Abstract
Nucleosome remodeling complexes comprise several large families of chromatin modifiers that integrate multiple epigenetic control signals to play key roles in cell type-specific transcription regulation. We previously isolated a methyl-binding domain protein 2 (MBD2)-containing nucleosome remodeling and deacetylation (NuRD) complex from primary erythroid cells and showed that MBD2 contributes to DNA methylation-dependent embryonic and fetal β-type globin gene silencing during development in vivo. Here we present structural and biophysical details of the coiled-coil interaction between MBD2 and p66α, a critical component of the MBD2–NuRD complex. We show that enforced expression of the isolated p66α coiled-coil domain relieves MBD2-mediated globin gene silencing and that the expressed peptide interacts only with a subset of components of the MBD2–NuRD complex that does not include native p66α or Mi-2. These results demonstrate the central importance of the coiled-coil interaction and suggest that MBD2-dependent DNA methylation-driven gene silencing can be disrupted by selectively targeting this coiled-coil complex.
Keywords: epigenetics, gene regulation
DNA methylation involves the enzymatic addition of a methyl group at the C5 position of symmetrically opposed cytosine bases in a double-stranded cytosine-guanosine sequence (CpG). Regions of increased CpG content (CpG islands) often are found associated with promoters and, when methylated, silence expression of the associated gene (1, 2). Although most CpG islands are largely unmethylated in normal adult tissues, a subset of CpG islands is methylated in specific tissue subtypes, stages of differentiation, and development. Importantly, hypermethylation and silencing of tumor suppressor genes represents a pro-oncogenic change found in a wide range of malignancies (3). These observations have raised interest in DNA methylation as both an important genetic regulatory mechanism and a potential therapeutic target for either re-expression of developmentally silenced genes or reversing tumor suppressor gene silencing in cancer (4, 5).
The methyl cytosine binding proteins include a family that specifically recognizes the methylated CpG sequence through an ∼60 amino acid methyl-binding domain (MBD). There are five members of the MBD family in mammals: methyl CpG-binding protein 2 (MeCP2), the first to be identified (6), and MBD1 through MBD4 (7). We and others have isolated and characterized an MBD2-containing nucleosome remodeling and deacetylation (NuRD) complex (referred to as “MBD2–NuRD”) that binds methylated DNA and regulates transcription of the associated gene (8–10). The MBD2–NuRD complex comprises at least one homolog of six core proteins: MBD2, retinoblastoma protein-associated protein (RbAp46 or -48) Mi-2(α or β), p66(α or β), histone deacetylase (HDAC1 or 2), and metastasis associated (MTA1 or -2) (Fig. 1A). However, the specific interactions involved in the formation of the MBD2–NuRD complex have not been delineated clearly; information that is key to understanding (i) how and in what stoichiometric ratios of the different proteins the complex forms, (ii) the functional roles of each of the component proteins, and (iii) how to target specific protein–protein interactions effectively for therapeutic benefit.
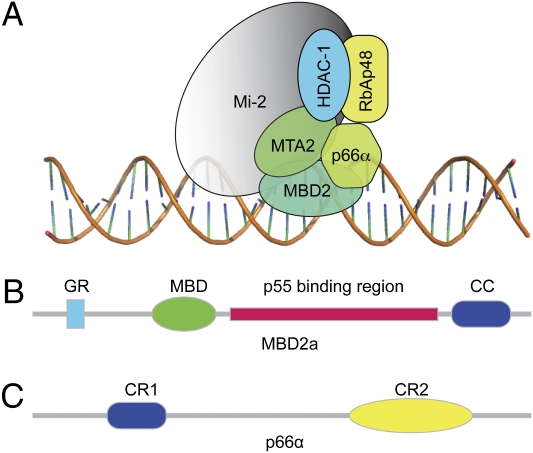
Fig. 1.
MBD2–NuRD complex. (A) Schematic representation of the six core components found in the MBD2–NuRD complex. (B) Domain organization of MBD2a. The p55 binding region was identified first in Drosophila as interacting with RbAp46/48 homolog p55; CC, ∼30 amino acids predicted to form a coiled coil; GR, glycine-arginine repeat region; MBD, methyl-binding domain. (C) Domain organization of p66α. CR1 is a coiled-coil domain; CR2 includes a GATA zinc finger domain.
In the work presented here, we investigated the interaction between MBD2 and p66α, critical components of the MBD2–NuRD complex. The human MBD2a protein (411 amino acids) contains an N-terminal glycine-arginine repeat region, a methyl-binding domain, a putative transcription repression domain, a region implicated in binding the RbAp46/48 homolog p55 in Drosophila (11), and a coiled-coil region (Fig. 1B). The p66α/β proteins are 66-kDa transcriptional repressors found in NuRD complexes that contain two highly conserved regions: an N-terminal coiled-coil domain (CR1) and a C-terminal GATA-like zinc finger domain (CR2) (Fig. 1B) (12, 13). Previous work has shown that the CR1 region of p66α is responsible for association with the MBD2–NuRD complex (12) and directly binds MBD2 (13).
We show that the coiled-coil regions from p66α and MBD2 proteins in isolation are largely monomeric helices that bind to form a stable heterodimeric complex in solution. The structure of this complex reveals a canonical anti-parallel coiled-coil with unique ionic/hydrogen bonding interactions that provide specificity to the interaction. Based on this structure, we show that the p66α coiled-coil peptide can bind to the native MBD2 protein in cells and functionally relieve MBD2-mediated gene silencing in tissue culture models of embryonic/fetal globin regulation. Further, we show that the isolated p66α coiled-coil peptide interacts with a subset of the MBD2–NuRD complex lacking the native p66α and Mi-2 components. These results suggest a model for MBD2–NuRD silencing of embryonic/fetal globin genes and show that blocking the native MBD2–p66α coiled-coil interaction can be used to relieve MBD2-dependent gene silencing.
Results
p66α and MBD2 Coiled-Coil Domains Form a Stable Heterodimeric Complex.
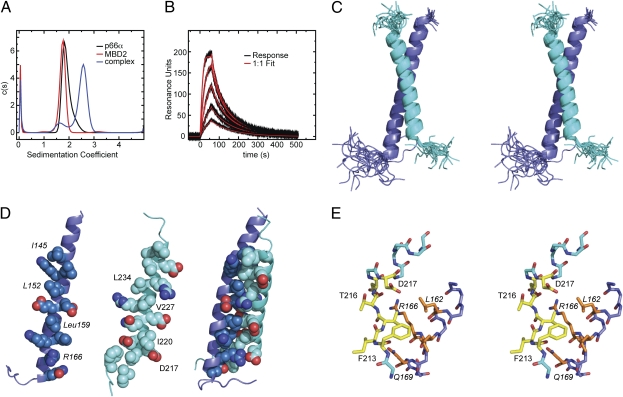
After cloning and expressing the coiled-coil domains from p66α and MBD2, we sought to establish whether these domains interact either to form homomeric complexes or with each other to form a heteromeric complex. When mixed at a 1:1 molar ratio, the p66α and MBD2 coiled-coil domains elute as a single peak earlier from gel filtration chromatography than either protein alone (Fig. S1). By analytical ultracentrifugation analysis (Fig. 2A), each protein behaves as a monomer in solution; when mixed, the two proteins bind to form a heterodimeric complex: MBD2 = 24.6 kDa observed based on sedimentation velocity analysis (22.0 kDa expected); p66α = 24.1 kDa observed (23.0 kDa expected); MBD2–p66α = 43.5 kDa observed (45.0 kDa expected). Surface plasmon resonance analysis of p66α coiled-coil binding to MBD2 could be fit quite well by a 1:1 binding model (Fig. 2B) with a relatively rapid on rate and slow off rate resulting in a high-affinity interaction (Kd = 12.4 nM, association rate constant (ka) = 2.4 × 106 M−1 s−1, dissociation rate constant (kd) = 0.031 s−1).
Fig. 2.
Binding analysis and solution structure of the p66α–MBD2 coiled-coil complex. (A) Analytical ultracentrifugation was performed on p66α and MBD2 coiled-coil domains individually and as a 1:1 molar mixture. The sedimentation velocity was fit using a continuous size distribution [c(s)], and the effective molecular weight was determined from the resulting sedimentation coefficients using SEDFIT software (sedfitsedphat.nibib.nih.gov). (B) Binding kinetics were analyzed for p66α coiled-coil binding to MBD2 coiled-coil coupled to a Sensor Chip CM5 on a Biacore T100 instrument (GE Healthcare). The data were fit to a 1:1 binding model using Biacore evaluation software. (C) Stereo view of the aligned 20 lowest-energy structures is shown as cartoon representations of p66α (blue) and MBD2 (cyan) coiled-coil domains. (D) The “knobs” and “holes” of the p66α and MBD2 coiled domain are shown using a sphere representation for each chain individually, with the contact surface facing the viewer, and as a complex. For reference, “knobs” and select residues are labeled. (E) Stereo view stick representation is shown for the main-chain atoms encompassing the N-terminal region extended to helical transition of MBD2 and the contacting C-terminal region of p66α. Key residues from MBD2 (yellow) and p66α (orange) are shown and labeled (p66α in italics). The figure was generated using the Pymol program (Delano Scientific LLC).
Upon binding, both proteins show broadened line widths and widespread chemical shift changes leading to improved dispersion in the 2D 15N-heteronuclear single quantum coherence (HSQC) spectra. An NMR titration experiment shows evidence of slow exchange between the bound and free states (Fig. S1). For both the isolated monomeric peptides and the complex, the 15N-NOESY HSQC spectra contain strong NOE cross-peaks characteristic of helical peptides [HN-HN(i ± 1) and HN-Hα(i-3)], and chemical shift analysis predicts that each is well structured and adopts helical ϕ/ψ backbone torsion angles (as predicted by the program TALOS+; Fig. S1) (14). Circular dichroism measurements show that the isolated monomeric peptides, p66α in particular, and the complex adopt a helical secondary structure (p66α, 72%; MBD2, 27%; MBD2–p66α complex, 60%; Fig. S1). As expected for coiled-coil complexes, the MBD2–p66α complex undergoes a cooperative unfolding transition that, when fit to a simple two-state thermodynamic model (15), yields a melting temperature (Tm) of 65.1 °C and enthalpy of unfolding transition (ΔHm) of 33.2 kcal/mol. Interestingly, as free peptides, p66α and MBD2 appear to undergo a similar cooperative transition at lower temperatures (p66α: Tm = 31.6 °C, ΔHm = 18.3 kcal/mol; MBD2: Tm = 13.0 °C, ΔHm = 13.2 kcal/mol). Taken together, these observations support a model in which the isolated p66α and MBD2 coiled-coil domains are largely preformed monomeric helices that interact to form a stable heterodimeric complex.
Solution Structure of the MBD2–p66α Coiled-Coil Complex.
The solution structure of the heterodimeric complex was well defined (Table S1), with a backbone rmsd for the ordered residues of 0.45 Å. The solvent-exposed residues contribute to a relatively large rmsd for all heavy atoms from the same region (1.2 Å), whereas the interface residues are well defined (backbone plus interface heavy atoms rmsd of 0.93 Å). The complex between MBD2 and p66α adopts an anti-parallel coiled-coil structure (Fig. 2 C–E) in which MBD2 residues Asp217-Met239 and p66α residues Pro138-Ile170 (p66α residues are italicized throughout the text and figures) form stable α-helices upon binding. The average interhelix angle is 154.1 ± 1.0° with an average interhelix distance of 9.3 ± 0.3 Å [averaged over the 20 lowest energy structures using the program interhlx (16)].
The helices interact through a series of highly conserved hydrophobic side chains to form the classic “knobs” (p66α: Ile145, Leu152, Leu159; MBD2: Ile220, Val227, Leu234) that fit into “holes” (p66α: between residues Leu148 and Lys149, Glu155 and Glu156, and Leu162 and Lys163; MBD2: between residues Gln223 and Glu224, Val230 and Arg231, and Ala237 and Leu238) in the arrangement of an anti-parallel coiled-coil (Fig. 2C). The interaction buries a total of 1,337 Å2 of solvent accessible surface area, 648 Å2 from MBD2 and 690 Å2 from p66α [as calculated using AREAIMOL from the CCP4 Software Suite (17, 18)]. In addition to the canonical hydrophobic packing, several close ionic interactions are formed that probably contribute to the specificity of this interaction. The hole formed between p66α Glu155 and Glu156 residues permits side-chain ionic interactions with MBD2 Arg226 and Arg231, respectively. Another close ionic interaction forms between p66α Lys163 and MBD2 Glu224. The pattern of hydrophobic knobs protruding from p66α terminates with the charged residue Arg166 (Fig. 2D), which forms a close ionic interaction and potential hydrogen bonding with Asp217 of MBD2.
The ionic interaction between Arg166 and Asp217 marks a transition from the extended structure to a helical secondary structure for MBD2. This transition is stabilized by a classic helix N-cap formed by a side-chain hydrogen bond between Thr216 and the backbone amide of Asp219, which in turn can form a side-chain hydrogen bond with the backbone amide of Thr216 (Fig. 2E). The extended N-terminal sequence allows Phe213 to interact with the hydrophobic Leu162 and Leu165 residues on the exposed side of p66α. The side chains of Val215 and Leu162 interact, and the side chain of Gln169 can form hydrogen bonds with the backbone of Val215 to stabilize this extended conformation of MBD2.
To test the relative importance of the different contacting residues, a series of truncation and point mutants of the p66α coiled-coil domain were generated and evaluated for binding to MBD2 coiled coil by gel filtration chromatography. Fragments incorporating either N- (residues 144–158) or C-terminal (residues 152–168) portions of p66α failed to form stable complexes with the MBD2 coiled coil. On the other hand, the central 25 amino acids (residues 144–168), incorporating most of the intermolecular contacts, does bind to form a stable complex (Kd ∼72 nM; Fig. S2). Mutating Arg166 to Glu or both Glu155 and Glu156 to Arg did not completely abolish binding as assayed by gel filtration chromatography. The affinity of these mutants was determined by surface plasmon resonance, which showed a marked decrease of almost three orders of magnitude for both Arg166Glu and Glu155Arg/Glu156Arg p66α mutations (Arg166Glu KD ∼8 μM, Glu155Arg/Glu156Arg Kd ∼17 μM; Fig. S2). The Arg166Glu/Glu155Arg/Glu156Arg triple mutant fails to bind MBD2 coiled coil even at a protein concentration of 30 μM (Fig. S2). Together, these findings confirm the importance of these ionic interactions as well as the intact full-length coiled-coil domain.
The key contacting residues are highly conserved across species and among homologous proteins for both MBD2 and p66α (Fig. S3). In particular, residues involved in stabilizing the N-cap of MBD2 and marking the transition from extended to helical structure are conserved from Drosophila to human and include both MBD2 and MBD3. The central glutamate residues from p66α (Glu155 and Glu156) are highly conserved, as are the interacting arginine residues Arg226 and Arg231. This high degree of conservation suggests the homologous proteins should form stable heterodimeric complexes in a similar manner. Accordingly, gel filtration analysis shows that the coiled-coil regions from MBD3, MBD3L1, and MBD3L2 all form stable complexes with the p66α coiled coil (Fig. S2). In contrast, the coiled-coil domains from MBD2 and MBD3 do not form a stable complex by gel filtration analysis (Fig. S2).
Expression of the p66α Coiled-Coil Domain Mimics Knockdown of MBD2 in Augmenting Fetal/Embryonic Globin Expression in Adult Erythroid Cell Tissue Culture.
Based on the structure of this stable, high-affinity coiled-coil complex, we postulated that enforced expression of the coiled-coil domain from p66α could competitively disrupt the native MBD2–NuRD complex and interfere with MBD2-mediated gene silencing. We have shown previously that MBD2 contributes to silencing of both chicken embryonic ρ-globin gene expression (8, 19) and human fetal γ-globin gene expression in a human β-globin yeast artificial chromosome (β-YAC) transgenic mouse model (20). An MBD2–NuRD complex was isolated from primary chicken erythroid cells, and MBD2 and other components of this complex were shown to bind and silence a methylated ρ-globin gene in stably transfected murine erythroid cells; furthermore, MBD2 was shown to bind to the methylated and silenced ρ-globin gene in primary erythroid cells (8). Accordingly, we chose to study the effects of overexpressing the p66α coiled-coil domain in corresponding tissue culture models of both chicken ρ-globin expression in MEL cells (8) and human γ-globin expression in murine hematopoietic chemical inducer of dimerization (CID) cells containing β-YAC (21). Notably, although MBD2 binds directly to the silent chicken embryonic ρ-globin locus (and hence has a direct effect) (8), it does not appear to bind directly to the human β-globin gene locus (suggesting an indirect effect) (20).
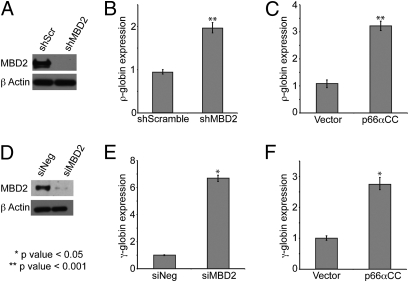
MEL-ρ is a modified mouse erythroleukemia (MEL) cell line that carries a stably integrated methylated chicken ρ-globin minilocus flanked by chicken HS-4 insulator elements. This locus has been identified previously as a direct binding target for an MBD2–NuRD complex, thus making it a valid reporter for MBD2-dependent gene silencing (8). Consistent with previous observations, stable knockdown of MBD2 by shRNA (Fig. 3A) leads to a twofold increase in ρ-globin expression (Fig. 3B). A pCMV-Tag2B vector containing the coding sequence for a Flag-tagged p66α coiled-coil domain and an empty vector control were transfected into MEL-ρ cells in which the methylation of the ρ-globin gene had been confirmed by bisulfite sequencing of the stably integrated gene (Fig. S4) and stable transfectants had been isolated. After transfection, both p66α coiled-coil mRNA and protein were expressed in vivo as detected by semiquantitative RT-PCR and Western dot blot analyses, respectively (Fig. S4). The small size of the peptide expressed, ∼4 kDa, hindered its detection using standard Western blot. Western dot blot, which has been shown to have 4,000-fold higher sensitivity for small peptides (22), was used as an alternative to detect expression. In the presence of the p66α coiled-coil peptide, embryonic ρ-globin gene expression was increased 2.5-fold compared with clones that expressed the vector-derived Flag peptide alone (Fig. 3C). Thus, the degree of ρ-globin gene de-repression in the presence of the p66α coiled-coil domain peptide was equivalent to that in cells in which MBD2 was stably knocked down by >90%.
Fig. 3.
The p66α coiled-coil peptide augments fetal/embryonic globin expression. (A–C) shMBD2 and shScramble pSuperior vector (n = 6), p66α-pCMVTag2B plasmid (n = 3), and an empty vector control (n = 3) were stably transfected into MEL-ρ cells. (D–F) Negative control (siNeg) or MBD2-specific (siMBD2) siRNA, p66α-pCMVTag2B plasmid, and an empty vector control (n = 3) were transiently transfected into CID β-YAC bone marrow progenitor cells. (A and D) Western blot analysis shows efficient knockdown of MBD2. qPCR-determined relative γ-globin mRNA levels show that MBD2 knockdown (B and E) and enforced expression of p66α coiled-coil domain (C and F) augment ρ-globin and γ-globin gene expression. Error bars indicate SE.
To extend this observation to human γ-globin gene regulation, similar studies were carried out in CID-dependent mouse bone marrow cells carrying β-YAC (21). The human γ-globin gene is highly repressed in these adult phenotype erythroid cells, which express high levels of β-globin RNA. MBD2 siRNA treatment reduced expression of MBD2 in CID cells by ∼80% (Fig. 3D) accompanied by a sevenfold increase in human γ-globin RNA expression (Fig. 3E), without affecting expression of the erythroid-specific α-globin, spectrin, and aminolevulinic acid dehydratase (ALAD) genes (Fig. S5). Knockdown of MBD2 in CD34+ precursor-derived primary human erythroid cells resulted in an ∼fivefold increase in γ-globin gene expression (Fig. S5), supporting the validity of the CID cell model and the importance of the MBD2–NuRD complex in autonomous silencing of this gene in normal adult erythroid cells. Transient expression of the p66α coiled-coil domain, as monitored using semiquantitative PCR and Western dot blot (Fig. S4), consistently yielded a 2.5- to 3.0-fold increase in γ-globin expression, but expression of the p66α coiled-coil triple-mutant peptide, which fails to bind MBD2, did not augment expression of γ-globin (Fig. 3F, and Fig. S5). Enforced expression of these same peptides does not appreciably affect expression of the α-globin, β-globin, spectrin, or ALAD erythroid-specific genes (Fig. S5). These results demonstrate that the isolated p66α coiled-coil domain can disrupt MBD2-mediated gene silencing in a model of developmental human fetal γ-globin gene regulation.
Enforced Expression of the p66α Coiled-Coil Domain Disrupts the Recruitment of Endogenous p66α and Mi-2 to the MBD2–NuRD Complex.
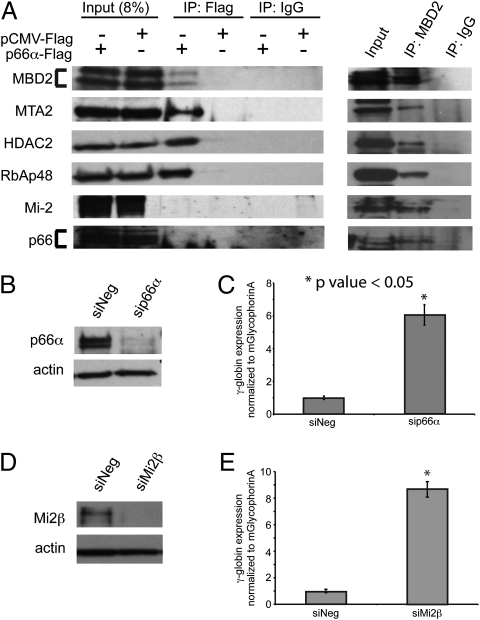
Studies, including our own (8), suggest that MBD2–NuRD complexes exist as relatively stable preformed complexes in the cell nucleus. To test for interactions of the p66α coiled-coil domain with MBD2 and the other components of the MBD2–NuRD complex in intact cells, the pCMV-Tag2B vector containing the p66α coiled-coil sequence and an empty vector control were transfected into high-transfection-efficiency 293T cells. Immunoprecipitation with anti-Flag antibody coprecipitated native MBD2a, MTA2, HDAC2, and RbAp48, but neither p66α/β nor Mi-2α/β was detected by Western blot analysis (Fig. 4A). These results confirm that the expressed p66α peptide stably interacts with native MBD2a and select components of NuRD in vivo. Immunoprecipitation with anti-MBD2 antibody from untransfected 293T cells was used as a positive control, and, as expected, all major components of the MBD2–NuRD complex were detected by Western blot analysis (Fig. 4A). Immunoprecipitation of the p66α coiled-coil domain also coprecipitated MBD3, as one would predict from the high degree of homology and our binding studies (see above). However, knockdown of MBD3 by ∼75% did not augment γ-globin expression in CID-dependent β-YAC bone marrow progenitor cells (Fig. S6), indicating that this interaction is not functionally relevant to globin gene silencing in this model system. In contrast, knockdown of p66α or Mi-2β augments expression of human γ-globin mRNA in the CID-dependent β-YAC bone marrow progenitor cells. Transient knockdown of p66α by ∼80% was accompanied by a sixfold increase in γ-globin mRNA (Fig. 4 B and C), and transient knockdown of Mi-2β by ∼80% resulted in an eightfold induction of γ-globin mRNA in these cells (Fig. 4 D and E). Together, these results demonstrate that the isolated p66α coiled-coil domain can disrupt gene silencing by blocking the recruitment of native p66α/β and Mi-2β to an MBD2-containing MBD2–NuRD complex.
Fig. 4.
The p66α coiled-coil domain disrupts recruitment of endogenous p66α and Mi-2 to the MBD2–NuRD complex. The p66α-pCMVTag2B plasmid and an empty vector control were transiently transfected into high-transfection-efficiency 293T cells. (A) Immunoprecipitation and Western blot analysis of transfected cells (anti-Flag, anti-MBD2, and IgG control antibodies) show that native MBD2a, MTA2, HDAC2, and RbAp48 coprecipitate with the Flag-p66α coiled-coil domain and MBD2, whereas p66α/β and Mi-2 coprecipitate with MBD2 but not with the Flag–p66α coiled-coil domain. (B–E) Negative control (siNeg), p66α-specific (sip66α), and Mi-2β-specific (siMi-2β) siRNA (n = 3) were transiently transfected into CID β-YAC bone marrow progenitor cells. Western blot analyses show efficient knockdown of both p66α (B) and Mi-2β (D) protein. qPCR analysis shows that p66α knockdown (C) and Mi-2β knockdown (E) augment γ-globin mRNA levels. Error bars represent SE.
Discussion
We previously isolated and characterized an MBD2-containing NuRD complex from a primary erythroid cell source and showed that MBD2 plays an important role in silencing transcription of both the embryonic ρ-globin gene in chickens and the fetal γ-globin gene in human β-YAC transgenic mice (8, 20). This complex has been implicated by a number of laboratories in binding to methylated DNA and silencing transcription of synthetic gene constructs (9). We also have shown that DNA methylation-mediated silencing of the embryonic ρ-globin gene by MBD2 is not dependent on histone deacetylase activity (19, 23). Similarly, it has been shown that histone deacetylase inhibitors alone do not relieve silencing of the γ-globin gene in adult β-YAC transgenic mice (24). To understand further the structure of the MBD2–NuRD complex and its function in globin gene silencing, we have studied the interaction between p66α and MBD2 proteins in the complex. These experiments have provided molecular details of the stable anti-parallel coiled-coil complex between MBD2 and p66α, details that led to in vivo studies of the p66α coiled-coil peptide as a potential inhibitor of MBD2 function. The latter investigation revealed a critical role for Mi-2β in globin gene silencing.
NMR analysis of the p66α-MBD2 coiled-coil complex shows that each protein adopts a regular helical structure to form a canonical anti-parallel coiled-coil interface with an observed disassociation constant in the low nanomolar region (12.4 nM). Although coiled-coil complexes can be promiscuous, forming a variety of oligomeric states and anti-parallel vs. parallel arrangements, the p66α and MBD2 coiled-coil domains behave largely as helical monomers in solution only to form a stable heterodimer when mixed (25). This observation argues against the idea that MBD2 and MBD3 proteins form homodimeric complexes and/or bind to one another through their coiled-coil domains and supports the observation that MBD2 and MBD3 form mutually exclusive NuRD complexes with unique functions (26).
A series of highly conserved close ionic interactions stabilize the p66α–MBD2 interaction and probably contribute to the specificity of binding. These residues are conserved across species and among different homologs in the same species. Interestingly, a conserved arginine residue disrupts the hydrophobic surface of p66α, which coincides with an extended N-terminal conformation of MBD2 developing several stabilizing hydrophobic and ionic/polar interactions. This unique feature of the p66α–MBD2 coiled-coil complex should preclude interaction with other amphipathic helices, permitting a high degree of specificity for what otherwise is a fairly common binding motif.
Because p66α and MBD2 bind through a small peptide complex involving two stable helices, we postulated that the p66α coiled-coil domain in isolation could bind native MBD2 and function as a competitive inhibitor. As predicted, enforced expression of this domain in two different cell models of globin gene regulation augmented fetal/embryonic globin gene expression to a similar degree as knockdown of MBD2. Immunoprecipitation shows that the p66α peptide interacts with MBD2a in vivo as well as with other components of MBD2–NuRD such as RbAp46, MTA2, HDAC1, and MBD2. However, recruitment of p66α/β and Mi-2 proteins to the MBD2–NuRD complex was disrupted by the p66α coiled-coil peptide. Likewise knockdown of either p66α or Mi-2 augments human γ-globin expression to at least the same extent as knockdown of MBD2. These results are consistent with the previous findings that treatment with HDAC inhibitors does not relieve silencing of the chicken embryonic ρ-globin gene (19, 23) or the human γ-globin gene in β-YAC transgenic mice (24), indicating that DNA methylation-mediated gene silencing does not depend on the enzymatic function of HDAC proteins in the MBD2–NuRD complex. These findings led to several interesting conclusions: (i) the MBD2–p66α coiled-coil interaction is functionally important for silencing transcription; (ii) the remaining sequence of p66α performs a critical role in the MBD2–NuRD complex (i.e., recruitment of Mi-2 which in turn is critical for globin gene silencing); and (iii) specifically targeting the coiled-coil interaction can block MBD2 function, presumably by interfering with the formation of its intact associated NuRD complex containing the Mi-2 chromatin-remodeling protein.
These studies have led us to propose a model of how the p66α coiled-coil domain inhibits function of the MBD2–NuRD complex (Fig. 5). The isolated coiled-coil domain of p66α binds competitively to MBD2, preventing recruitment of full-length native p66α and Mi-2 into the MBD2–NuRD complex. Because MBD2 contains the methylated DNA-binding domain, this p66α- and Mi-2–deficient complex still may bind methylated DNA and in this manner could act as a dominant negative inhibitor of binding by an intact functional MBD2–NuRD. Alternatively, the p66α coiled-coil peptide may sequester the majority of MBD2 into a nonfunctional complex, thus preventing formation of sufficient MBD2–NuRD to bind and silence methylated gene targets. This model explains how a small peptide sequence can effectively block gene silencing by the large native MBD2–NuRD complex. The results here also point to a critical role for Mi-2 and p66α in the MBD2-mediated silencing of embryonic/fetal globin genes in adult phenotype erythroid cells.
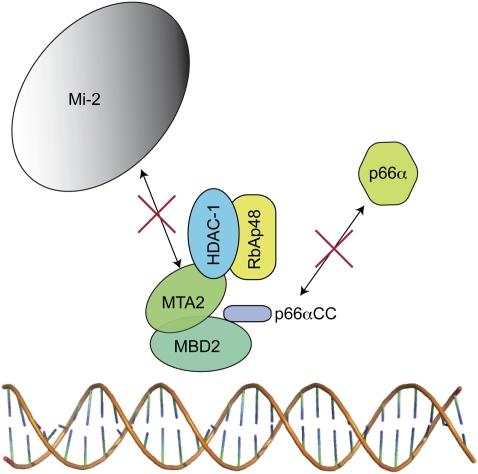
Fig. 5.
Model of the inhibition of the MBD2–NuRD complex by the p66α coiled-coil peptide. A schematic model showing that the p66α coiled-coil peptide binds to four of the six core components from MBD2–NuRD (MTA2, RbAp48, HDAC-1, and MBD2) but prevents recruitment of native p66α and Mi-2 proteins.
In summary, we have determined the structure of the p66α–MBD2 coiled-coil protein complex integral to the formation of the MBD2–NuRD complex. We have shown that the p66α coiled-coil domain can competitively inhibit function of this complex in vivo in cultured erythroid cells. Therefore, this coiled-coil interaction plays a functionally important role in silencing of globin genes through DNA methylation. This observation raises the possibility that targeting the p66α-MBD2 interaction could inhibit DNA methylation-dependent silencing as a therapeutic strategy for β-hemoglobinopathies and other diseases including cancer.
Methods
Structure and Binding Analyses.
The coiled-coil regions of human MBD2b and p66α were cloned and expressed as thioredoxin fusion proteins using a pET32a vector [modified to incorporate a thrombin cleavage site immediately N-terminal to the cloned sequence (27)] and purified using standard techniques. The p66α and MBD2 coiled-coil domains were analyzed by gel filtration chromatography, analytical ultracentrifugation, surface plasmon resonance, and circular dichroism. Standard NMR experiments for resonance assignments and structure determination, including residual dipolar coupling measurements, were collected on a Varian 500 MHz Unity+ spectrometer at 25 °C. The structure of the complex was calculated by simulated annealing using the Xplor-NIH (28) software package (SI Methods).
RNAi-Knockdown Studies.
Cells were transfected with 1 μM of MBD2, p66α, and Mi-2α and -β gene-specific siRNAs, and AllStars siNeg scramble control (Qiagen). RNA isolation, cDNA synthesis, and quantitative PCR (qPCR) were performed as described previously (20). For protein analysis, 2–4 × 106 cells were lysed with 4.5% SDS buffer containing protease inhibitor mixture (Pierce), and Western blot analysis was performed as described previously (SI Methods) (19).
Enforced Expression Studies.
The wild-type and triple-mutant (Arg166Glu/Glu155Arg/Glu156Arg) p66α coiled-coil domains (amino acids 138–178) were cloned into the pCMVTag2B (Stratagene) vector in frame with an N-terminal Flag-tag sequence. These vectors (including an empty vector control) were transfected into either CID β-YAC bone marrow progenitor cells or MEL-ρ cells, and expression was confirmed by PCR and dot blot analyses (SI Methods).
Immunoprecipitation.
293T cells were transfected with p66α-pCMV-Tag2B plasmid and empty vector control (10 μg) by the calcium phosphate precipitation method (29) and harvested at 48 h. Cells were lysed and immunoprecipitated with anti-Flag M2 agarose affinity gel (Sigma), anti-MBD2 (Santa Cruz), and mouse and goat IgG (Santa Cruz) controls according to the Sigma Flag-IPT kit protocol.
Primers and Antibodies.
Primer sequences and antibodies are provided in Tables S2 and S3.
Statistics.
All experiments were performed three or more times. Results are shown as mean ± SE. For comparison between data sets, unpaired two-tailed t tests were performed.
Supplementary Material
Acknowledgments
We thank Dan Conrad and Joel Mathews for assistance with surface plasmon resonance analysis, Ellis Bell for assistance with circular dichroism analysis and Ken Peterson and Tony Blau for the CID cells. This work was supported by a Junior Faculty Scholar Award from the American Society of Hematology (to D.C.W.), American Cancer Society Grant IRG-73-001 (to D.C.W.), and National Institutes of Health Grant R01 DK029902 (to G.D.G). NMR data were acquired using NMR instrumentation in the Virginia Commonwealth University Structural Biology Core, and surface plasmon resonance data were acquired in the Virginia Commonwealth University Flow Cytometry Core, both supported, in part, by funding from National Cancer Institute Cancer Center Core Support Grant P30 CA016059.
Footnotes
The authors declare no conflict of interest.
Data deposition: The coordinates for the p66α–MBD2 coiled-coil complex structural model have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2L2L). The NMR assignments have been deposited in the BioMagRes Bank, www.bmrb.wisc.edu (accession no. 17138).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015341108/-/DCSupplemental.
References
- 1.Illingworth RS, Bird AP. CpG islands—‘a rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 2.Straussman R, et al. Developmental programming of CpG island methylation profiles in the human genome. Nat Struct Mol Biol. 2009;16:564–571. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ginder GD, Gnanapragasam MN, Mian OY. The role of the epigenetic signal, DNA methylation, in gene regulation during erythroid development. Curr Top Dev Biol. 2008;82:85–116. doi: 10.1016/S0070-2153(07)00004-X. [DOI] [PubMed] [Google Scholar]
- 5.Brock MV, Herman JG, Baylin SB. Cancer as a manifestation of aberrant chromatin structure. Cancer J. 2007;13:3–8. doi: 10.1097/PPO.0b013e31803c5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 7.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kransdorf EP, et al. MBD2 is a critical component of a methyl cytosine-binding protein complex isolated from primary erythroid cells. Blood. 2006;108:2836–2845. doi: 10.1182/blood-2006-04-016394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, et al. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- 11.Marhold J, Brehm A, Kramer K. The Drosophila methyl-DNA binding protein MBD2/3 interacts with the NuRD complex via p55 and MI-2. BMC Mol Biol. 2004;5:20. doi: 10.1186/1471-2199-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Q, et al. Identification and functional characterization of the p66/p68 components of the MeCP1 complex. Mol Cell Biol. 2002;22:536–546. doi: 10.1128/MCB.22.2.536-546.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brackertz M, Gong Z, Leers J, Renkawitz R. p66α and p66β of the Mi-2/NuRD complex mediate MBD2 and histone interaction. Nucleic Acids Res. 2006;34:397–406. doi: 10.1093/nar/gkj437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koepf EK, Petrassi HM, Sudol M, Kelly JW. WW: An isolated three-stranded antiparallel β-sheet domain that unfolds and refolds reversibly; evidence for a structured hydrophobic cluster in urea and GdnHCl and a disordered thermal unfolded state. Protein Sci. 1999;8:841–853. doi: 10.1110/ps.8.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yap KL, Ames JB, Swindells MB, Ikura M. Vector geometry mapping. A method to characterize the conformation of helix-loop-helix calcium-binding proteins. Methods Mol Biol. 2002;173:317–324. doi: 10.1385/1-59259-184-1:317. [DOI] [PubMed] [Google Scholar]
- 17.Saff EB, Kuijlaars ABJ. Distributing many points on a sphere. The Mathematical Intelligencer. 1997;19:5–11. [Google Scholar]
- 18.Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 19.Singal R, Wang SZ, Sargent T, Zhu SZ, Ginder GD. Methylation of promoter proximal-transcribed sequences of an embryonic globin gene inhibits transcription in primary erythroid cells and promotes formation of a cell type-specific methyl cytosine binding complex. J Biol Chem. 2002;277:1897–1905. doi: 10.1074/jbc.M105580200. [DOI] [PubMed] [Google Scholar]
- 20.Rupon JW, Wang SZ, Gaensler K, Lloyd J, Ginder GD. Methyl binding domain protein 2 mediates γ-globin gene silencing in adult human betaYAC transgenic mice. Proc Natl Acad Sci USA. 2006;103:6617–6622. doi: 10.1073/pnas.0509322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blau CA, et al. γ-Globin gene expression in chemical inducer of dimerization (CID)-dependent multipotential cells established from human β-globin locus yeast artificial chromosome (β-YAC) transgenic mice. J Biol Chem. 2005;280:36642–36647. doi: 10.1074/jbc.M504402200. [DOI] [PubMed] [Google Scholar]
- 22.Duchesne L, Fernig DG. Silver and gold nanoparticle-coated membranes for femtomole detection of small proteins and peptides by Dot and Western blot. Anal Biochem. 2007;362:287–289. doi: 10.1016/j.ab.2006.12.022. [DOI] [PubMed] [Google Scholar]
- 23.Ginder GD, Whitters MJ, Pohlman JK. Activation of a chicken embryonic globin gene in adult erythroid cells by 5-azacytidine and sodium butyrate. Proc Natl Acad Sci USA. 1984;81:3954–3958. doi: 10.1073/pnas.81.13.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pace B, Li Q, Peterson K, Stamatoyannopoulos G. α-Amino butyric acid cannot reactivate the silenced γ gene of the β locus YAC transgenic mouse. Blood. 1994;84:4344–4353. [PubMed] [Google Scholar]
- 25.Steinmetz MO, et al. Molecular basis of coiled-coil formation. Proc Natl Acad Sci USA. 2007;104:7062–7067. doi: 10.1073/pnas.0700321104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Guezennec X, et al. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol. 2006;26:843–851. doi: 10.1128/MCB.26.3.843-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai M, et al. Solution structure of the phosphoryl transfer complex between the signal-transducing protein IIAGlucose and the cytoplasmic domain of the glucose transporter IICBGlucose of the Escherichia coli glucose phosphotransferase system. J Biol Chem. 2003;278:25191–25206. doi: 10.1074/jbc.M302677200. [DOI] [PubMed] [Google Scholar]
- 28.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 29.Kingston RE, Chen CA, Okayama H. Curr Protoc Cell Biol. 2003. Calcium phosphate transfection. 10.1002/0471143030.cb2003s19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.