Abstract
The Origin Recognition Complex (ORC) is a six-subunit protein important for the initiation of DNA replication in eukaryotic cells. Orc6 is the smallest and the least conserved among ORC subunits. It is required for the DNA replication but also has a function in cytokinesis in metazoan species, however, the mechanisms of Orc6 action in these processes are not clear. Here we report a structure of the middle domain of human Orc6. This domain has an overall fold similar to the corresponding helical domain of transcription factor TFIIB. Based on these findings, a model of Orc6 binding to DNA is produced. We have identified amino acids of Orc6 which are directly involved in DNA binding. Alterations of these amino acids abolish DNA binding ability of Orc6 and also result in reduced levels of DNA replication in vitro and in cultured cells. Our data indicate that Orc6 is one of the DNA binding subunits of ORC in metazoan species. We propose that Orc6 may participate in positioning of ORC at the origins of DNA replication similar to the role of TFIIB in positioning transcription preinitiation complex at the promoter.
The hexameric Origin Recognition Complex (ORC) is an important and conserved component for eukaryotic DNA replication (1). Extensive studies both in yeast and higher eukaryotes laid the foundation for understanding the functions of this important key initiation factor (2–5). ORC binds to the origin DNA and plays a critical role in the recruitment of additional replication factors to the origin resulting in the formation of the prereplicative complex (pre-RC) (2, 3, 6, 7). The structural and biochemical studies of recent years begin to clarify the complex nature of the ORC–DNA interface by determining the DNA binding motifs of ORC involved in origin recognition, ORC’s architecture, and interactions between ORC and other members of pre-RC (8–11).
The Orc6 protein is the least conserved and most enigmatic of all ORC subunits, and amino acid alignments between budding yeast and metazoan proteins do not show statistically significant homologies. In budding yeast, Orc6 is essential for viability but is not required for DNA binding in vitro (12, 13). In Xenopus and humans, Orc6 protein does not seem to be tightly associated with other core ORC subunits (14–17). By contrast, Orc6 is an integral part of Drosophila ORC and is essential for both DNA binding and replication activity both in vitro and in vivo (18–20). Drosophila Orc6 has a DNA binding activity on its own, with a specificity that can account for the poly(dA) preference of ORC binding (19). Recently, an evidence appeared for the role of Orc6 in pre-RC formation in budding yeast, where Orc6 directly interacts with Cdt1 during repeated Mcm2–7 loading (21, 22). Orc6 in budding yeast is significantly larger than in metazoan species and it remains to be investigated whether Orc6 is important for pre-RC formation in higher eukaryotes. Nevertheless, despite such diversity of functions across different species, in all studied organisms, Orc6 is required for DNA replication and the latest data provide compelling evidence that Orc6 is critical for ORC function. Moreover, in both Drosophila and human cells, Orc6 has been implicated in coordinating cytokinesis with pre-RC formation and chromosome segregation, a role that it performs independently of the rest of the complex (5, 23). Therefore, solving the structure of Orc6 would be very beneficial to study the functions of this important protein. Our earlier model predicted that the N-terminal replication domain of metazoan Orc6 might have a fold similar to the helical domain of transcription factor TFIIB (19, 24). To test this model and to characterize the functions of Orc6 further, we determined the structure of human Orc6 middle region (Orc6-M), which includes amino acids 94–187.
Results and Discussion
Overall Structure of Orc6-M and Structure Comparison of Orc6-M with TFIIB.
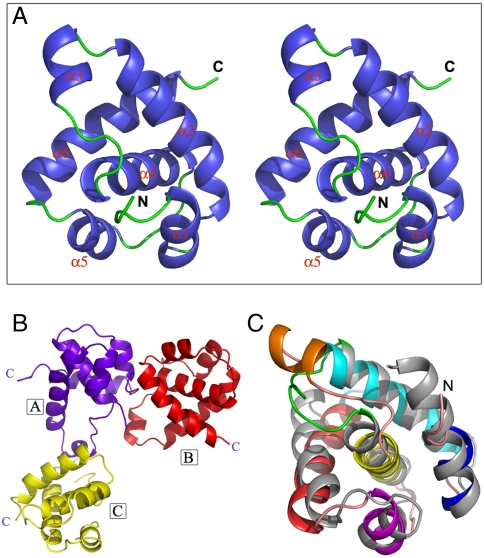
The full-length human Orc6 protein proved to be difficult to crystallize. If our model of structural homology between Orc6 and TFIIB (19) is correct, Orc6 core replicative domain would consist of two smaller globular domains connected with a short linker region. Therefore, we attempted to crystallize these domains separately. HsOrc6 protein fragment comprised of amino acids 94–187 (Orc6-M) produced crystals that diffracted to 2.5 Å. The structure of the protein was determined by single isomorphous replacement with anomalous signal (SIRAS) method. The final Rwork was refined to 22.6% and Rfree was 26.6% (Fig. 1A and Table 1). A figure showing the quality of the electron density map is shown in Fig. S1. The crystal belongs to P4 space group. There are three similar monomers (rmsd nearly 0.8 Å to each other, backbone comparison) in one asymmetry unit. Monomers are located asymmetrically to each other, as a reverse “L” shape (Fig. 1B): monomer A locates at the turn corner of the L, whereas B and C monomers locate at the two ends of the L. Thus, A interacts with both B and C, whereas B does not have direct interaction with C. This kind of special arrangement leads to different interaction surfaces for A to interact with B and C. The structure of one molecule shows that the protein consists of six helices and several loops (Fig. 1A). By performing a Dali search (http://www2.ebi.ac.uk/dali/), we found that the overall folding of Orc6-M is very similar to the second helical domain (residues 1206–1300) in the carboxyl terminal of TFIIB (PDB ID code 1AIS) (rmsd ∼ 2.4 Å, backbone comparison) (Fig. 1C) (25). Most of the helices in the two proteins have equivalent locations and directions in the structures. One major difference is that TFIIB helical domain 2 has only five helices, whereas there is an additional short helix (helix 3) formed in a loop region between helix 2 and helix 4 in Orc6-M. This loop region in Orc6-M is six residues longer than its counterpart in TFIIB. The C-terminal domain of TFIIB contains two structurally similar repeated helical motifs. The structural analysis of these TFIIB repeats in a complex with the DNA-bound TATA-binding protein (TBP) revealed that these repeats of TFIIB are important for interacting with TBP and the TBP-bound TATA box DNA (25). Interestingly, two fragments of Orc6 (1–93 and 94–187) are homologous with 17% sequence identity (Fig. S2), suggesting that Orc6 may also consist of two similar motifs as is observed for TFIIB as is also predicted by our model (19). This result confirmed our previous prediction that Orc6 may have an overall structure similar to TFIIB helical domains and also provided the detailed structural information for human Orc6 protein.
Fig. 1.
Overall structure of human Orc6-M. (A) Stereo view of Orc6-M overall structure. (B) Spatial arrangement of the three Orc6-M monomers in an asymmetric unit. The three molecules are indicated in different colors and labeled with boxed A, B and C. The C termini of these molecules are also indicated. (C) Superimposition of Orc6-M to TFIIB second helical domain (gray). Orc6-M is blue (helix 1), cyan (helix 2), golden (helix 3), yellow (helix 4), magenta (helix 5), and red (helix 6). The loop in TFIIB helical domain corresponding to Orc6-M helix 3 is green.
Table 1.
Data statistic and structure refinement
| Orc6 native | Orc6-Pt derivative | |
| Data collection | ||
| Beamline | Rigaku FR-E | Rigaku FR-E |
| Wavelength, Å | 1.5418 | 1.5418 |
| Space group | P4 | P4 |
| Cell dimensions | ||
| a, b, c, Å | 104.57, 104.57, 30.94 | 104.69, 104.69, 30.80 |
| α, β, γ, ° | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution, Å | 50.00–2.50 (2.59–2.50)* | 50.00–3.1 (3.21–3.10)* |
| Rmerge | 0.067 (0.515)* | 0.086 (0.509)* |
| I/σI | 38.9 (4.9)* | 23.9 (3.6)* |
| Completeness, % | 99.9 (100.0)* | 99.9 (100.0)* |
| Redundancy | 7.1 (7.1)* | 7.3 (6.7)* |
| Phasing | ||
| No. heavy atoms | 3 | |
| FOM (initial) | 0.314 | |
| Refinement | ||
| Resolution, Å | 50.0–2.50 | |
| Total no. reflection/free | 85,740/12,005 | |
| Rwork/Rfree | 0.226/0.266 | |
| No. atoms | ||
| Protein | 2,101 | |
| Water | 69 | |
| Avg B factors, Å2 | 58.2 | |
| rmsd | ||
| Bond lengths, Å | 0.013 | |
| Bond angles, ° | 1.40 | |
| Ramachandran Plot | ||
| Favored, % | 91.2 | |
| Allowed, % | 8.8 |
FOM, figure of merit.
*Values in parentheses are for highest-resolution shell.
Model of Orc6-M Association with DNA.
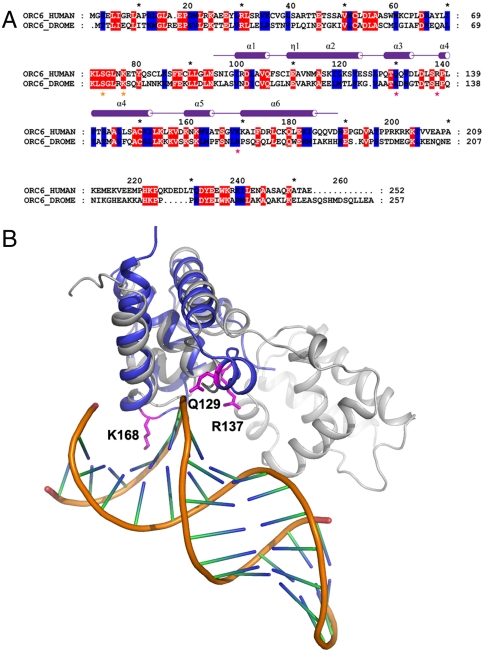
TFIIB is an important transcription factor that can interact with DNA and that actively participates in a formation of transcription preinitiation complex (pre-IC). TFIIB is also a target of multiple regulatory factors; the interaction of TFIIB with many of these factors is achieved through the helical domains of the protein (26). We have also shown previously that Drosophila Orc6 has a DNA binding ability (19). We asked whether human Orc6 is able to interact with DNA. Because Orc6 has a fold resembling helical domains of TFIIB, we anticipate that Orc6 may interact with DNA in a manner similar to that of TFIIB. By performing structure comparison, we built an Orc6–DNA interaction model based on TFIIB–DNA complex structure (PDB ID code 1AIS) (Fig. 2B). We superimposed our Orc6 fragment structure to TFIIB–DNA complex resulting in a model to analyze the possible interaction mode of Orc6 to DNA. High structure similarity of these two proteins makes this model very informative. We identified several charged residues of Orc6 that are located within a region in a close proximity with DNA, including amino acids Q129 (helix 3), R137 (loop region between helices 3 and 4), and K168 (helix 6) (Fig. 2 A and B). The locations of these residues suggest that Orc6 may interact with both major (K168) and minor (Q129 and R137) grooves of DNA.
Fig. 2.
Superimposition of Human Orc6-M to structure of TFIIB–DNA complex (PDB ID code 1AIS). (A) Sequence alignment between human and Drosophila Orc6. Amino acids mutated in human and Drosophila Orc6 are indicated with stars colored magenta in this experiment, whereas amino acids mutated in Drosophila Orc6 in ref. 19 are indicated with stars colored orange. (B) Superimposition of human Orc6-M (blue) to structure of TFIIB–DNA complex (PDB ID code 1AIS). TFIIB is colored gray and the dsDNA are colored gold and green-blue short lines. The three residues, K168, R137, and Q129 in Orc6 are shown in magenta.
Orc6 Is a DNA Binding Subunit of ORC.
In human cells, Orc6 localizes in the nucleus together with other ORC subunits and also associates with chromatin (27, 28). Based on our structural studies, we asked if human Orc6, similar to its Drosophila homolog, could interact with DNA directly. To examine whether the human Orc6 subunit was able to bind DNA on its own, a series of gel shift experiments were carried out. Human lamin B2 and Drosophila ori-β DNA fragments with origins of DNA replication were used as probes for electrophoretic mobility shift assays (EMSA). Human Orc6 was expressed in Escherichia coli and purified through several chromatographic steps (see Experimental Procedures). We found that a recombinant Orc6 formed a distinct complex with both human lamin B2 (Fig. 3A, lanes 1–4) and Drosophila ori-β (Fig. S3) fragments.
Fig. 3.
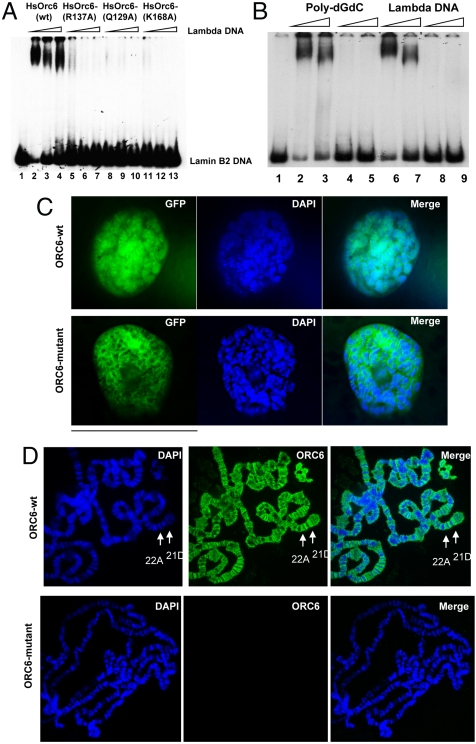
DNA binding ability of HsOrc6. (A) DNA binding of human Orc6 to radiolabeled origin DNA fragment Lamin B2 was monitored by EMSA. Orc6-WT (lanes 2–4), Orc6-R137A (lanes 5–7), Orc6-Q129A (lanes 8–10), and Orc6-K168A (lanes 11–13) mutant proteins (50 ng each) were incubated with human Lamin B2 origin DNA fragment in the presence of increasing amounts of competitor lambda DNA. The amount of competitor was 50, 75, and 100 ng. No protein was added to DNA fragment in lane 1. (B) DNA binding of human wild-type (lanes 2, 3 and 6, 7) and triple mutant (lanes 4, 5 and 8, 9) Orc6 proteins (50 ng) to the lamin B2 DNA in the presence of 50–100 ng of poly(dG-dC) (lanes 2–5) or lambda (lanes 6–9) DNA. (C) GFP-fused human Orc6-WT (Upper), Orc6-triple mutant (Lower) were expressed in Drosophila salivary glands and analyzed as described in Experimental Procedures. (D) Human Orc6 prefers less condensed interband regions in Drosophila polytene chromosomes. Arrows indicate examples of heavily stained bands in regions 21D and 25A.
To analyze the binding ability of human Orc6 to DNA in more detail, we used a mutagenesis approach. Several amino acids were identified in close proximity to DNA, which might contribute to DNA binding and recognition. Of particular interest were Q129, R137, and K168 residues of Orc6 (Fig. 2B). These amino acids were mutated to alanines by site-directed mutagenesis. The corresponding motif in TFIIB interacts with DNA specifically with the assistance of K1245, R1283, and R1285 residues (24, 29). Obtained Orc6 mutant proteins were expressed in E. coli as His-tag fusions and purified. Silver stained gels of the Orc6 proteins, used in these studies, are shown in Fig. 4A and Fig. S4. Mutant human Orc6-Q129A, Orc6-R137A, and Orc6-K168A and mutant Orc6 containing all three mutations were analyzed for DNA binding in EMSA reactions. As shown in Fig. 3 A and B and Fig. S3, these alanine mutations completely abolished the Orc6 binding to human lamin B2 and Drosophila ori-β fragments, suggesting that protein interactions with DNA were affected. This inability to bind with DNA was not due to the structural distortion of the mutant proteins. Mutants expressed and purified normally, undistinguishable from the wild-type Orc6 protein. CD analysis of the Orc6 protein containing triple alanine mutations did not reveal significant differences as compared to the wild-type protein (Fig. S5). Thus, we conclude that human Orc6 can interact with DNA directly, and Q129, R137, and K168 residues are involved in this interaction. We also tried to identify the high-affinity Orc6 binding site within lamin B2 DNA by DNase I footprinting. However, we were unable to detect a discrete binding site using either E. coli or baculovirus expressed Orc6. Instead we found that the addition of increasing amounts of Orc6 protein caused the entire DNA fragment to be protected, as was observed in Drosophila ORC and Orc6 footprints (19, 30). Interestingly, Orc6 was also able to bind ssDNA (Fig. S6). The ability to bind ssDNA was also reported earlier for budding yeast ORC (31).
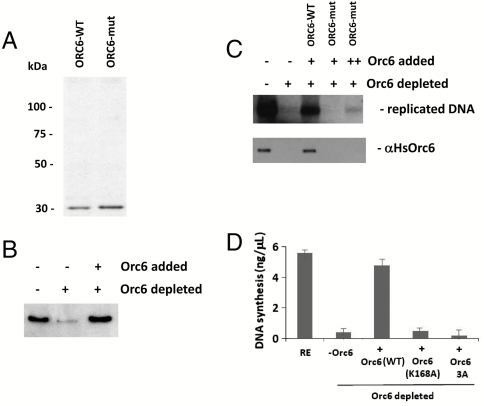
Fig. 4.
In vitro DNA replication in Xenopus extracts is Orc6 dependent. (A) Silver stained gel of purified wild-type human Orc6 (lane 1) and Orc6 triple mutant containing Q129A, R137A, and K168A mutations (lane 2). (B) Western immunoblotting analysis of Xenopus egg extract (lane 1) depleted of Xenopus Orc6 using the mix of antibodies against human and Drosophila Orc6 proteins (lane 2). Recombinant human wild-type Orc6 protein is added to the extract in lane 3. Rabbit polyclonal antibodies raised against human Orc6 were used in the Western immunoblotting experiment. (C) In vitro DNA replication in Orc6-depleted Xenopus extracts can be rescued by the addition of recombinant wild-type human Orc6 but not with human Orc6 triple mutant. Lower panel shows the association of Orc6 protein with sperm chromatin in the same experiment. In a parallel experiment (D), DNA replication efficiency was measured by trichloroacetic acid precipitation. Quantification in different experiments shown was by measuring the incorporation of radiolabeled dNTP into sperm chromatin from three independent reactions done in parallel, with error bars indicating the standard deviation.
Human Orc6 associates with chromatin in human cell lines (27, 28). Moreover, human Orc6 partially restores DNA replication when expressed in Drosophila mutant cells carrying the deletion of orc6 gene (20). In order to visualize the binding of human Orc6 to the chromosomes in vivo, we expressed it in Drosophila salivary glands. Nuclei of Drosophila salivary glands contain polytene chromosomes that can be easily visualized with microscopy because of their giant size and well-determined structure. In our experiment, GFP-fused wild-type and mutant human orc6 genes were subcloned into the P(UAST) vector, which allows for GAL-4-induced expression in Drosophila. Fly stocks were set up for each fusion construct. GFP–Orc6 expression was induced in salivary glands of third instar larvae, as described (19), to test chromosome binding of human Orc6 in this heterologous system. Fig. 3C shows the localization of GFP-fused wild-type and mutant human Orc6 expressed under the control of Sgs3 promoter in the nucleus of Drosophila salivary glands. GFP-fused wild-type Orc6 protein was tightly associated with polytene chromosomes (Fig. 3C, Upper). In contrast, human Orc6 mutant carrying triple alanine substitutions of Q129, R137, and K168 failed to associate with chromosomes (Fig. 3C, Lower) in agreement with in vitro DNA binding experiments shown in Fig. 3B.
To detect human Orc6 preferences on chromosomes, if any, immunostained polytene chromosomes from the Drosophila strains expressing described human GFP–Orc6 variants were stained with anti-Orc6 antibodies. Similar to the overexpressed Drosophila wild-type Orc6, human Orc6 did not follow the DNA distribution along chromosomes but preferred less condensed, AT-rich interband regions (Fig. 3D, Upper). No staining on polytene chromosomes was observed when triple mutant Orc6 protein was expressed (Fig. 3D, Lower).
Orc6 Is Important for DNA Replication.
Using Xenopus in vitro DNA replication assay (32, 33), we have shown that the purified recombinant human wild-type Orc6 could efficiently restore DNA replication activity in Xenopus egg extracts depleted of endogenous Orc6 (Fig. 4). In these experiments, endogenous Xenopus Orc6 protein was depleted from the extracts using antibodies raised against human and Drosophila Orc6 proteins (Fig. 4B). Next, Orc6-depleted extracts were supplemented with recombinant proteins (Fig. 4 A and B) to verify the activity of the human wild-type and mutant Orc6 in DNA replication. As presented in Fig. 4C (Upper) and D, wild-type but not triple mutant protein was able to rescue DNA replication in Orc6-depleted Xenopus extract. In parallel chromatin association experiments (Fig. 4C, Lower), no Orc6 was detected on DNA when Orc6 triple mutants were added to Orc6-depleted extracts consistent with the inability of this mutant to bind DNA and restore DNA replication.
We and others have shown that the ablation of Orc6 resulted in decreased BrdU incorporation and cell death in both Drosophila and human cells (27, 34). We asked what effect would Orc6 mutants, deficient for DNA binding, have if expressed in the cultured cells. The expression of either wild-type or mutant Orc6 in human cells did not result in any changes to the levels of DNA replication. We speculate that the reason for this is that Orc6 in human cells is not associated tightly with the core ORC and may only assemble into six-subunit ORC at the origins of DNA replication during G1 phases of the cell cycle. Therefore, the expressed Orc6 mutant deficient for DNA binding is unable to bind DNA and is also unable to interact with the core ORC (1–5) in solution. As a result, we were not able to detect dominant-negative effect of expressing human Orc6 mutant in human cultured cells. To explore this question further, we designed mutations in Drosophila Orc6 based on the protein sequence alignment (Fig. 2A). Drosophila Orc6 is tightly associated with the ORC complex and is important for DNA binding of ORC (19). Mutations of three residues in Drosophila Orc6—E128A, H136A, and R167A—corresponding to residues Q129, R137, and K168 in human Orc6, respectively, were chosen for analysis. We found that the expression of Drosophila Orc6 triple-mutant carrying mutations E128A, H136A, R167A in Drosophila L2 cells results in significantly less replication in cultured cells after ablation of the endogenous protein during RNAi experiment. The results are present in Table S1 and Fig. S7. The endogenous protein was removed by RNAi using dsRNA complementary to the 3’UTR of Orc6, and Orc6 wild-type and mutant proteins were expressed under control of inducible MT promoter as in ref. 19 (Fig. S7). Interestingly, the ablation of Orc6 from these cells resulted in mislocalization of other ORC subunits in many cells (judged by the staining of cells with antibodies against Orc2) (Fig. S7B), indicating that Orc6 is important for DNA binding of the whole ORC in Drosophila cells in agreement with our earlier studies (19, 34). At the next step, mutant Orc6 proteins were transiently expressed in L2 cells as described (34). Multiple experiments were performed to quantify the level of BrdU incorporation as an indicator of DNA replication in cells transfected with wild-type and mutant Orc6 constructs. We counted a number of cells with GFP-tagged mutant proteins and then counted a number of cells replicating DNA based on BrdU incorporation on the same slides. Only 6.7% of cells were incorporating BrdU after Orc6 dsRNA treatment (Table S1). In contrast, BrdU was incorporated in 36% of the total number of cells expressing wild-type GFP–Orc6 after 10–12 h of incubation (Table S1), comparable to the number of cells incorporating BrdU in nontreated cells. The expression of each of individual amino acid mutants resulted in at least a 50% decrease in DNA replication in the cells judged by BrdU incorporation. However, the expression of triple-Orc6 mutant resulted in a three- to fourfold decrease of BrdU incorporation. Only 8.8% of GFP-positive cells were found to contain BrdU, when the Orc6 triple amino acid mutant was overexpressed in L2 cells (Table S1).
Our previous analysis of Drosophila Orc6 based on structure prediction and molecular modeling allowed identification of two amino acids, Ser72 and Lys76, which are essential for the DNA binding ability of Orc6 (19). Alterations of these amino acids severely compromised the functions of reconstituted Drosophila ORC protein in DNA binding and in DNA replication in vitro. In vivo, mutant Orc6 proteins do not associate with chromosomes and have dominant-negative effects when expressed in Drosophila tissue culture cells. Cells with overexpressed mutant proteins have a reduced replication activity, as judged by BrdU incorporation. The residues we examined for human Orc6 in this report are located within the second helical domain, whereas the residues Ser72 and Lys76 in Drosophila Orc6, which are also found to be required for DNA binding, are predicted to be located in the first helical domain. Combining these results, we propose that both helical domains in Orc6 are essential for DNA binding.
Replication and transcription are two cellular fundamental processes; they also share some similar properties for DNA recognition, initiation, and elongation. Orc6 is an essential component for replication initiation, although its functions vary in different eukaryotic species. In yeast Saccharomyces cerevisiae, Orc6 is an integral part of the complex but is not involved in DNA binding. Budding yeast Orc6 interacts with Cdt1 protein and facilitates the loading of Mcm2–7 helicase. It remains to be investigated whether this function of Orc6 is conserved in metazoan species. Drosophila Orc6 protein is also tightly associated with other ORC subunits; however, it does not bear any homologies with yeast protein. In Drosophila, Orc6 participate in DNA recognition and may serve in positioning of ORC at the origin. Human Orc6, although homologous with Drosophila protein, loosely associates with core ORC subunits, however, it is required for replication, possesses DNA binding ability, and associates with DNA both in vitro and in vivo.
TFIIB protein recognizes and binds DNA sequence at the promoter regions, helps in positioning of the transcription pre-IC and further recruits some components of transcription machinery (24, 29). In the TBP-TFIIB–DNA complex structure, the two helical domains in TFIIB can interact with both TBP and DNA. These interactions are shown to be important for transcription initiation complex assembly and also define initiation starting sites for transcription (24, 29). Our results indicate that Orc6 has two structural repeats similar to TFIIB helical motifs, suggesting that functions of these two proteins may be similar during DNA recognition and an assembly of larger complexes at the promoter or origin regions. Orc6 may position ORC and resulting pre-RC at the origin, therefore defining the site where DNA replication initiates. TFIIB is also a target of a considerable number of regulatory factors, containing activation domains from various classes. Similarly, Orc6 might also be a target of multiple activators and/or repressors, which help in determining the time and localization of pre-RC formation and origin firing. Numerous reports indicate that transcription factors can interact with ORC and help in defining the sites of the initiation of DNA replication (2, 5). Several examples include E2F and Myb transcription factors, which interact with ORC and target it to the origin at chorion amplification locus in Drosophila (35, 36). Recent studies revealed the role of HMG1A and WD repeat-containing LRWD1 proteins in targeting of ORC to the chromatin in human cells (28, 37). Interestingly, human Orc6 and HMG1A proteins interact directly and may both contribute to the targeting of ORC to the sites of replication initiation and to the pre-RC assembly.
Based on our data, we would like to suggest a model describing the role of Orc6 in origin recognition and initiation of DNA replication in general. Orc6 may function in targeting and positioning of ORC to the origins of DNA replication. Orc6 binds DNA directly, but it does not recognize a specific sequence. Rather it has an affinity for structural and topological features associated with poly(dA) stretches such as a minor groove structure (19). The importance of the topological state of DNA for the entire Drosophila ORC–DNA binding activity has been shown before (30). In vivo, Drosophila Orc6 associates with AT-rich, early replicating, interband regions of polytene chromosomes. Genome-wide analysis of the entire ORC distribution in Drosophila revealed its preferential localization to AT-rich transcriptionally active chromosomal sites, many of which coincide with early replication origins (38, 39). Orc6 is tightly associated with the core ORC1–5 in Drosophila, and mutant Drosophila ORC missing Orc6 subunit does not interact with DNA (18). In this case, the binding of Drosophila ORC to the origin DNA is initially mediated by its Orc6 subunit, which works as an anchor and targets ORC to the origins of DNA replication. This initial binding step is followed by ATP-dependent binding of the entire ORC. In contrast, human Orc6 protein is loosely associated with core ORC1–5 complex, which can bind DNA in the absence of Orc6 subunit. In this case, the Orc6 protein may act, according to our model, as an assembly factor or combine with ORC1–5 to create a functional DNA binding competent six-subunit ORC at the origins of DNA replication, possibly with the assistance of other chromatin- and ORC-associated proteins such as HMG1A and LRWD1. As a result, six-subunit ORC is assembled at the origin at the end of mitosis and activated to ensure correct and timely origin licensing. Our results will provide a platform for further studies on the functions of Orc6 and ORC in general during eukaryotic cell cycle.
Experimental Procedures
Protein Expression and Purification.
The DNA fragment corresponding to amino acids 94–187 of human Orc6 protein (Orc6-M) was cloned into the sites of BamHI and XhoI in pGEX-4T-2 (GE Healthcare). Plasmid with inserted target fragment was transformed into E. coli BL21(DE3). Cells were grown at 37 °C in LB medium containing 0.1 mg/mL ampicillin until the A600 reached about 0.6. Then the culture was adjusted to 0.5 mM IPTG to induce protein production and incubation was continued for 3–4 h at 37 °C. Cells were harvested, collected, and resuspended in 1× PBS. After cells lysis by ultrasonication, the debris was removed by centrifugation at 30,700 × g for 30 min at 4 °C. The supernatant then was loaded on a Glutathione Sepharose 4B (GE Healthcare) column equilibrated with 1× PBS and washed thoroughly. Fusion protein was cleaved on column by thrombin. The elution was further purified by gel filtration through a Hiload 16/60 Superdex-75 or Superdex-200 column (GE Healthcare) equilibrated with a buffer containing 20 mM Tris•HCl (pH 8.0), 150 mM NaCl, 1 mM DTT, and 1 mM EDTA. SDS-PAGE analysis was carried out to identify the elution peak corresponding to the target protein. Peak fractions were pooled up and concentrated to at least 10 mg/mL.
Recombinant wild-type and mutant Orc6 proteins were cloned and expressed as a fusion protein with a 6His-tag at their amino terminus. They were first purified by nickel affinity column (Novagen, Inc.) and then further purified by gel filtration column similar to that of Orc6-M.
Protein Crystallization and Data Collection.
Hanging drop vapor diffusion method was used to screen protein crystals. Crystals used for X-ray diffraction were obtained in 100 mM MES pH 6.5, 200 mM Li2SO4, 30% PEG4000 at 16 °C. For heavy atom derivatives, a little K2PtCl4 powder was directly added to the drop containing optimum crystals. Native and derivative datasets were both collected in-house at Cu Kα radiation wavelength at 100 K on an R-AXIS IV++ Image Plate detector (Rigaku). All images were processed with the HKL2000 program suite (40). The crystals belong to the P4 space group and have cell dimensions of a = b = 104.571 and c = 30.936 Å.
Structure Determination.
The structure of Orc6-M was solved by the method of SIRAS using the Pt derivative data. SHELXD (41) as part of the HKL2MAP (42) package was used to locate platinum atom positions and three heavy atoms were found in one asymmetric unit. Phasing and initial model building was carried out by using PHENIX (43). The initial model contains 176 (136 with side-chain) residues. Model building and manual adjustment of the model were carried out using the program COOT (44), and the model was refined by PHENIX and Refmac5 (45). The final model contained 276 residues (with three monomers in one asymmetric unit) and had an Rwork factor of 22.6% and an Rfree factor of 26.6% (see Table 1 for detailed statistics). Stereochemical quality of the structure was also checked by using PROCHECK (46). Of all the residues, 91.2% locate in the favorite region and none in the disallowed region. Structural figures were prepared using PyMOL.
DNA Binding Assays.
EMSA reactions were performed essentially as described (19, 20). Each reaction contained approximately 50 ng of purified Orc6 protein, 50–200 ng of competitor (see figure legends for details), and approximately 1 ng of end-labeled specific DNA probes. Lamin B2 (520 bp) and ori-β (810 bp) fragments were used as specific probes in EMSA reactions. Reactions were set up on ice, incubated at room temperature for 30 min, and loaded on a 4% native polyacrylamide gel. The size of synthetic competitor DNAs (Amersham Biosciences) used in competition EMSA experiments was from 500 to 3,000 bp. Chromatin binding was performed as described previously (18).
DNA replication assay in Xenopus extracts was performed essentially as previously described (32, 33).
BrdU Incorporation and GFP–Orc6 Expression in L2 Cells.
Orc6 wild type and mutants were fused with GFP at the N terminus, subcloned into pMT/V5-B vector, and transfected into Drosophila L2 cells according to the manufacturer’s (Invitrogen) recommendations. The MT promoter was induced by 0.5 mM CuSO4 and cells were incubated with BrdU overnight at a final concentration of 10 μM. Cells were fixed with 2% formaldehyde, treated with DNase, incubated with anti-BrdU antibodies (Becton Dickinson), and subjected to immunofluorescent microscopy as described (19, 20).
GFP–Orc6 Expression in Salivary Glands.
Fused, wild-type GFP–Orc6 and GFP–Orc6 mutants were cloned under the UAS promoter into the pUAST vector and injected into w1118 fly embryos. Homozygous fly stocks were set up. To induce GFP–ORC expression, UAS-GFP-ORC flies were crossed to flies bearing GAL4 driven by sgs3 promoter (Bloomington stock w1118; P{w+mC = Sgs3-GAL4·PD}TP1). Sgs3 promoter induces GAL4 in the salivary glands of third instar larvae. Salivary glands of these larvae were dissected in PBS, stained with DAPI, and mounted in 60% glycerol, 4% N-propyl-gallate. Slides were sealed and live images were taken using a fluorescent microscope within 2 h. Immunostaining of chromosomes was performed as described (19).
Supplementary Material
Acknowledgments.
We thank R.-M. Xu for advice and valuable discussions and J. Zhou for assistance in data collection and structure determination. This work was supported by the Ministry of Science and Technology 863 Project (2006AA02A314), Ministry of Science and Technology 973 Project (2007CB914303 and 2011CB910304), National Science Foundation of China Grants 30925011 and 31030024 (to Y.L.), and by National Institutes of Health Grants GM69681, GM69681-S1, and GM97052 (to I.N.C.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and diffraction data have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3M03).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013676108/-/DCSupplemental.
References
- 1.Bell SP, Kobayashi R, Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262:1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- 2.Bell SP. The origin recognition complex: From simple origins to complex functions. Genes Dev. 2002;16:659–672. doi: 10.1101/gad.969602. [DOI] [PubMed] [Google Scholar]
- 3.Machida YJ, Hamlin JL, Dutta A. Right place, right time, and only once: Replication initiation in metazoans. Cell. 2005;123:13–24. doi: 10.1016/j.cell.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 4.DePamphilis ML. Cell cycle dependent regulation of the origin recognition complex. Cell Cycle. 2005;4:70–79. doi: 10.4161/cc.4.1.1333. [DOI] [PubMed] [Google Scholar]
- 5.Chesnokov IN. Multiple functions of the origin recognition complex. Int Rev Cytol. 2007;256:69–109. doi: 10.1016/S0074-7696(07)56003-1. [DOI] [PubMed] [Google Scholar]
- 6.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 7.Diffley JF. Regulation of early events in chromosome replication. Curr Biol. 2004;14:R778–786. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 8.Chastain PD, 2nd, Bowers JL, Lee DG, Bell SP, Griffith JD. Mapping subunit location on the Saccharomyces cerevisiae origin recognition complex free and bound to DNA using a novel nanoscale biopointer. J Biol Chem. 2004;279:36354–36362. doi: 10.1074/jbc.M403501200. [DOI] [PubMed] [Google Scholar]
- 9.Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol. 2005;12:965–971. doi: 10.1038/nsmb1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z, et al. The architecture of the DNA replication origin recognition complex in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2008;105:10326–10331. doi: 10.1073/pnas.0803829105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarey MG, Botchan M, Nogales E. Single particle EM studies of the Drosophila melanogaster origin recognition complex and evidence for DNA wrapping. J Struct Biol. 2008;164:241–249. doi: 10.1016/j.jsb.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li JJ, Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- 13.Lee DG, Bell SP. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol Cell Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhar SK, Dutta A. Identification and characterization of the human ORC6 homolog. J Biol Chem. 2000;275:34983–34988. doi: 10.1074/jbc.M006069200. [DOI] [PubMed] [Google Scholar]
- 15.Vashee S, Simancek P, Challberg MD, Kelly TJ. Assembly of the human origin recognition complex. J Biol Chem. 2001;276:26666–26673. doi: 10.1074/jbc.M102493200. [DOI] [PubMed] [Google Scholar]
- 16.Gillespie PJ, Li A, Blow JJ. Reconstitution of licensed replication origins on Xenopus sperm nuclei using purified proteins. BMC Biochem. 2001;2:15. doi: 10.1186/1471-2091-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vashee S, et al. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003;17:1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chesnokov I, Remus D, Botchan M. Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc Natl Acad Sci USA. 2001;98:11997–12002. doi: 10.1073/pnas.211342798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balasov M, Huijbregts RP, Chesnokov I. Role of the Orc6 protein in origin recognition complex-dependent DNA binding and replication in Drosophila melanogaster. Mol Cell Biol. 2007;27:3143–3153. doi: 10.1128/MCB.02382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balasov M, Huijbregts R, Chesnokov I. Functional analysis of an Orc6 mutant in Drosophila. Proc Natl Acad Sci USA. 2009;106:10672–10677. doi: 10.1073/pnas.0902670106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semple JW, et al. An essential role for Orc6 in DNA replication through maintenance of pre-replicative complexes. EMBO J. 2006;25:5150–5158. doi: 10.1038/sj.emboj.7601391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S, de Vries MA, Bell SP. Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2–7 loading. Genes Dev. 2007;21:2897–2907. doi: 10.1101/gad.1596807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasaki T, Gilbert DM. The many faces of the origin recognition complex. Curr Opin Cell Biol. 2007;19:337–343. doi: 10.1016/j.ceb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 24.Tsai FT, Sigler PB. Structural basis of preinitiation complex assembly on human pol II promoters. EMBO J. 2000;19:25–36. doi: 10.1093/emboj/19.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosa PF, Ghosh G, DeDecker BS, Sigler PB. The 2.1-A crystal structure of an archaeal preinitiation complex: TATA-box-binding protein/transcription factor (II)B core/TATA-box. Proc Natl Acad Sci USA. 1997;94:6042–6047. doi: 10.1073/pnas.94.12.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng W, Roberts S. TFIIB and the regulation of transcription by RNA polymerase II. Chromosoma. 2007;116:417–429. doi: 10.1007/s00412-007-0113-9. [DOI] [PubMed] [Google Scholar]
- 27.Prasanth SG, Prasanth KV, Stillman B. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science. 2002;297:1026–1031. doi: 10.1126/science.1072802. [DOI] [PubMed] [Google Scholar]
- 28.Thomae AW, et al. Interaction between HMGA1a and the origin recognition complex creates site-specific replication origins. Proc Natl Acad Sci USA. 2008;105:1692–1697. doi: 10.1073/pnas.0707260105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikolov DB, et al. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 30.Remus D, Beall EL, Botchan MR. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC–DNA binding. EMBO J. 2004;23:897–907. doi: 10.1038/sj.emboj.7600077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee DG, Makhov AM, Klemm RD, Griffith JD, Bell SP. Regulation of origin recognition complex conformation and ATPase activity: Differential effects of single-stranded and double-stranded DNA binding. EMBO J. 2000;19:4774–4782. doi: 10.1093/emboj/19.17.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- 33.Ranjan A, Gossen M. A structural role for ATP in the formation and stability of the human origin recognition complex. Proc Natl Acad Sci USA. 2006;103:4864–4869. doi: 10.1073/pnas.0510305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chesnokov IN, Chesnokova ON, Botchan M. A cytokinetic function of Drosophila ORC6 protein resides in a domain distinct from its replication activity. Proc Natl Acad Sci USA. 2003;100:9150–9155. doi: 10.1073/pnas.1633580100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beall EL, et al. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature. 2002;420:833–837. doi: 10.1038/nature01228. [DOI] [PubMed] [Google Scholar]
- 36.Bosco G, Du W, Orr-Weaver TL. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat Cell Biol. 2001;3:289–295. doi: 10.1038/35060086. [DOI] [PubMed] [Google Scholar]
- 37.Shen Z, et al. A WD-repeat protein stabilizes ORC binding to chromatin. Mol Cell. 2010;40:99–111. doi: 10.1016/j.molcel.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacAlpine DM, Rodriguez HK, Bell SP. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacAlpine DM, Bell SP. A genomic view of eukaryotic DNA replication. Chromosome Res. 2005;13:309–326. doi: 10.1007/s10577-005-1508-1. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 41.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 42.Pape T, Schneider T. HKL2MAP: A graphical user interface for macromolecular phasing with SHELX programs. J Applied Crystallogr. 2004;37:843–844. [Google Scholar]
- 43.Adams P, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 46.Laskowski R, MacArthur M, Moss D, Thornton J. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.