain alone could rescue the behavioral phenotypes of pitp-1 mutants, whereas the fragment that lacked the PITP domain did not rescue the defects (Fig. 5D). These results are consistent with the idea that PITP-1 regulates behavior by transporting PtdIns. Consistent with the proposed actions in presynaptic sites, the N-terminal fragment that rescued behavioral defects showed subcellular localizations virtually identical to the full-length PITP-1 (Fig. S4 L–N).
Discussion
Regulation of Chemosensory Preferences at the Sensory Neuron Synapses.
The pitp-1 mutations reduced salt-attraction behavior and caused a concomitant decrease in the NaCl downstep-evoked Ca2+ response in AIB interneurons without affecting the response in ASE sensory neurons. On the other hand, salt-attraction defect was rescued by the expression of pitp-1 in ASE neurons, suggesting its function for the output of the sensory neurons, an idea consistent with synaptic localization of PITP-1 protein. Activation of DAG signals restored attraction behavior in pitp-1 mutants, leading us to propose that PITP-1 has a role in transport of PtdIns for DAG production, which ensures efficient neurotransmission from the ASE presynapses and thereby supports full attraction behavior (Fig. 5E).
The salt-repulsion behavior also seems to be regulated at the ASE presynapses by PITP-1. As prior sensory experience of salt is required to cause plasticity of salt chemotaxis (8), reduced sensory signaling can lead to impaired plasticity. However, sensory transduction process is not affected in pitp-1 mutants with respect to Ca2+ responses in the ASER cell body in both naive and NaCl-conditioned states. Therefore, we infer that phospholipids produced from PtdIns may directly contribute to the plasticity in the axon (Fig. 5E). In addition, PITP-1 may regulate plasticity through PtdIns-derived signals other than DAG, as dgk-1 mutations did not suppress the plasticity defects. It was previously shown that a PI3K signaling is required for chemotaxis plasticity, and genetic epistasis analysis suggests that the PI3K signaling may negatively regulate DAG signals (10). Thus, reduced availability of presynaptic PtdIns may lead to decreased PtdIns(3,4,5)P3 production in addition to reduced DAG production; the combined result could be a low level of DAG production that is not further down-regulated during NaCl conditioning because of impaired production of PtdIns(3,4,5)P3. In summary, we propose that the chemotaxis switch between attraction and repulsion is regulated by PITP-1 and multiple PtdIns-derived signals at the ASE presynapses.
The presynapses of sensory neurons are suggested to be the regulatory sites for chemosensory preferences in this study and previous studies (4, 10). Mutations in gcy-28 and daf-18 lead to avoidance behaviors in place of the attractive behaviors normally directed by the AWCON olfactory neuron and the ASER neuron, respectively (4, 10). These behavioral changes are rescued by activation of DAG signals in sensory neurons (4, 10). As an analogy to the neuromuscular junction, the effects of enhanced DAG signals are attributed to facilitation of neurotransmission, leading to the hypothesis that presynaptic changes are important for the regulation of chemosensory preferences. This notion is further supported by the identification of PITP-1 as a presynaptic component required for chemotaxis plasticity.
Then what is the nature of the presynaptic changes underlying the behavioral switch from attraction to repulsion? ASE sensory neurons synapse onto several classes of interneurons and other sensory neurons (6). Although speculative at present, a subset of these synapses seem to be inactivated in the course of NaCl conditioning because the switch is blocked by activating DAG/PKC signals in ASE, probably through enhancing synaptic transmission from ASE (9). Selectively silencing certain synapses but leaving other synapses active in a single neuron may require a mechanism for synapse-specific regulation (Fig. S6 C and D), which to our knowledge has not been demonstrated in this organism. The punctate localizations of PITP-1 in the presynaptic regions may imply the independent pool of PtdIns in each synapse, and each pool may undergo synapse-specific regulation of DAG and PtdIns(3,4,5)P3 production. Alternatively, the presynaptic changes may occur at the level of the axon as a whole and synaptic transmission at all of the synapses of ASE can be equally reduced (Fig. S6E). Even in this case, it is possible that a subset of interneurons selectively become unresponsive, assuming varying sensitivities of interneuron classes to the synaptic input from ASE. The precise sets of interneurons mediating the switched behaviors and the mechanism to differentially activate them are issues for future studies.
Synaptic Roles of PITPs in Neuronal and Behavioral Plasticity.
The requirement of PITP-1 for DAG production parallels the role of RdgB in providing PtdIns for production of second-messenger DAG in Drosophila photoreceptors (19). In mammalian cells, PITPNM1 is mainly localized to the Golgi and regulates DAG levels at Golgi membranes (37). Furthermore, the immunolocalization studies in Drosophila detected expression of RdgB in the lamina and medulla, the target neuropiles of photoreceptor axons (20), suggesting that RdgB may localize to the presynaptic sites of photoreceptor neurons. Thus, the presynaptic localization of PITP-1 and its role in supporting synaptic production of DAG may be conserved through evolution.
The C. elegans gustatory neuron ASER can be a suitable model to elucidate the synaptic function of PITPs. The rdgB flies have defective light responses and photoreceptors that degenerate (19). In contrast, ASER does not require PITP-1 for the sensory response. We reason that this difference is caused by the requirement of inositol lipids in sensory transduction: the phototransduction in fly photoreceptors is mediated by hydrolysis of PtdIns(4,5)P2 by a phospholipase C (19), whereas the sensory transduction in ASER is mediated by guanylyl cyclases and cGMP-gated channels (26, 27) and does not require inositol lipids.
The expression of pitp-1 is not limited to ciliated sensory neurons, but widely distributed in the nervous system. Consistent with our observations, RdgB is expressed not only in sensory neurons of visual and olfactory organs, but also in integration centers of the head, such as mushroom bodies, which mediate olfactory memory in flies (20). One of the mammalian class IIA proteins PITPNM2 is expressed in hippocampus in addition to retina (38), whereas another ortholog PITPNM1 is widely expressed in the brain (www.brain-map.org). Thus, orthologs in flies and mammals are expressed in brain regions involved in memory formation. It will be interesting to ask whether these orthologs have a role in learning and memory. There are two genes encoding class I PITPs (Y71G12B.17 and Y54F10AR.1) in the C. elegans genome (www.wormbase.org), and these proteins may act cooperatively with PITP-1 for cellular distribution of PtdIns. Interestingly, the mammalian class I protein PITPNA is localized to presynaptic terminals of differentiated cultured hippocampal neurons (39). Thus, further analysis including these genes will be necessary to explore the importance of synaptic regulation of phospholipid signaling in behavioral plasticity.
Materials and Methods
Strains and Culture.
C. elegans strains were cultivated at 20 °C under standard conditions (40) except that the Escherichia coli strain NA22 was used as a food source in most experiments unless otherwise noted. Strains used were wild-type Bristol N2, EG1642 lin-15(n765) X; oxEx166, EG1470 oxEx229, pitp-1(pe1209) III, pitp-1(tm1500) III, pitp-1(pe1297) III, and dgk-1(sy428) X.
Behavioral Assays.
Assays of chemotaxis to NaCl was performed as previously described (10). Briefly, a gradient of NaCl was created on an assay plate [9-cm diameter Petri dish poured with 10 mL of 2% agar, 5 mM potassium phosphate (pH 6.0), 1 mM CaCl2, 1 mM MgSO4] by placing an 0.13 mL of agar plug, which contained 100 mM NaCl at 1.5 cm from the edge of the plate 18 to 24 h before assay. To examine plasticity of chemotaxis to NaCl, washed animals were incubated with 1 mL of assay buffer [5 mM potassium phosphate (pH 6.0), 1 mM CaCl2, 1 mM MgSO4, 0.05% gelatin] with (NaCl-conditioned) or without (mock-conditioned) 20 mM NaCl for 1 h. One microliter each of 0.5 M NaN3 was spotted to the peak of NaCl gradient and to the opposite position of the plate just before placing animals. Fifty to 200 animals were placed at the center of the assay plate and allowed to run for 30 min. A chemotaxis index was calculated as [(number of animals within a 2-cm radius from the NaCl peak) − (number of animals within a 2-cm radius of control spot)]/[(total number of animals) − (number of animals near the center of the plate)].
For each datapoint, at least five trials were independently performed and the results were statistically validated as indicated in the figure legends.
Genetic screens and identification of pitp-1(pe1209), targeted gene deletion by MosDEL, other behavioral assays, heat-shock experiments, in vivo Ca2+ imaging, plasmid constructions, and germ-line transformation are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the National Bioresource Project for the Nematode for the pitp-1(tm1500) strain; all other nematode strains, except pe1209 and pe1297, used in this study were provided by the Caenorhabditis Genetics Center. We also thank N. Chronis for design of olfactory chip, C. I. Bargmann for AIBp::G-CaMP and ASHp::G-CaMP, J. Nakai for GCaMP2 cDNA, S. Tokuoka for useful discussion, T. Karashima for constructs and technical advice, and T. Adachi and K. Yamada for comments on the manuscript. This work was supported by Global Center of Excellence Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms) and Grant-in-Aid for Scientific Research (B) and on Innovative Areas (Systems Molecular Ethology), Ministry of Education, Culture, Sports, Science, and Technology, Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016232108/-/DCSupplemental.
References
- 1.Luo L, Flanagan JG. Development of continuous and discrete neural maps. Neuron. 2007;56:284–300. doi: 10.1016/j.neuron.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 2.Barth M, Schultze M, Schuster CM, Strauss R. Circadian plasticity in photoreceptor cells controls visual coding efficiency in Drosophila melanogaster. PLoS ONE. 2010;5:e9217. doi: 10.1371/journal.pone.0009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kandel ER. The molecular biology of memory storage: A dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 4.Tsunozaki M, Chalasani SH, Bargmann CI. A behavioral switch: cGMP and PKC signaling in olfactory neurons reverses odor preference in C. elegans. Neuron. 2008;59:959–971. doi: 10.1016/j.neuron.2008.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singer JH, et al. Functional properties of synaptic transmission in primary sense organs. J Neurosci. 2009;29:12802–12806. doi: 10.1523/JNEUROSCI.3346-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous-system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 7.de Bono M, Maricq AV. Neuronal substrates of complex behaviors in C. elegans. Annu Rev Neurosci. 2005;28:451–501. doi: 10.1146/annurev.neuro.27.070203.144259. [DOI] [PubMed] [Google Scholar]
- 8.Saeki S, Yamamoto M, Iino Y. Plasticity of chemotaxis revealed by paired presentation of a chemoattractant and starvation in the nematode Caenorhabditis elegans. J Exp Biol. 2001;204:1757–1764. doi: 10.1242/jeb.204.10.1757. [DOI] [PubMed] [Google Scholar]
- 9.Adachi T, et al. Reversal of salt preference is directed by the insulin/PI3K and Gq/PKC signaling in Caenorhabditis elegans. Genetics. 2010;186:1309–1319. doi: 10.1534/genetics.110.119768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomioka M, et al. The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron. 2006;51:613–625. doi: 10.1016/j.neuron.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 11.Tanizawa Y, et al. Inositol monophosphatase regulates localization of synaptic components and behavior in the mature nervous system of C. elegans. Genes Dev. 2006;20:3296–3310. doi: 10.1101/gad.1497806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okochi Y, Kimura KD, Ohta A, Mori I. Diverse regulation of sensory signaling by C. elegans nPKC-epsilon/eta TTX-4. EMBO J. 2005;24:2127–2137. doi: 10.1038/sj.emboj.7600697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biron D, et al. A diacylglycerol kinase modulates long-term thermotactic behavioral plasticity in C. elegans. Nat Neurosci. 2006;9:1499–1505. doi: 10.1038/nn1796. [DOI] [PubMed] [Google Scholar]
- 14.Bargmann CI, Horvitz HR. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991;7:729–742. doi: 10.1016/0896-6273(91)90276-6. [DOI] [PubMed] [Google Scholar]
- 15.Bessereau JL, et al. Mobilization of a Drosophila transposon in the Caenorhabditis elegans germ line. Nature. 2001;413:70–74. doi: 10.1038/35092567. [DOI] [PubMed] [Google Scholar]
- 16.Cockcroft S, Carvou N. Biochemical and biological functions of class I phosphatidylinositol transfer proteins. Biochim Biophys Acta. 2007;1771:677–691. doi: 10.1016/j.bbalip.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 17.Harris WA, Stark WS. Hereditary retinal degeneration in Drosophila melanogaster. A mutant defect associated with the phototransduction process. J Gen Physiol. 1977;69:261–291. doi: 10.1085/jgp.69.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hotta Y, Benzer S. Genetic dissection of the Drosophila nervous system by means of mosaics. Proc Natl Acad Sci USA. 1970;67:1156–1163. doi: 10.1073/pnas.67.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trivedi D, Padinjat R. RdgB proteins: Functions in lipid homeostasis and signal transduction. Biochim Biophys Acta. 2007;1771:692–699. doi: 10.1016/j.bbalip.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 20.Vihtelic TS, Goebl M, Milligan S, O'Tousa JE, Hyde DR. Localization of Drosophila retinal degeneration B, a membrane-associated phosphatidylinositol transfer protein. J Cell Biol. 1993;122:1013–1022. doi: 10.1083/jcb.122.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frøkjaer-Jensen C, et al. Targeted gene deletions in C. elegans using transposon excision. Nat Methods. 2010;7:451–453. doi: 10.1038/nmeth.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- 23.Maricq AV, Peckol E, Driscoll M, Bargmann CI. Mechanosensory signalling in C. elegans mediated by the GLR-1 glutamate receptor. Nature. 1995;378:78–81. doi: 10.1038/378078a0. [DOI] [PubMed] [Google Scholar]
- 24.Hart AC, Sims S, Kaplan JM. Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature. 1995;378:82–85. doi: 10.1038/378082a0. [DOI] [PubMed] [Google Scholar]
- 25.Hall DH, Hedgecock EM. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell. 1991;65:837–847. doi: 10.1016/0092-8674(91)90391-b. [DOI] [PubMed] [Google Scholar]
- 26.Coburn CM, Bargmann CI. A putative cyclic nucleotide-gated channel is required for sensory development and function in C. elegans. Neuron. 1996;17:695–706. doi: 10.1016/s0896-6273(00)80201-9. [DOI] [PubMed] [Google Scholar]
- 27.Komatsu H, Mori I, Rhee JS, Akaike N, Ohshima Y. Mutations in a cyclic nucleotide-gated channel lead to abnormal thermosensation and chemosensation in C. elegans. Neuron. 1996;17:707–718. doi: 10.1016/s0896-6273(00)80202-0. [DOI] [PubMed] [Google Scholar]
- 28.Nakai J, Ohkura M, Imoto K. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat Biotechnol. 2001;19:137–141. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- 29.Chalasani SH, et al. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature. 2007;450:63–70. doi: 10.1038/nature06292. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki H, et al. Functional asymmetry in Caenorhabditis elegans taste neurons and its computational role in chemotaxis. Nature. 2008;454:114–117. doi: 10.1038/nature06927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammond GR, Schiavo G. Polyphosphoinositol lipids: Under-PPInning synaptic function in health and disease. Dev Neurobiol. 2007;67:1232–1247. doi: 10.1002/dneu.20509. [DOI] [PubMed] [Google Scholar]
- 32.Lackner MR, Nurrish SJ, Kaplan JM. Facilitation of synaptic transmission by EGL-30 Gqalpha and EGL-8 PLCbeta: DAG binding to UNC-13 is required to stimulate acetylcholine release. Neuron. 1999;24:335–346. doi: 10.1016/s0896-6273(00)80848-x. [DOI] [PubMed] [Google Scholar]
- 33.Miller KG, Emerson MD, Rand JB. Goalpha and diacylglycerol kinase negatively regulate the Gqalpha pathway in C. elegans. Neuron. 1999;24:323–333. doi: 10.1016/s0896-6273(00)80847-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuki M, Kunitomo H, Iino Y. Goalpha regulates olfactory adaptation by antagonizing Gqalpha-DAG signaling in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2006;103:1112–1117. doi: 10.1073/pnas.0506954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nurrish S, Ségalat L, Kaplan JM. Serotonin inhibition of synaptic transmission: Galpha(0) decreases the abundance of UNC-13 at release sites. Neuron. 1999;24:231–242. doi: 10.1016/s0896-6273(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 36.Colbert HA, Bargmann CI. Odorant-specific adaptation pathways generate olfactory plasticity in C. elegans. Neuron. 1995;14:803–812. doi: 10.1016/0896-6273(95)90224-4. [DOI] [PubMed] [Google Scholar]
- 37.Litvak V, Dahan N, Ramachandran S, Sabanay H, Lev S. Maintenance of the diacylglycerol level in the Golgi apparatus by the Nir2 protein is critical for Golgi secretory function. Nat Cell Biol. 2005;7:225–234. doi: 10.1038/ncb1221. [DOI] [PubMed] [Google Scholar]
- 38.Lu C, Vihtelic TS, Hyde DR, Li T. A neuronal-specific mammalian homolog of the Drosophila retinal degeneration B gene with expression restricted to the retina and dentate gyrus. J Neurosci. 1999;19:7317–7325. doi: 10.1523/JNEUROSCI.19-17-07317.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cosker KE, et al. Regulation of PI3K signalling by the phosphatidylinositol transfer protein PITPalpha during axonal extension in hippocampal neurons. J Cell Sci. 2008;121:796–803. doi: 10.1242/jcs.019166. [DOI] [PubMed] [Google Scholar]
- 40.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.