Abstract
Hembase (http://hembase.niddk.nih.gov) is an integrated browser and genome portal designed for web-based examination of the human erythroid transcriptome. To date, Hembase contains 15 752 entries from erythroblast Expressed Sequenced Tags (ESTs) and 380 referenced genes relevant for erythropoiesis. The database is organized to provide a cytogenetic band position, a unique name as well as a concise annotation for each entry. Search queries may be performed by name, keyword or cytogenetic location. Search results are linked to primary sequence data and three major human genome browsers for access to information considered current at the time of each search. Hembase provides interested scientists and clinical hematologists with a genome-based approach toward the study of erythroid biology.
INTRODUCTION
Diseases involving erythroid cells afflict millions of people worldwide. Those clinical syndromes encompass all forms of anemia, erythroleukemia and malaria. Erythroid cells normally have the fundamental role of delivering oxygen from the lungs to the other body tissues. The production of red blood cells occurs by a process called erythropoiesis whereby erythroid progenitor cells proliferate and differentiate into erythroid precursor cells and mature erythrocytes. Normally, this process is highly dependent upon and regulated by a hormone produced by the kidneys called erythropoietin. Based on the mapping of the human genome and the development of information databases, a broad description of genes transcribed during human erythropoiesis is now known. Hembase was created to provide scientists and clinicians with genome-based access to that information.
HEMBASE ENTRIES
Hembase uses the FileMakerPro 5.0 database engine for the collection and organization of information regarding mRNA sequences of genes relevant for the study of erythroid biology. A total of 15 752 EST files derived from mRNA gene libraries from developmentally staged, primary human erythroblasts are included. Those EST were obtained from high-throughput sequencing (1) of three separate erythroblast libraries. Entries containing the ‘ad’ or ‘ax’ prefixes were obtained from highly purified populations of erythroid progenitor cells, and those identified with a ‘cl’ prefix from more mature erythroid precursor cells. Several hundred referenced genes described in the literature as having importance in erythroid biology have also been included to provide a more complete description of the human erythroid transcriptome. Each file entry contains information regarding the clone ID, GenBank accession numbers, nucleotide sequences and genomic location as defined by BLAST comparisons (2). Those BLAST comparisons with significant homology to RefSeq (3) mRNA sequences in GenBank are described using RefSeq annotation.
FORMATTING BY CYTOGENETIC BAND
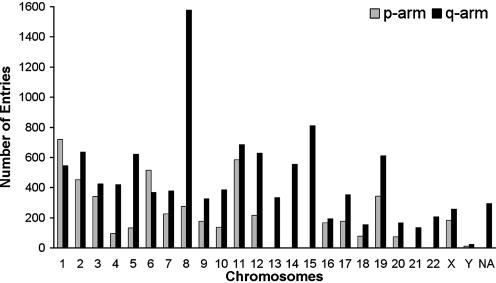
Traditionally, those genes with relevance defined by clinical studies have been referenced in the literature according to their cytogenetic location. In order to integrate those cytogenetic descriptions with physical map information, a cytogenetic format was adopted for each Hembase entry. Cytogenetic bands were defined according to the physical base pair position of the human genome using information from the UCSC genome browser (4). The position of each Hembase entry within that physical map was collected by BLAT alignments (5); 296 entries could not be positioned due to their small size, repeat structures or incomplete genome sequencing (Fig. 1). Finally, those entries that aligned within a discrete range on the human genome were sorted into groups that correspond to the specified cytogenetic bands.
Figure 1.
Distribution of 16 132 Hembase entries (15 752 EST + 380 referenced sequences) according to their location in the human genome.
HEMBASE SEARCH
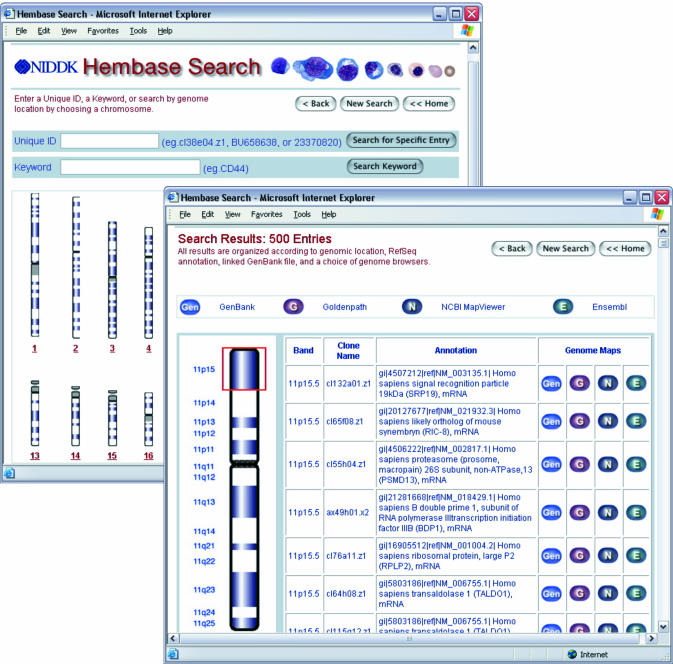
By associating each database entry with a specific cytogenetic band, Hembase provides an organizational framework of erythroid transcription based entirely upon the human genome. Hembase users can search the database using a unique clone ID from GenBank or a keyword contained in the RefSeq annotation. Alternatively, a graphic-base selection is possible according to genome location (Fig. 2). The chromosome images were generated using the Perl scripts version 1.2 from the Colored Chromosomes Project (6).
Figure 2.
An example of the Hembase search and results pages after selection of the 11p15 cytogenetic band. Search results are displayed according to cytogenetic position, clone name and RefSeq annotation. Hyperlink buttons on the right are directed to primary sequence files in GenBank, and clone-specific locations in the three major human genome browsers.
The search results display is formatted as a concise table with an accompanying cytogenetic graphic that highlights the browsed region (Fig. 2). Each table contains a cytogenetic location, clone name, RefSeq annotation and hyperlinks to primary sequence data or the three major human genome browsers currently in the public domain. Those hyperlinks are directed to GenBank (7), NCBI LocusLink (3), UCSC Genome Browser (4) and Ensembl (8). Vast quantities of additional data may then be attained through internet links initiated from the genome browser displays. This hyperlink strategy was utilized to avert undue aging of the information contained in the primary Hembase files. Hembase therefore functions as browser and genome portal for current information pertaining to erythroid biology rather than as a separate depository of that information.
AVAILABILITY AND FUTURE OF HEMBASE
All the sequence information present in Hembase is avail able worldwide (http://hembase.niddk.nih.gov) without registration or fee. All EST sequences may be downloaded using the NCBI Unigene Library Browser (9). Regular updates in Hembase are planned in parallel with expected refinements in the description of the human genome. Suggestions regarding possible improvements in the Hembase format including links to other sources of data relevant for scientists or clinicians interested in erythroid biology are welcome.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Sandy Desautels, Rick Roland and Tatiana Shima for assistance with the design and construction of the Hembase website, and Tiffany Trice for annotation updates. Jim Kent kindly provided information regarding the physical and cytogenetic map comparisons.
REFERENCES
- 1.Gubin A.N., Njoroge,J.M., Bouffard,G.G. and Miller,J.L. (1999) Gene expression in proliferating human erythroid cells. Genomics, 59, 168–177. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 3.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karolchik D., Baertsch,R., Diekhans,M., Furey,T.S., Hinrichs,A., Lu,Y.T., Roskin,K.M., Schwartz,M., Sugnet,C.W., Thomas,D.J. et al. (2003) The UCSC Genome Browser Database. Nucleic Acids Res., 31, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kent W.J. (2002) BLAT—the BLAST-like alignment tool. Genome Res., 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Böhringer S., Gödde,R., Böhringer,D., Schulte,T. and Epplen,J.T. (2002) A software package for drawing ideograms automatically. Online J. Bioinformatics, 1, 51–59. [Google Scholar]
- 7.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J. and Wheeler,D.L. (2003) GenBank. Nucleic Acids Res., 31, 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clamp M., Andrews,D., Barker,D., Bevan,P., Cameron,G., Chen,Y., Clark,L., Cox,T., Cuff,J., Curwen,V. et al. (2003) Ensembl 2002: accommodating comparative genomics. Nucleic Acids Res., 31, 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wheeler D.L., Church,D.M., Federhen,S., Lash,A.E., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Sequeira,E., Tatusova,T.A. et al. (2003) Database Resources of the National Center for Biotechnology. Nucleic Acids Res., 31, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]