Abstract
NASC operates an Affymetrix ‘GeneChip’ (microarray) service for the Arabidopsis thaliana community. All data produced by the service are publicly available through our microarray data base ‘NASCArrays’ published at http://affymetrix.arabidopsis.info. The data are accessible through text searching and a series of data mining tools. All data are annotated with sample preparation details, and the original Affymetrix data are available for download. The database aims to be MIAME supportive and provide a coordinated resource for re searchers interested in the transcriptome of Arabidopsis. Using this database, data produced will be shared with other databases worldwide.
INTRODUCTION
NASC’s Affymetrix service
Arabidopsis thaliana is a commonly used model organism for the study of the biology of flowering plants (1). Since the completion of the Arabidopsis genome in 2000 (2), various tools have been made available to exploit the availability of genome-wide sequence data, such as large-scale insertion mutation collections, proteomics, SNP detection and microarrays.
Microarrays allow the monitoring of the expression of many genes in parallel (3). The availability of a completed genome sequence for an organism allows microarrays to be constructed that contain features representing large numbers of genes—potentially allowing the measurement of gene expression for every gene in a species on one slide.
The Affymetrix system of high-density oligonucleotide arrays (‘GeneChips’) is one such microarray system (4,5). Two GeneChips from Affymetrix are represented within our database, a smaller and older GeneChip design which represents ∼8300 genes, and a second more recent ‘ATH1’ GeneChip design, which represents most of the Arabidopsis genome as annotated by TIGR in their database in December 2001.
Since February 2002, the Nottingham Arabidopsis Stock Centre (NASC) has operated a transcriptomics service for the Arabidopsis community using the Affymetrix system. Users of the service are entirely responsible for the growth of their plants and preparation of the RNA extracts, which are then sent to NASC. NASC then performs all further microarray steps from labelling through to data preparation.
The NASCArrays database
Although the users of the transcriptomics service have the option of a short (maximum 6 month) confidentiality period before data release, all data that are produced by the service are made available to the public without condition. NASCArrays, NASC’s microarray database, is the primary method of distributing the data the Affymetrix service produces. It can be accessed by choosing Affymetrix Data from our Affymetrix website http://affymetrix.arabidopsis.info.
The Affymetrix equipment produces expression data for every gene represented on the microarray. This data are fed into the database, and are coordinated with user-supplied annotation on how the original RNA samples were prepared. The service insists that users supply annotation for each sample. NASC aims to hold full MIAME (6)-supportive data for its experiments, and as such good annotation from individual users is vital. The database currently contains 40 experiments made up of ∼400 GeneChips but the number is increasing rapidly.
DATABASE FEATURES
Annotation pages and Search
The front page of the database contains a list of experiments that are available in the database, the search tools and links to the data mining tools. If you select any of the experiments from the list the basic experiment page is shown.
On the top of the experiment page is an abstract for the experiment, and contact details for the experimenter. Beneath this is a list of the GeneChips (referred to as ‘slides’) used in the experiment. Each slide has information about how the sample that was hybridized to the slide was prepared by the user, and how the slide was handled when it was processed at NASC.
There are currently two searches available from the front page. The experiment search provides a keyword search of experiments like a search engine. The slide search selects slides that match criteria entered exactly—for instance all slides that were produced from samples treated in a certain way.
Data available
At many points in the database, the output data from the microarrays can be downloaded. NASC uses Affymetrix MAS 5.0 software for scanning and analysis of Affymetrix microarrays. NASCArrays stores four data points for each gene per GeneChip from this software: Signal, StatPairsUsed, PresentCall and Detection P-value. These are reproduced with some rudimentary annotation for the probes on the GeneChip when users download data from the database.
Data can be downloaded for one or many slides at once. Data are supplied as a comma separated values (CSV) file that can be read by many spreadsheet programs. Data can be downloaded over the web, or emailed to the user.
As an option, users can download data ‘for clustering’. These are data specially formatted for EPCLUST (7). Using this feature allows users to easily perform clustering analysis using data from NASCArrays.
Data mining tools
In NASCArrays, a series of ‘Data mining tools’ are available. These allow researchers to use a ‘gene-centric’ rather than ‘array-centric’ approach to finding data of interest. Many researchers have ‘genes of interest’—these tools allow researchers to find experiments that are related to their gene of interest. The NASCArrays tools allow users to pick a gene of interest using the probe set reference number as given by Affymetrix, a gene symbol, an Arabidopsis Genome Initiative (AGI) identifier for a gene or a Complete Arabidopsis Transcriptome MicroArray (CATMA) code.
Spot history
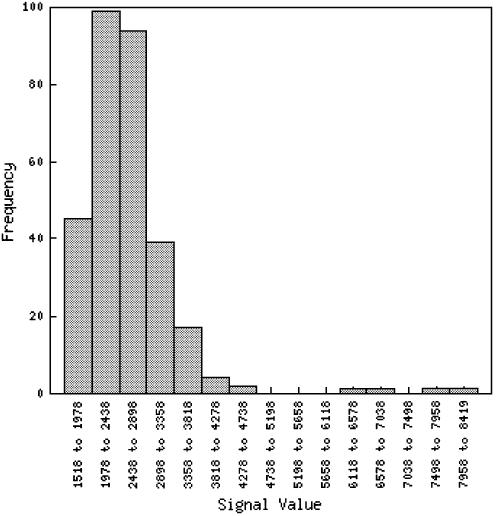
The spot history is a tool that is available in other microarray databases (8). It shows the distribution of expression of a gene of interest over all experiments in the database. Results are displayed in the form of a histogram. Each bar in the histogram can be selected, and the slides that made up that range in the histogram will be shown. Researchers can thus easily locate slides that have unusual values of expression for a given gene. (Fig. 1 is a histogram from the spot history tool. This histogram shows the distribution of gene expression for At3g08580.)
Figure 1.
A histogram from the spot history tool. This histogram shows the distribution of gene expression for At3g08580.
Two-gene scatter plot
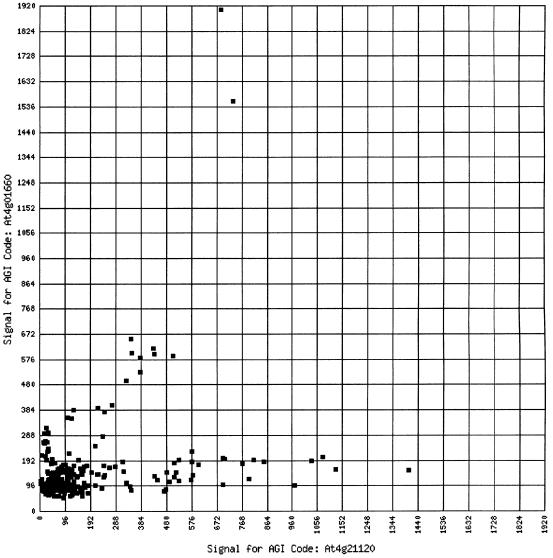
The two-gene scatter plot is a tool for rudimentary comparison of the expression profile of two genes. It takes the form of a scatter plot, with the expression values of each gene along the two axes. Using this tool allows researchers to quickly see if there are trends between the two genes. Any given point on the plot can be selected, and the slide corresponding to that point will be shown (Fig. 2).
Figure 2.
A sample scatter plot from the two-gene scatter plot.
Gene swinger
The ‘gene swinger’ is a tool that allows researchers to sort all experiments based on which experiments a gene of interest shows most variability in. Upon choosing a gene of interest, the tool calculates the standard error for that gene for each experiment, and displays the experiments starting with the highest variability first. Experiments that are highly variable with respect to a gene should be of interest to researchers working on that gene.
Bulk gene download
If researchers have a series of genes they are interested in, they can download the expression over all experiments for these genes using Bulk gene download.
The selection
The selection is a feature analogous to the shopping cart on a web shopping site. It allows users to choose an arbitrary selection of slides and perform actions on them. For instance, both the spot history and two-gene scatter plot can operate on just the slides held in the selection. Data can be downloaded for just the slides in the selection—for instance a user could fill the selection with slides based on a certain ecotype, and then download all of these data as one file.
FURTHER DATA DISSEMINATION
AffyWatch
Some researchers want the original files that come from the Affymetrix software. Other researchers want to have all of the data for populating their own in-house databases. NASC operates a subscription CD service called ‘AffyWatch’. This enables users to receive all data produced by NASC’s Affymetrix service in CD format.
Other databases
As part of NASC’s commitment to data sharing, data will also be donated to other databases. Currently data are donated in MAGE-ML (9) format to ‘ArrayExpress (10)’, the European Bioinformatics Institute’s microarray database, where it will be further distributed to GEO at the NCBI (11). In the future, the data will also be shared with TAIR’s microarray facility (12).
REFERENCES
- 1.Meinke D.W., Cherry,M., Dean,C., Rounsley,S.D. and Koornneef,M. (1998) Arabidopsis thaliana: a model plant for genome analysis. Science, 282, 662–682. [DOI] [PubMed] [Google Scholar]
- 2.Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 3.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a cDNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 4.Lockhart D.J., Dong.H., Byrne,M.C., Follettie,M.T., Gallo,M.V., Chee,M.S., Mittmann,M., Wang,C., Kobayashi,M., Horton,H. et al. (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol., 14, 1675–1680. [DOI] [PubMed] [Google Scholar]
- 5.Lipshutz R.J., Fodor,S.P.A., Gingeras,T.R. and Lockhart,D.J. (1999) High density synthetic oligonucleotide arrays. Nature Genet., 21, 20–24. [DOI] [PubMed] [Google Scholar]
- 6.Brazma A., Hingamp,P., Quackenbush,J., Sherlock,G., Spellman,P., Stoeckert,C., Aach,J., Ansorge,W., Ball,C.A., Causton,H.C. et al. (2001) Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nature Genet., 29, 365–371. [DOI] [PubMed] [Google Scholar]
- 7.Vilo J., Kapushesky,M., Kemmeren,P., Sarkans,U. and Brazma,A. (2003) Expression Profiler. In Parmigiani,G., Garrett,E.S., Irizarry,R. and Zeger,S.L. (eds), The Analysis of Gene Expression Data: Methods and Software. Springer Verlag, New York, NY. [Google Scholar]
- 8.Gollub J., Ball,C.A., Binkley,G., Demeter,J., Finkelstein,D.B., Herbert,J.M., Hernandez-Boussard,T., Jin,H., Kaloper,M., Matese,J.C. et al. (2003) The Stanford Microarray Database: data access and quality assessment tools. Nucleic Acids Res., 31, 94–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spellman P.T., Millar,M., Stewart,J., Troup,C., Sarkans,U., Chervitz,S., Bernhart,D., Sherlock,G., Ball,C., Lepage,M. et al. (2002) Design and implementation of microarray gene expression markup language (MAGE-ML). Genome Biol., 3, RESEARCH0046.1–0046.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brazma A., Parkinson,H., Sarkans,U., Shojatalab,M., Vilo,J., Abeygunwardene,N., Holloway,E., Kapushesky,M., Kemmeren,P. et al. (2003) ArrayExpress—a public repository for microarray gene expression data at the EBI. Nucleic Acids Res., 31, 68–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgar R., Domrachev,M. and Lash,A.E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res., 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhee S.Y., Beavis,W., Beradini,T.Z., Chen,G., Dixon,D., Doyle,A., Garcia-Hernandez,M., Huala,E., Lander,G., Montoya,M. et al. (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res., 31, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]