Abstract
Alu elements are short interspersed elements (SINEs) ∼300 nucleotides in length. More than 1 million Alus are found in the human genome. Despite their being genetically functionless, recent findings suggest that Alu elements may have a broad evolutionary impact by affecting gene structures, protein sequences, splicing motifs and expression patterns. Because of these effects, compiling a genomic database of Alu sequences that reside within protein-coding genes seemed a useful enterprise. Presently, such data are limited since the structural and positional information on genes and Alu sequences are scattered throughout incompatible and unconnected databases. AluGene (http://Alugene.tau.ac.il/) provides easy access to a complete Alu map of the human genome, as well as Alu-associated information. The Alu elements are annotated with respect to coding region and exon/intron location. This design facilitates queries on Alu sequences, locations, as well as motifs and compositional properties via a one-stop search page.
INTRODUCTION
Alu sequences are short interspersed elements (SINEs), typically 300 nucleotides in length, which account for more than 10% of the human genome (1). Alus have a dimeric structure and are ancestrally derived from the gene specifying 7SL RNA, an abundant cytoplasmic component of the signal recognition particle that mediates the translocation of secreted proteins across the endoplasmic reticulum (2). Alu elements multiply within the genome through RNA polymerase III-derived transcripts in a process termed retroposition.
Alu sequences can be divided into five subfamilies of related elements based upon key diagnostic nucleotide positions shared by subfamily members (3). Several overlapping subfamilies of Alu repeats of different evolutionary ages have been identified. These observations have led to the suggestion that Alu subfamilies have originated through successive waves of fixation from sequential small subsets of active Alu sequences. The oldest Alu-related elements are the monomeric FAM, FRAM and FLAM sequences. The oldest Alu dimeric subfamilies are Alu-Jo and Alu-Jb, estimated to be ∼80 million years old. The intermediately aged Alu subfamilies belong to the Alu-S class, which is divided into subfamilies Sx, Sp, Sq, Sg and Sc. These subfamilies are estimated to be 30–50 million years old. The youngest subfamilies belong to the Alu-Y class, which are less than 15 million years old (2). Because of their newness, some Alu-Y elements have neither reached fixation nor been lost, and they exist in a polymorphic state. Most Alu repeats in the human genome belong to the Alu-S class, with Alu-Sx being the commonest (2).
Despite their being genetically functionless, recent findings suggest that Alu elements have a broad evolutionary impact. Parts of Alu elements may become inserted into mature mRNAs by way of splicing in a process called ‘exonization’. Presumably, the exonization process is facilitated by sequence motifs within Alu that resemble splice sites (4–6). Indeed, more than 5% of the alternatively spliced exons in the human genome are Alu derived. All Alu-containing internal exons studied so far were found to be alternatively spliced (6). It was, thus, concluded that mutations resulting in constitutively spliced exonic Alus would result, in the vast majority of cases, in the creation of defective genes causing deleterious effects on fitness. An example of such an occurrence is the addition of a new Alu-derived exon in conjunction with exon skipping in the β-glucuronidase gene resulting in a mild form of Sly syndrome (7). Another example of splicing-mediated diseases caused by Alu is the insertion of an Alu element into intron 18 of the Factor VIII gene, which leads to exon 19 skipping and results in a severe form of hemophilia A (8). Alu-mediated homologous unequal recombination may also result in genetic defects, as in the case of iduronate-2-sulfatase in which an Alu-mediated exon 8 deletion results in Hunter syndrome (9).
Alu insertions may sometimes create a new function or modify an existing one. One such example concerns tissue localization of casein kinase 2 (CK2) (10). CK2 is a 2α + 2β tetrameric enzyme that phosphorylates serine and threonine residues, and is essential for the viability of eukaryotic cells. A novel isoform of the α subunit was recently found to be highly expressed in the liver. Examination of the isoform sequence CK2α′, revealed a translated Alu-containing cassette exon incorporated into the mature mRNA. This C-terminal sequence was found to be essential in the determination of the nuclear localization of the CK2α′′ isoform (10). Alus were also found to be involved in expression regulation. For instance, Alu repeats in the distal promoter region of the human colesteryl ester transfer protein (CETP) were found to act as repressive regulatory elements of the activity of the promoter (11). Alus have also been found to be involved in apoptosis. An alternatively spliced Alu-like exon was found to be essential for the ability of the Bcl-rambo β protein to promote etoposide- and taxol-induced cell death (12). Interestingly, Alus are found to be highly clustered in genes that are involved in metabolism, transport and signaling processes, while they are less abundant in genes encoding structural proteins or information-pathway components. This non-random distribution was claimed to support the hypothesis that Alus may not always be useless (junk) DNA (13,14). An alternative explanation may be that Alu insertions (or other mutations) may be deleterious, and hence more strongly selected against if they affect genes involved in information storage and processes.
Given the ‘anecdotal’ evidence concerning Alu involvement in gene structure and expression, it seems worthwhile to construct a genomic compilation of Alus that reside within protein-coding genes. Such a database may be important in our efforts to elucidate the rules governing Alu exonization, gene regulation by Alu sequences and Alu-associated risk factors for mutation and pathogenesis. Presently, such data are limited since the information on gene and Alu location, as well as their characteristic and relative positions, is disjointed and scattered throughout incompatible and unconnected files in the various human genome databases. AluGene aims to provide an easy access to the complete Alu map within the human genome, as well as associated information, such as GC content and subfamily affiliation.
DATABASE DESIGN
The AluGene database was implemented using the Select Query Language (SQL) from the MySQL database server (http://www.mysql.com/). The database merges three main constituents: (i) a map of mRNAs juxtaposed on the human genome, (ii) a map of Alu sequences and (iii) a comparison (alignment) of each Alu element with the consensus sequence of the subfamily to which it belongs. Currently, the database relies on the May 2003 version of the NCBI human genome sequence (http://www.ncbi.nlm.nih.gov/), and will be updated with each new release. We used Perl scripts to extract positions and sequences of genes and their coding sequence from the GenBank files. The contig coordinates and orientations were stored as mapping data for each gene. Additional descriptive data for the mRNAs, such as LocusLink entry and protein entry, were also stored in the database. The locations of exons and intron were stored in distinct tables.
We used the RepeatMasker software (http://ftp.genome.washington.edu/RM/RepeatMasker.html) to search for Alu sequences in human genomic contigs. For each Alu entry, its location on the contig, orientation and sequence were stored in the database. Genomic locations of Alus and genes were calculated from their position in the contigs and the relative location of the contig within the chromosome according to the seq_contig.md file in the NCBI. By using ClustalW (15), each Alu sequence was aligned to the consensus sequence of its subfamily (A. F. A. Smit and P. Green, unpublished data). Indels were interpreted as insertions or deletions, respectively, according to the absence or presence of non-null nucleotides in the aligned positions in the consensus sequence.
Data
The AluGene database contains a map of the human transcriptome, and a map and properties of Alu sequences in the human genome. The transcriptome is split into three sets (tables): mRNA, intron and exon. The mRNA records are linked to other genetic databases through four different accession keys provided by the NCBI: Refseq ID, GI number, Interim ID and LocusLink ID. In addition, each mRNA record was identified by its genomic location. Intron and exon records were linked to their corresponding mRNA through the GI number.
The data for each Alu entry include DNA sequence, genomic location, Alu subfamily to which the Alu entry belongs, an alignment of the entry to the consensus sequence of the subfamily and a list of differences from the consensus sequence of the subfamily. The Alu data also include GC content and length of the poly(A) tail, i.e. features that have been shown to affect the role that Alu may play in the genome (16,17). The identification of the poly(A) tail might be problematic since in addition to the terminal poly(A) sequence, Alu elements contain an internal poly(A). Thus, as far as partial Alu insertions are concerned, the internal poly(A) may be confused with the terminal one. Using pairwise alignments of each Alu to its subfamily consensus sequence, we were able to ascertain that the poly(A) at the 3′ of an Alu sequence instance is indeed a tail.
It is, of course, not our purpose here to provide an all-exhaustive statistical description of the ‘Aluome’. In the following, we provide a couple of illustrative enumerative statistics that can be gleaned from AluGene. The total number of Alu elements in the currently sequenced human genome is 1 169 291. Forty five percent (45%) of all Alus are contained within genes; the rest lie within intergenic regions. There are 28 049 transcripts in AluGene, of which 17 781 (63%) contain at least one Alu element. Within 1 kb regions upstream of transcription initiation sites, there are 9212 Alus. These Alus are found in locations that potentially may affect expression levels of the downstream genes.
Search
The AluGene database can be accessed freely at http://Alugene.tau.ac.il/. Its main goal is to facilitate the search for Alus that are located either within genes or in their immediate proximity. By merging the genomic locations of genes and Alus, it is possible to find overlapping areas between the two, such as Alus residing within exons. Moreover, using additional information concerning Alus, it is possible to study the Alu subset that affects processes such as exonization, expression regulation, etc. AluGene enables specific queries about certain genes or loci, as well as a wide range of queries designed by the user. The outcome of the specific queries can be either a schematic map, or an alignment of the nucleotide sequences. In the alignment queries, the Alus are aligned either to the genomic locus in which they are embedded or to the consensus sequence of their subfamily. Thus, Alus can be searched by their location in the genome, or by location in genes (in exons, or introns, or in splice sites), and by properties such as GC content and length of A-tail. Moreover, AluGene enables search of Alu by sequence motifs, i.e. retrieve all Alus that contain a certain sequence pattern. The schematic map generated by AluGene presents the extent and locations of genes and Alus in the area defined by the user query. Viewing extended information about the elements, i.e. exons, Alus, etc., can be done by placing the cursor on the map symbols.
ILLUSTRATIVE EXAMPLES
An interesting option in AluGene is the ‘Do it yourself’ query. One such example concerns the observation that Alu elements may contain active splice sites (4–6). In such cases, part of the Alu element will be found in an intron, and the other part in an exon. Let us use this option, for instance, to identify all Alus that contain splice sites on chromosome 21. The query in SQL can be viewed in the ‘Examples’ section of the ‘Do it yourself’ query by clicking on the button ‘Alus spanning splice-sites in chromosome 21’. In other words, for all the Alus on chromosome 21, we look for those that begin upstream of a splice site (either 5′ or 3′) and end downstream of it.
The result of this query was the identification of four transcripts on chromosome 21 for which evidence for Alu exonization exists. Of these, one is annotated as a hypothetical gene with no additional information, and three were identified. These are: (i) NM_015833 (ADARB1), a 13-exon gene coding the enzyme responsible for pre-mRNA editing; (ii) NM_015834, a transcript variant of ADARB1; and (iii) NM_032589 (DSCR8), a 4-exon gene the Down syndrome critical region.
If we repeat the same query for chromosome 20 we obtain three transcripts, one of which is an unannotated open reading frame (ORF) and two are identified. These are: (i) NM_031483 (ITCH), a 26-exon gene encoding a protein that interacts with atrophin-1, and (ii) NM_016082 (CDK5RAP1), a 15-exon gene encoding a neuronal CDC2-like kinase that is involved in the regulation of neuronal differentiation.
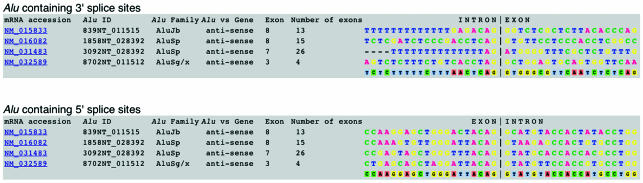
The above-described Alus can be used to study sequences that mediate Alu exonization. AluGene provides a specific query type for this purpose, the ‘Exonization’ alignment query. The result of this query is a splice-site alignment of the Alus in the search criteria. Figure 1 shows such an alignment, produced for the four Alus from the annotated genes in chromosomes 20 and 21. As seen in the alignment of the 3′ splice sites of these Alus (top), an AG dinucleotide always appears at the end of the intron upstream Alu exons. Indeed, such an AG pair has recently been reported to be essential for Alu exonization (4–6). The alignment of the 5′ splice sites (bottom) also shows several conserved nucleotide positions. For example, the last two nucleotides of the exon (AG), as well as the first, third and fifth positions of the intron (G, A and G, respectively) are conserved in all four Alu-derived splice sites. These positions are in agreement with the general consensus of 5′ splice sites (18).
Figure 1.
Output of an ‘exonization’ query. The query was conducted using genes NM_015833, NM_032589 from chromosome 21 and NM_016083, NM_031483 from chromosome 20. Alus containing 3′ splice sites are shown at the top; Alus containing 5′ splice sites are at the bottom. Twenty nucleotides either side of splice sites are shown (the window width was a parameter of the query). The results include (from left to right): accession number, Alu ID in AluGene, Alu subfamily, orientation of the Alu versus the gene, exon number, total exons in the gene and sequences of Alus at the splice sites. The consensus sequences for each multiple sequence alignment are shown in the bottom-most rows.
FUTURE DEVELOPMENTS
AluGene is an ongoing project whose scopes and uses will be extended in the future. One of the first extensions will involve the addition of information concerning gene products. Proteins will be classified according to biochemical pathways or genetic disorders caused by their incapacitation. Such a task may be accomplished by the linking of AluGene to the Online Mendelian Inheritance in Man (OMIM) database (http://www.ncbi.nlm.nih.gov/omim/). Such a link may be especially useful to study Alu insertions that result in pathological manifestation. Currently the exons in AluGene are derived only from RefSeq, and do not contain exons that are supported solely by EST data. Therefore, an additional expansion may include EST-based exons using dbEST (http://www.ncbi.nlm.nih.gov/dbEST/). Information about Alus within RNA-specifying genes is expected to be included within AluGene as well. Other possible additions may include single nucleotide polymorphisms within Alus, as well as a taxonomic expansion into simian and scandentian genomes as these become available. The taxonomic expansion is expected to add vAluable information on the evolution of Alu sequences.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Ran Blekhman, Giddy Landan and Itay Mayrose for their help. T.D. was supported in part by a scholarship in Complexity Science from the Yeshaia Horvitz Association.
REFERENCES
- 1.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 2.Mighell A.J., Markham,A.F. and Robinson,P.A. (1997) Alu sequences. FEBS Lett., 417, 1–5. [DOI] [PubMed] [Google Scholar]
- 3.Kapitonov V. and Jurka,J. (1996) The age of Alu subfamilies. J. Mol. Evol., 42, 59–65. [DOI] [PubMed] [Google Scholar]
- 4.Lev-Maor G., Sorek,R., Shomron,N. and Ast,G. (2003) The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science, 300, 1288–1291. [DOI] [PubMed] [Google Scholar]
- 5.Nekrutenko A. and Li,W.H. (2001) Transposable elements are found in a large number of human protein-coding genes. Trends Genet., 17, 619–621. [DOI] [PubMed] [Google Scholar]
- 6.Sorek R., Ast,G. and Graur,D. (2002) Alu-containing exons are alternatively spliced. Genome Res., 12, 1060–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vervoort R., Gitzelmann,R., Lissens,W. and Liebaers,I. (1998) A mutation (IVS8+0.6kbdelTC) creating a new donor splice site activates a cryptic exon in an Alu-element in intron 8 of the human β-glucuronidase gene. Hum. Genet., 103, 686–693. [DOI] [PubMed] [Google Scholar]
- 8.Ganguly A., Dunbar,T., Chen,P., Godmilow,L. and Ganguly,T. (2003) Exon skipping caused by an intronic insertion of a young Alu Yb9 element leads to severe hemophilia A. Hum. Genet., 113, 348–352. [DOI] [PubMed] [Google Scholar]
- 9.Ricci V., Regis,S., Di Duca,M. and Filocamo,M. (2003) An Alu-mediated rearrangement as cause of exon skipping in Hunter disease. Hum. Genet., 112, 419–425. [DOI] [PubMed] [Google Scholar]
- 10.Hilgard P., Huang,T., Wolkoff,A.W. and Stockert,R.J. (2002) Translated Alu sequence determines nuclear localization of a novel catalytic subunit of casein kinase 2. Am. J. Physiol. Cell Physiol., 283, C472–C483. [DOI] [PubMed] [Google Scholar]
- 11.Le Goff W., Guerin,M., Chapman,M.J. and Thillet,J. (2003) A CYP7A promoter binding factor site and Alu repeat in the distal promoter region are implicated in regulation of human CETP gene expression. J. Lipid Res., 44, 902–910. [DOI] [PubMed] [Google Scholar]
- 12.Yi P., Zhang,W., Zhai,Z., Miao,L., Wang,Y. and Wu,M. (2003) Bcl-rambo β, a special splicing variant with an insertion of an Alu-like cassette, promotes etoposide- and Taxol-induced cell death. FEBS Lett., 534, 61–68. [DOI] [PubMed] [Google Scholar]
- 13.Makalowski W. (2003) Not junk after all. Science, 300, 1246–1247. [DOI] [PubMed] [Google Scholar]
- 14.Grover D., Majumder,P.P., Rao,C.B., Brahmachari,S.K. and Mukerji,M. (2003) Non-random distribution of Alu elements in genes of various functional categories: Insight from analysis of human chromosomes 21 and 22. Mol. Biol. Evol., 20, 1420–1424. [DOI] [PubMed] [Google Scholar]
- 15.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roy-Engel A.M., Salem,A.H., Oyeniran,O.O., Deininger,L., Hedges,D.J., Kilroy,G.E., Batzer,M.A. and Deininger,P.L. (2002) Active Alu element ‘A-tails’: Size does matter. Genome Res., 12, 1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jurka J., Krnjajic,M., Kapitonov,V.V., Stenger,J.E. and Kokhanyy,O. (2002) Active Alu elements are passed primarily through paternal germlines. Theor. Popul. Biol., 61, 519–530. [DOI] [PubMed] [Google Scholar]
- 18.Horowitz D.S. and Krainer,A.R. (1994) Mechanisms for selecting 5′ splice sites in mammalian pre-mRNA splicing. Trends Genet., 10, 100–106. [DOI] [PubMed] [Google Scholar]