Abstract
It is well-documented that the methylation of histone H3 lysine 4 (H3K4) and of H3K9 are mutually exclusive, an epigenetic phenomenon conserved from yeast to humans. How this opposed methylation modification is accomplished and coordinated in mammalian cells is poorly understood. Here we report that the H3K9 trimethyl demethylase JMJD2B is an integral component of the H3K4-specific methyltransferase, the mixed-lineage leukemia (MLL) 2 complex. We show that the JMJD2B/MLL2 complex is copurified with estrogen receptor α (ERα) and is required for ERα-regulated transcription. We demonstrate that H3K9 demethylation and H3K4 methylation are coordinated in ERα-activated transcription such that H3K9 demethylation is a prerequisite for H3K4 methylation. Significantly, depletion of JMJD2B impairs the estrogen-induced G1/S transition of the cell cycle in vitro and inhibits breast tumorigenesis in vivo. Interestingly, JMJD2B itself is an ERα target gene, and forms a feed-forward regulatory loop in regulation of the hormone response. Our results provide a molecular basis for the coordinated H3K4 methylation/H3K9 demethylation in transcription activation, link the trimethyl demethylase JMJD2B to euchromatin functions, and provide a mechanism for JMJD2B in breast carcinogenesis.
Keywords: histone methylation, breast cancer
Recent studies indicate that, analogous to other covalent histone modifications such as acetylation, histone methylation is reversible. However, in contrast to histone acetylation, which is generally associated with active transcription, histone methylation can be associated with either activation or repression of transcription, depending on what effector protein is recruited (1). Even within the same lysine residue, the biological consequences of methylation seem to be variable. For example, methylation of histone H3 lysine 9 (H3K9), long considered a hallmark of heterochromatin (2–8), was recently found to also be present at the transcribed regions of active mammalian genes (9, 10), suggesting that certain methyl marks can have multiple functions in the cell.
Given the complexity of histone methylation modification, the integration and/or coordination of methylation/demethylation in a particular biological process becomes an issue of great importance in further understanding the role of histone methylation in nucleosome functioning. Specifically, actively transcribed genes, including the ones that are regulated by estrogen receptor (ER) (11–13), are marked by methylation at histone H3K4, but at the same time by demethylation at H3K9; H3K4 and H3K9 methylation levels are mutually exclusive, and this relationship is conserved from fission yeast to humans. How opposed H3K4 methylation and H3K9 demethylation are achieved and coordinated in transcription activation in mammalian cells is not fully understood.
H3K4 methylation is deposited by a family of histone methyltransferases (HMT) that share a conserved SET (Su(var), E(z), and Trithorax) domain. In mammalian cells, multiple HMTs have been characterized: SET1A and SET1B (the mammalian orthologs of yeast Set1), five mixed-lineage leukemia (MLL)-family HMTs (MLL1–5), SMYD3, and SET7/9 (14). The increased complexity of the enzymatic machineries suggests that H3K4 methylation is biologically important and subject to sophisticated controls in higher eukaryotes. Indeed, many H3K4 methyltransferases in mammalian cells are essential genes (15–17).
The demethylation state of H3K9 is believed to be maintained by several members of the JmjC domain family of demethylases. Specifically, it has been reported that, in euchromatin, JHDM2A catalyzes the removal of mono- and dimethylation at H3K9 (H3K9me1/2) (18) and JMJD2C acts to erase trimethylation at H3K9 (H3K9me3) (19), whereas JMJD2B has been shown to demethylate H3K9me3 at pericentric heterochromatin in mammalian cells (20, 21). A role for JMJD2B in euchromatin function has not yet been defined.
Here we report that the H3K9 trimethyl demethylase JMJD2B is physically associated with and an integral component of the H3K4 methyltransferase MLL2 complex. We demonstrate that the JMJD2B/MLL2 complex interacts with ERα and is required for the cellular function of ERα. We show that JMJD2B and the MLL2 complex interact to define the methylation status of H3K4 and H3K9 in ERα-activated transcription, in which H3K9 demethylation precedes H3K4 methylation. In addition, we find that JMJD2B itself is transcriptionally targeted by ERα and may thus form a feed-forward regulatory loop in promoting hormonally responsive breast carcinogenesis.
Results and Discussion
JMJD2B Is Physically Associated with the MLL2 Complex.
To further understand the biological activity of JMJD2B and to investigate its role, if any, in euchromatin functions, we generated a mammary carcinoma MCF-7 cell line that stably expresses FLAG-tagged JMJD2B (FLAG-JMJD2B). Immunopurification of JMJD2B from MCF-7 cell extracts with anti-FLAG and analysis of the JMJD2B-containing protein complex by mass spectrometry revealed that JMJD2B is associated with multiple components of the MLL2 complex (MLL2, ASH2L, RbBP5, and WDR5) as well as with several other proteins (hnRNP U, HSP70, and ribosomal protein S9). Interestingly, four peptides matching ERα were also identified in the JMJD2B-containing protein complex (Fig. 1A and SI Appendix, Supplemental File 1).
Fig. 1.
Physical interaction between JMJD2B and the MLL2 complex. (A) Immunoaffinity purification of JMJD2B-containing protein complexes. Cellular extracts from MCF-7 cells stably expressing FLAG or FLAG-JMJD2B were immunopurified with anti-FLAG-affinity beads and eluted with FLAG peptides. The eluates were resolved on SDS/PAGE and silver-stained. The protein bands were retrieved and analyzed by mass spectrometry. Detailed results from the mass spectrometric analysis are provided in SI Appendix, Supplemental File 1. (B) Association of JMJD2B with the MLL2 complex. Whole-cell lysates from MCF-7 cells were immunoprecipitated with antibodies against the indicated proteins. Immunocomplexes were then immunoblotted using antibodies against the indicated proteins. (C) Cofractionation of protein complexes by FPLC. Nuclear extracts from MCF-7 cells were fractionated on Superose 6 size-exclusion columns. Chromatographic elution profiles and immunoblotting analysis of the chromatographic fractions are shown. The elution positions of calibration proteins with known molecular masses are indicated, and an equal volume from each fraction was analyzed. The immunoprecipitates were incubated with in vitro recombinant nucleosomes in histone methylation assay buffer or incubated with in vivo isolated native mononucleosomes in histone demethylation assay buffer. The reaction mixtures were analyzed by Western blotting using antibodies against the indicated histone marks or proteins. (D) Silver staining and Western blot analysis of the JMJD2B-containing complex fractionated by Superose 6 gel filtration. Chromatographic elution profiles and immunoblotting analysis of the chromatographic fractions are shown. The elution positions of calibration proteins with known molecular masses are indicated, and an equal volume from each fraction was analyzed.
To confirm the in vivo association between JMJD2B and the MLL2 complex, coimmunoprecipitation experiments were performed with MCF-7 cell extracts. Immunoprecipitation with antibodies against JMJD2B followed by immunoblotting with antibodies against WDR5, RbBP5, ASH2L, or MLL2 demonstrated that JMJD2B was coimmunoprecipitated with all of these components of the MLL2 complex, but not with JARID1B (PLU-1), another member of the JmjC domain-containing demethylase, although JARID1B could be effectively immunoprecipitated with antibodies against HDAC1 (22) (Fig. 1B Left). Reciprocally, immunoprecipitation with antibodies against the components of the MLL2 complex and immunoblotting with antibodies against JMJD2B also revealed that WDR5, RbBP5, ASH2L, and MLL2, but not MLL1 and MLL4, were coimmunoprecipitated with JMJD2B, although MLL1 and MLL4 could be efficiently coimmunoprecipitated with RbBP5, a subunit shared by all of the MLL1–4 complexes (17, 23, 24) (Fig. 1B Right). A weak interaction between JMJD2B and MLL3 was also detected (Fig. 1B Lower Right). Collectively, these results support the suggestion that JMJD2B is physically associated with the MLL2 complex in vivo.
To further validate the in vivo interaction between JMJD2B and the MLL2 complex, protein-fractionation experiments were carried out by FPLC with Superose 6 columns and a high-salt-extraction and size-exclusion approach. The results indicate that native JMJD2B from MCF-7 cells eluted with an apparent molecular mass much greater than that of the monomeric protein (Fig. 1C). Significantly, the elution pattern of JMJD2B largely overlapped that of the protein components of the MLL2 complex including WDR5, RbBP5, ASH2L, and MLL2 (Fig. 1C). Moreover, the chromatographic profiles of JMJD2B and the MLL2 complex were compatible with their associated enzymatic activities. Specifically, both H3K9me3 demethylation and H3K4 methylation activities were detected when the corresponding chromatographic fractions were incubated with in vivo isolated native mononucleosomes or in vitro assembled recombinant nucleosomes, respectively, and then analyzed by Western blotting with antibodies against H3K9me3 and H3K4me3 (Fig. 1C). In addition, consistent with the copurification of ERα with the JMJD2B-containing protein complex (Fig. 1A), the chromatographic profile of ERα also overlapped that of JMJD2B and the core components of the MLL2 complex (Fig. 1C). Interestingly, size-exclusion chromatography demonstrated JMJD2B, ASH2L, RbBP5, and WDR5 not only coeluted with the 667--2,000 kDa complex containing MLL2 and ERα but were also found in a fraction below 440 kDa devoid of MLL2 and ERα, suggesting that JMJD2B, ASH2L, RbBP5, and WDR5 could exist in a subcomplex to execute a distinct function in the absence of MLL2 and ERα. Furthermore, analysis of FLAG-JMJD2B affinity eluate by FPLC with Superose 6 gel filtration revealed that, although purified FLAG-JMJD2B also existed in its free form (Fig. 1D, fraction 34), a multiprotein complex did exist, which peaked in fraction 18 containing WDR5, RbBP5, ASH2L, MLL2, ERα, and JMJD2B (Fig. 1D). Collectively, these results further support the observation that JMJD2B is associated with the MLL2 complex in vivo.
JMJD2B/MLL2 Complex Is Associated with ERα and Is Required for ERα-Activated Gene Transcription.
The copurification of ERα in the JMJD2B-containing complex (Fig. 1A) and the coelution of ERα with the JMJD2B/MLL2 complex (Fig. 1 C and D) suggest that JMJD2B/MLL2 might play a role in the ERα signaling pathway. To investigate this hypothesis, we first verified the physical association of the JMJD2B/MLL2 complex with ERα by coimmunoprecipitation experiments. Immunoprecipitation of MCF-7 cell extracts with antibodies against MLL2, WDR5, RbBP5, ASH2L, or JMJD2B followed by immunoblotting with antibodies against ERα revealed that all components of the JMJD2B/MLL2 complex including MLL2, but not MLL3, were coimmunoprecipitated with ERα (Fig. 2A Left). Reciprocally, immunoprecipitation with antibodies against ERα followed by immunoblotting with antibodies against MLL2, WDR5, RbBP5, ASH2L, or JMJD2B demonstrated that ERα was associated with all of these proteins in vivo (Fig. 2A Right).
Fig. 2.
JMJD2B/MLL2 complex is associated with ERα and is required for ERα-activated gene transcription. (A) Association of the JMJD2B/MLL2 complex with ERα in MCF-7 cells. Whole-cell lysates were immunoprecipitated with antibodies against the indicated proteins. Immunocomplexes were then immunoblotted using antibodies against the indicated proteins. (B) The effect of MLL2 and JMJD2B knockdown on the expression of endogenous ERα target genes. MCF-7 cells were treated with control siRNA or siRNA specific for JMJD2B or MLL2. Cells were deprived of estrogen for 3 d before treatment with E2 for 6 h. Total RNAs were then extracted from the cells for measurement of the mRNA expression of the indicated genes by real-time RT-PCR, with the expression of GAPDH as a normalizer. Each bar represents the mean ± SD for triplicate experiments. (C) The knockdown effect of JMJD2B and MLL2 was validated by real-time RT-PCR and/or Western blotting. P values were determined by Student's t test. *P < 0.001, #P < 0.01.
Next, we tested the effect of the JMJD2B/MLL2 complex on ERα-regulated transcription. To this end, MCF-7 cells were transfected with specific siRNA molecules to knock down the expression of MLL2 or JMJD2B. The cells were deprived of estrogen for at least 3 d and treated with 17β-estradiol (E2). Real-time RT-PCR measurements revealed that knockdown of either MLL2 or JMJD2B severely inhibited E2-stimulated expression of endogenous ERα target genes, including TFF1, EBAG9, cathepsin D, and GREB1 (Fig. 2B), although to variable extents. The efficiency and specificity of MLL2 and JMJD2B knockdown were examined by real-time quantitative RT-PCR and/or Western blotting analysis (Fig. 2C). These results indicate that the JMJD2B/MLL2 complex is required for ERα-activated gene transcription.
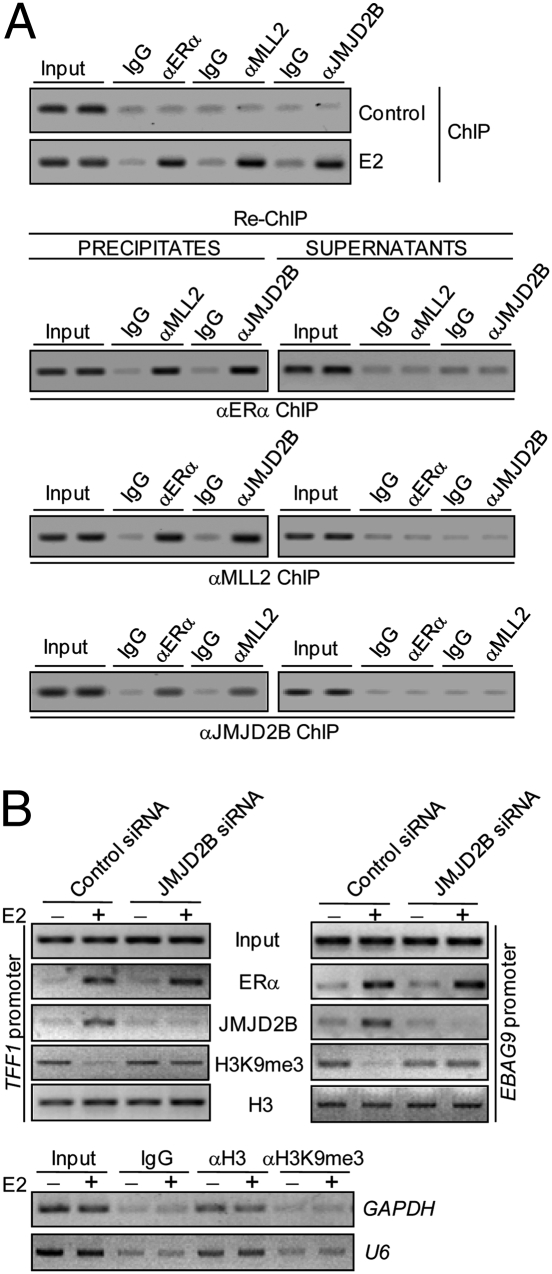
To further support this observation, we examined the recruitment of the JMJD2B/MLL2 complex on ERα target gene promoters in vivo upon E2 stimulation. Chromatin immunoprecipitation (ChIP) assays were performed on MCF-7 cells that were deprived of estrogen for at least 3 d before treatment with E2 for 1 h. These experiments revealed that, similar to ERα, MLL2 and JMJD2B were recruited to the promoter region of the TFF1 gene upon E2 stimulation (Fig. 3A). In addition, sequential ChIP or ChIP/Re-chromatin immunoprecipitation (Re-ChIP) (25–27) was performed to examine whether the presence of ERα, MLL2, and JMJD2B on the TFF1 promoter represents these proteins in the same protein complex. In these experiments, soluble chromatins were first immunoprecipitated with antibodies against ERα, and both the supernatants and immunoprecipitates were subsequently reimmunoprecipitated with antibodies against MLL2 or JMJD2B, and vice versa. The results of these experiments indicate that in precipitates, the TFF1 promoter that was immunoprecipitated with antibodies against ERα could be reimmunoprecipitated with antibodies against MLL2 or JMJD2B, whereas in the supernatants, the ERα antibody-immunoprecipitated TFF1 promoter could not be detected with antibodies against either MLL2 or JMJD2B (Fig. 3A). The same was true when the initial ChIP was done with antibodies against MLL2 or JMJD2B, in that the TFF1 promoter was detected in precipitates but not in supernatants following Re-ChIP with antibodies against ERα (Fig. 3A). Taken together, these experiments not only support the idea that TFF1 is targeted by the JMJD2B/MLL2 complex but also confirm that JMJD2B is physically associated with and is an integral component of the MLL2 complex in vivo.
Fig. 3.
Physical and functional association of JMJD2B/MLL2 complex with ERα target promoter. (A) Co-occupancy of the TFF1 promoter by JMJD2B and the components of the MLL2 complex. MCF-7 cells were deprived of estrogen for 3 d and then treated with E2 for 1 h. Soluble chromatin was prepared for ChIP and Re-ChIP assays with antibodies against the indicated proteins. (B) The effect of JMJD2B knockdown on H3K9 methylation on TFF1 and EBAG9 promoters. MCF-7 cells were treated with control siRNA or JMJD2B siRNA. Cells were deprived of estrogen for 3 d and then treated with E2 for 1 h. Soluble chromatin was prepared for ChIP assays using the indicated antibodies. GAPDH and U6 were used as negative controls.
As a general theme in transcription activation, H3K9 demethylation has also been reported in ERα-activated transcription (12). As stated before, JMJD2B has been demonstrated to be able to demethylate H3K9me3 at pericentric heterochromatin in mammalian cells (20, 21). In light of our observation that JMJD2B influences ERα-regulated transcription and its demethylase activity is essential for that effect, it is logical to postulate that JMJD2B may also function in euchromatin regions and act to erase the H3K9me3 mark in ERα-activated transcription. To investigate this hypothesis, the expression of JMJD2B was knocked down by its specific siRNA in MCF-7 cells, and the level of H3K9me3 at the promoters of ERα targets TFF1 and EBAG9 was examined in these cells with and without E2 treatment. As shown in Fig. 3B, E2 treatment, and hence the activation of ERα-mediated transcription, was associated with a marked decrease in the level of H3K9me3 on the promoters of ERα targets TFF1 and EBAG9. However, compared with control siRNA-transfected cells, cells with loss of function of JMJD2B displayed a substantial retention of H3K9me3 in the TFF1 and EBAG9 promoters upon ERα activation, although the total histone H3 levels and H3K9me3 status on other gene promoters were not affected (Fig. 3B). To extend this analysis, we examined the JMJD2B depletion effect on H3K9me3 status in ZR-75-1 and T47D cells by quantitative ChIP (qChIP). These results indicated that depletion of JMJD2B resulted in an increased level of H3K9me3 on the promoters of TFF1 and EBAG9 genes (SI Appendix, Fig. S1 A and B). These data support the notion that JMJD2B functions to erase the H3K9me3 mark in ERα-activated transcription.
Functional Coordination Between the MLL2 Complex and JMJD2B in ERα-Activated Transcription.
As stated above, H3K4 hypermethylation and H3K9 hypomethylation are prevalent histone marks that are associated with transcriptional activation; these sites are indeed so marked in ERα-activated transcription. Our observation that the H3K4 methyltransferase, the MLL2 complex, and the H3K9 demethylase, JMJD2B, are physically associated and coexist in the same protein complex raises a distinct possibility that methylation at H3K4 and demethylation at H3K9 are functionally connected, but it also raises an intriguing question about how these opposed enzymatic activities are coordinated.
To investigate the functional coordination between H3K4 methylation and H3K9 demethylation, loss-of-function experiments were performed for JMJD2B or MLL2 proteins in MCF-7 cells, and the recruitment of the JMJD2B/MLL2 complex proteins and the methylation status of H3K4 and H3K9 on ERα target gene promoters were examined by conventional ChIP as well as by qChIP. These experiments indicated that depletion of JMJD2B resulted in a diminished/reduced binding by MLL2, WDR5, and RbBP5 proteins on the promoters of TFF1 in MCF-7 cells, which was concomitant with an increase in H3K9me3 and a decrease in H3K4 methylation (SI Appendix, Fig. S2A). Surprisingly, however, although depletion of MLL2 resulted in a diminished methylation activity on H3K4, it had no effect on the recruitment of JMJD2B and on the demethylation of H3K9me3 (SI Appendix, Fig. S2B).
To further support this observation and to investigate whether or not H3K4 methylation is affected by H3K9me3 demethylation, mouse Jmjd2b (W-mJmjd2b or M-mJmjd2b) (20, 28) was transfected into JMJD2B-depleted (with stable transfection of JMJD2B-specific siRNA molecules) MCF-7 cells. These cells were deprived of estrogen for 3 d and then treated with E2. ChIP and qChIP experiments revealed that, although the recruitment of ERα, Jmjd2b, MLL2, WDR5, and RbBP5 on the TFF1 promoter could be detected in either W-mJmjd2b- or M-mJmjd2b-transfected/JMJD2B-depleted MCF-7 cells, both H3K9 demethylation and H3K4 methylation were severely impaired in M-mJmjd2b-transfected cells (SI Appendix, Fig. S2C). Together with the above-described loss-of-function experiments for JMJD2B and MLL2, these data suggest that the demethylation of H3K9 by JMJD2B might be a necessary precondition for H3K4 methylation by the MLL2 complex in coactivating ERα-mediated transcription. If this interpretation is correct, it means that demethylation of H3K9 is a prerequisite for and a preceding event to H3K4 methylation.
To further support this deduction, FLAG-JMJD2B and FLAG-MLL2/SET (SET domain-containing MLL2 fragment) were purified from mammalian cells with anti-FLAG immunoaffinity resin and incubated with synthesized unmethylated or K9-trimethylated histone H3 peptides for methylation/demethylation assays. Subsequent examination of the peptides by Western blotting indicated that the deposition of K4me1/2 by FLAG-MLL2/SET was impaired in the presence of K9me3. Addition of FLAG-JMJD2B, which demethylates K9me3, greatly facilitated K4 methylation by FLAG-MLL2/SET (SI Appendix, Fig. S2D Left). In addition, liquid assays by scintillation counting (SI Appendix, Fig. S2D Right) supported the data from Western blotting. These experiments clearly demonstrate that H3K9 demethylation is required for sufficient H3K4 methylation, supporting our model for the coordinated methylation regulation of H3K9 and H3K4 by the JMJD2B/MLL2 complex in ERα-regulated transcription. Although the exact mechanism for this coordination is currently unknown, it is conceivable that H3K9 methylation may present a stoichiometric hindrance in which substrate recognition at and around the active site of MLL2 might be compromised by the presence of a trimethyl mark at K9. Alternatively, demethylated H3K9 may be required for subsequent binding of the component(s) of the MLL2 complex to chromatin, which, in turn, may be necessary for exposure of the enzymatic center of MLL2. In any case, it appears that ERα-activated transcription may need to clean up the repression mark(s) before the addition of an activation signature(s).
JMJD2B Promotes Cell Proliferation in Vitro and Tumorigenesis in Vivo.
JMJD2B belongs to the JMJD2 family of JmjC domain-containing demethylases. Because the JMJD2C gene (also known as the GASC1 gene) is amplified in esophageal squamous cell carcinoma, JMJD2-family genes have been suggested to be cancer-associated genes (29). Because estrogen is a classical etiologic factor for breast cancer (30–32), the requirement of JMJD2B in ERα-regulated transcription implies that JMJD2B might play an important role in the cell proliferation and carcinogenesis of breast cancer. Indeed, bioinformatics analysis of the cancer microarray database ONCOMINE (Compendia Bioscience) (33) revealed that the expression of JMJD2B is closely and positively correlated with that of ERα in breast carcinoma samples (SI Appendix, Fig. S3). To investigate the functional connection between JMJD2B and ERα and the possible role of JMJD2B in breast cancer development, we first examined the effect of JMJD2B on the proliferation of MCF-7 cells. Colony formation assays demonstrated that depletion of JMJD2B was associated with a significant inhibition in the proliferation and thus the colony numbers of MCF-7 cells in response to E2 stimulation (Fig. 4A). Furthermore, flow cytometry analysis revealed that knockdown of JMJD2B resulted in a significant reduction in the cell population in S phase, from 33 to 23%, with a concomitant increase in the cell population in G1 phase, from 53 to 66%; in contrast, the cell population in G2/M phases was essentially unaffected (Fig. 4B). These data suggest that JMJD2B promotes estrogen-induced MCF-7 cell proliferation by facilitating the G1-to-S transition. Because estrogen stimulates mammary epithelial cells mainly by promoting the G1-to-S transition of the cell cycle (32, 34), these observations are consistent with the role of JMJD2B in coactivating ERα-regulated transcription. In addition, W-mJmjd2B, but not M-mJmjd2B, was able to rescue the inhibitory effect of JMJD2B knockdown on cell proliferation (Fig. 4B), further supporting the idea that JMJD2B functions as a demethylase in estrogen signaling.
Fig. 4.
JMJD2B promotes MCF-7 cell proliferation and tumorigenesis. (A) Knockdown of JMJD2B impedes the proliferation of MCF-7 cells. MCF-7 cells stably transfected with JMJD2B siRNA were grown in the presence or absence of E2 and subjected to colony formation assays. A representative colony formation assay is shown. Each bar represents the mean ± SD for triplicate experiments. (B) Knockdown of JMJD2B impairs the cell-cycle progression of MCF-7 cells. MCF-7 cells stably transfected with JMJD2B siRNA were grown in the presence or absence of E2 and subjected to flow cytometry assays. W-mJmjd2b or M-mJmjd2b was used to rescue the JMJD2B depletion effect. (C) Effect of JMJD2B depletion on the growth of transplanted tumors in nude mice. MCF-7 or MDA-MB-231 tumors with no change in JMJD2B expression or with JMJD2B knockdown were transplanted into ovariectomized athymic mice. Tumor growth was monitored under treatment with vehicle or E2 pellets. Tumors were measured weekly with vernier calipers and volume was calculated using the formula π/6 × length × width2. Each point represents the mean ± SD for different animal measurements (n = 6). The levels of JMJD2B and ERα proteins in MCF-7 tumors were examined by Western blotting. P values were determined by Student's t test. *P < 0.001, #P < 0.01.
To further establish the role of JMJD2B in promoting estrogen-stimulated cell proliferation and breast carcinogenesis, we transplanted three types of breast tumor, developed from ER+ MCF-7 cells, ER+ ZR-75-1 cells, or ER− MDA-MB-231 cells, onto ovariectomized athymic mice (BALB/c; Charles River). The transplanted tumors had either unchanged expression of JMJD2B (infected with lentivirus carrying a control siRNA) or specific knockdown of JMJD2B expression (infected with lentivirus carrying JMJD2B siRNAs). Tumor growth was monitored in mice that received no treatment or treatment with E2. In athymic mice that received a transplant of MCF-7 tumors with unchanged JMJD2B expression, estrogen stimulated tumor growth over 8 wk (Fig. 4C). In athymic mice that received tumor transplants with JMJD2B knockdown, the tumor growth stimulation by E2 was greatly attenuated (Fig. 4C). On the other hand, knockdown of JMJD2B had minimal effects on the growth of MDA-MB-231 tumors, regardless of the presence or absence of estrogen treatment (Fig. 4C). Collectively, these experiments strongly indicate that JMJD2B is a key effector of estrogen-stimulated growth of MCF-7 tumors.
JMJD2B Is Transcriptionally Targeted by ERα.
The positive correlation of the expression between ERα and JMJD2B in breast carcinoma samples suggests that JMJD2B might be a transcriptional target for ERα. Indeed, real-time RT-PCR analysis of MCF-7 cells revealed a dose-dependent E2 induction in the mRNA expression of JMJD2B but not in the genes encoding for other JmjC domain-containing histone lysine demethylases (SI Appendix, Fig. S4A). In addition, the induction of JMJD2B mRNA expression in MCF-7 cells was dependent on ERα, as knockdown of ERα expression led to a diminished effect of E2-induced JMJD2B mRNA expression (SI Appendix, Fig. S4B). Importantly, corresponding to the mRNA induction, the level of JMJD2B protein was also elevated in response to E2 treatment (SI Appendix, Fig. S4C).
Bioinformatics analysis of genome-wide ERα-binding site data (35) identified two putative half-estrogen response element (ERE) sites located in intron 1 of the JMJD2B gene (SI Appendix, Fig. S4D). ChIP assays confirmed E2-dependent recruitment of ERα to these sites in vivo in MCF-7 cells (SI Appendix, Fig. S4D). We thus constructed two luciferase reporters, ER_3291-LUC and ER_3292-LUC, driven by JMJD2B intron 1-derived sequences containing the putative half-ERE sites and tested their responsiveness to E2 treatment. The reporter assays revealed that E2 treatment resulted in a significant increase in the luciferase activity of ER_3291-LUC and ER_3292-LUC but not that of the half-ERE site-mutated counterparts (SI Appendix, Fig. S4D). Collectively, these data support the argument that JMJD2B is transcriptionally targeted by ERα.
In summary, our observations that JMJD2B is both physically and functionally associated with the MLL2 complex and that JMJD2B promotes cell proliferation in vitro and breast carcinogenesis in vivo support JMJD2B as a breast cancer-associated gene. The importance of JMJD2B in estrogen actions and breast carcinogenesis is further supported by our finding that JMJD2B itself is transcriptionally regulated by ERα. It is conceivable that, through the up-regulation of JMJD2B, ERα and JMJD2B form a feed-forward regulatory loop to enhance and amplify the hormone response under physiological as well as pathological conditions.
Materials and Methods
Preparation of Mononucleosomes and Recombinant Nucleosomes.
Mononucleosomes were prepared according to protocols described previously (36). Histone octamers were assembled as previously reported (37, 38), whereas pG5E4 plasmid DNA was used to reconstitute the recombinant nucleosomes in vitro (39).
Lentiviral Production.
The construction of an RNAi lentivirus system using pLL3.7 and other LentiLox vectors was carried out according to a protocol described online (http://web.mit.edu/jacks-lab/protocols/lentiviralproduction.htm). In brief, siRNA sequences targeting JMJD2B or control siRNA sequences were designed and cloned into the pLL3.7 shuttle vector. The recombinant construct, as well as three assistant vectors, pMDLg/pRRE, pRSV-REV, and pVSVG, were then transiently transfected into HEK 293T cells. Viral supernatants were collected 48 h later, clarified by filtration, and concentrated by ultracentrifugation at 50,000 × g for 16 h at 4 °C.
Tumor Xenografts.
MCF-7 and MDA-MB-231 breast cancer cells were plated and infected in vitro with mock or lentiviruses carrying control or JMJD2B RNAi at a multiplicity of infection (MOI) of 100. Forty-eight hours after infection, 5 ×106 viable cells in 200 μL PBS were injected into the mammary fat pads of 6- to 8-wk-old ovariectomized athymic mice (BALB/c; Charles River). Eight animals per group were used in each experiment. E2 pellets (0.72 mg per pellet, 60 d release; Innovative Research of America) or vehicle pellets were implanted 1 d before the tumor cell injection. Tumors were measured weekly using a vernier caliper and the volume was calculated according to the formula π/6 × length × width2. All studies were approved by the Animal Care Committee of Peking University Health Science Center.
Supplementary Material
Acknowledgments
This work was supported by Grants 30830032 and 30921062 (to Y.S.) and 81071673 (to L. Sun) from the National Natural Science Foundation of China and 973 Program Grants 2011CB504204 and 2007CB914503 (to Y.S.) and 2009CB918903 (to Z.Y.) from the Ministry of Science and Technology of China.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017374108/-/DCSupplemental.
References
- 1.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: Histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martens JH, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 4.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 5.Li F, et al. Lid2 is required for coordinating H3K4 and H3K9 methylation of heterochromatin and euchromatin. Cell. 2008;135:272–283. doi: 10.1016/j.cell.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brinkman AB, et al. Histone modification patterns associated with the human X chromosome. EMBO Rep. 2006;7:628–634. doi: 10.1038/sj.embor.7400686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rougeulle C, et al. Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol Cell Biol. 2004;24:5475–5484. doi: 10.1128/MCB.24.12.5475-5484.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Squazzo SL, et al. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 10.Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26:9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dreijerink KM, et al. Menin links estrogen receptor activation to histone H3K4 trimethylation. Cancer Res. 2006;66:4929–4935. doi: 10.1158/0008-5472.CAN-05-4461. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Bassets I, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janowski BA, et al. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 14.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 15.Briggs SD, et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milne TA, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 17.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 18.Yamane K, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 19.Wissmann M, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- 20.Fodor BD, et al. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshioka H, McCarrey JR, Yamazaki Y. Dynamic nuclear organization of constitutive heterochromatin during fetal male germ cell development in mice. Biol Reprod. 2009;80:804–812. doi: 10.1095/biolreprod.108.072603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett A, et al. Breast cancer associated transcriptional repressor PLU-1/JARID1B interacts directly with histone deacetylases. Int J Cancer. 2007;121:265–275. doi: 10.1002/ijc.22673. [DOI] [PubMed] [Google Scholar]
- 23.Steward MM, et al. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol. 2006;13:852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 24.Issaeva I, et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, et al. Differential gene regulation by the SRC family of coactivators. Genes Dev. 2004;18:1753–1765. doi: 10.1101/gad.1194704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 28.Whetstine JR, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 29.Katoh M, Katoh M. Identification and characterization of JMJD2 family genes in silico. Int J Oncol. 2004;24:1623–1628. [PubMed] [Google Scholar]
- 30.Key TJ, Verkasalo PK, Banks E. Epidemiology of breast cancer. Lancet Oncol. 2001;2:133–140. doi: 10.1016/S1470-2045(00)00254-0. [DOI] [PubMed] [Google Scholar]
- 31.Mesko TW, Dunlap JN, Sutherland CM. Risk factors for breast cancer. Compr Ther. 1990;16:3–9. [PubMed] [Google Scholar]
- 32.Shang Y. Molecular mechanisms of oestrogen and SERMs in endometrial carcinogenesis. Nat Rev Cancer. 2006;6:360–368. doi: 10.1038/nrc1879. [DOI] [PubMed] [Google Scholar]
- 33.Rhodes DR, et al. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doisneau-Sixou SF, et al. Estrogen and antiestrogen regulation of cell cycle progression in breast cancer cells. Endocr Relat Cancer. 2003;10:179–186. doi: 10.1677/erc.0.0100179. [DOI] [PubMed] [Google Scholar]
- 35.Carroll JS, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 36.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 38.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 39.Loyola A, Reinberg D. Histone deposition and chromatin assembly by RSF. Methods. 2003;31:96–103. doi: 10.1016/s1046-2023(03)00093-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.