Abstract
Proper periplasmic disulfide bond formation is important for folding and stability of many secreted and membrane proteins, and is catalyzed by three DsbA oxidoreductases in Neisseria meningitidis. DsbD provides reducing power to DsbC that shuffles incorrect disulfide bond in misfolded proteins as well as to the periplasmic enzymes that reduce apo-cytochrome c (CcsX) or repair oxidative protein damages (MrsAB). The expression of dsbD, but not other dsb genes, is positively regulated by the MisR/S two-component system. qRT-PCR analyses showed significantly reduced dsbD expression in all misR/S mutants, which was rescued by genetic complementation. The direct and specific interaction of MisR with the upstream region of the dsbD promoter was demonstrated by EMSA, and the MisR-binding sequences were mapped. Further, the expression of dsbD was found to be induced by dithiothrietol (DTT), through the MisR/S regulatory system. Surprisingly, we revealed that inactivation of dsbD can only be achieved in a strain carrying an ectopically located dsbD, in the dsbA1A2 double mutant or in the dsbA1A2A3 triple mutant, thus DsbD is indispensable for DsbA-catalyzed oxidative protein folding in N. meningitidis. The defects of the meningococcal dsbA1A2 mutant in transformation and resistance to oxidative stress were more severe in the absence of dsbD.
Keywords: Neisseria meningitidis, DsbD, MisRS, two-component regulatory system, DsbA
INTRODUCTION
Neisseria meningitidis, an exclusive pathogen of humans, remains a leading cause of meningitis and sepsis, usually in otherwise healthy individual (Stephens et al., 2007, Stephens, 2007). As a result of successful conjugate vaccines in reducing the incidence of meningitis caused by Streptococcus pneumoniae and Haemophilus influenzae infections, N. meningitidis has become a leading cause of bacterial meningitis in children and young adults in the United States (Schuchat et al., 1997, Whitney et al., 2003). Despite the sensitivity of meningococcus to many antibiotics, meningococcal disease still causes substantial mortality and morbidity (Bilukha & Rosenstein, 2005).
N. meningitidis inhabits the human nasopharynx of approximately 10% of the population and can also rapidly disseminate to cause invasive disease (Stephens & Zimmer, 2002, Tzeng & Stephens, 2000). To successfully colonize and survive as an obligate human pathogen, N. meningitidis responds to signals emanating from the human host, a task often carried out by environmental sensing two-component systems that generally consist of a sensor histidine kinase and a response regulator (Hoch, 2000). Upon sensing specific signals, the histidine kinase autophosphorylates and the phosphoryl group is subsequently transferred to the cognate response regulator. The phosphorylation status of the response regulator typically mediates affinity of the protein to the promoters of target genes. In contrast to other gram-negative bacteria such as E. coli, which encodes more than 30 two-component regulatory systems (Oshima et al., 2002, Yamamoto et al., 2005), the meningococcus contains only four predicted pairs (Tettelin et al., 2000, Parkhill et al., 2000), a characteristic common to obligate pathogens from restricted environments. Capsular polysaccharides, lipooligosaccharides (LOS), outer membrane receptors for iron acquisition and pili are among the important virulence factors critical for meningococcal colonization and pathogenesis (Tzeng & Stephens, 2000). The meningococcal MisR/MisS two-component system has been shown to influence phosphorylation of the LOS inner core (Tzeng et al., 2004), directly repress expression of the type I secretion proteins (Sannigrahi et al., 2009), and activate expression of the HmbR hemoglobin receptor and the HemO heme oxygenase, both important in heme utilization (Zhao et al., 2010). Inactivation of the response regulator MisR also results in sensitivity to antimicrobial peptides (Johnson et al., 2001, Tzeng et al., 2004) and virulence attenuation in a murine model of infection (Newcombe et al., 2004).
In gram-negative bacteria, many membrane and exported proteins are stabilized by intramolecular disulfide bridges between two cysteine residues which are introduced by the oxidoreductase, DsbA. The lack of disulfide bonds in periplasmic proteins results in protein misfolding and loss of function with ultimate removal due to increased sensitivity to proteolytic degradation in the periplasm. To complete the protein oxidation pathway, DsbA is reoxidized by the inner membrane protein DsbB, which utilizes the respiratory chain as electron acceptors. The protein oxidation pathway is partnered by an isomerization pathway consisting of an inner membrane protein, DsbD, which utilizes the cytoplasmic thioredoxinto reduce the disulfide isomerase DsbC. DsbC catalyses the shuffling of multiple disulfide bonds within a single substrate protein until the correct conformation is obtained (Collet & Bardwell, 2002, Kadokura et al., 2003). Dsb proteins are critical for physiological processes such as cytochrome c maturation (Kranz et al., 1998, Thony-Meyer, 2002), and are necessary for many important pathogenic phenotypes including attachment, invasion and intracellular survival (Yu, 1998, Raivio, 2005, Bringer et al., 2007).
N. meningitidis encodes three dsbA genes (Sinha et al., 2004, Tinsley et al., 2004): dsbA1 is meningococcal specific (Tinsley & Nassif, 1996), while dsbA2 and dsbA3 are also present in Neisseria gonorrhoeae. Similar to most other bacterial DsbA proteins, DsbA3 is a soluble periplasmic protein; however, DsbA1 and DsbA2 are found to be lipoproteins (Tinsley et al., 2004). Phenotypic analyses have shown that meningococcal dsbA1A2 double mutants have reduced transformation efficiency and adherence to endothelial cells (Tinsley et al., 2004, Sinha et al., 2008). Further, the defect of the dsbA1A2 mutant in natural competence has been linked to inefficient folding of PilE and PilQ (Tinsley et al., 2004, Sinha et al., 2008). Single copies of dsbB, dsbC and dsbD have been identified in the meningococcal genomes (Sinha et al., 2004, Tinsley et al., 2004).
We have previously carried out microarray analyses of a misR mutant to identify MisR-regulated genes and to define the minimal regulon (Tzeng et al., 2008). Our study has found that expression of dsbD is significantly down regulated in the misR mutant (Tzeng et al., 2008). As this is the first regulatory network impacting the expression of the isomerization arm of the Dsb system, we further characterized the mechanisms of transcriptional control of meningococcal dsbD. Consistent with its role as a transcriptional activator, a direct and specific interaction of MisR with the upstream region of the dsbD promoter was demonstrated.
Surprisingly, we found that DsbD is required for the viability of N. meningitidis, and can only be mutated in the presence of a second functional copy or when the major activity of the oxidative Dsb pathway was inactivated.
RESULTS
Expression of dsbD is controlled by the MisR/MisS two-component system
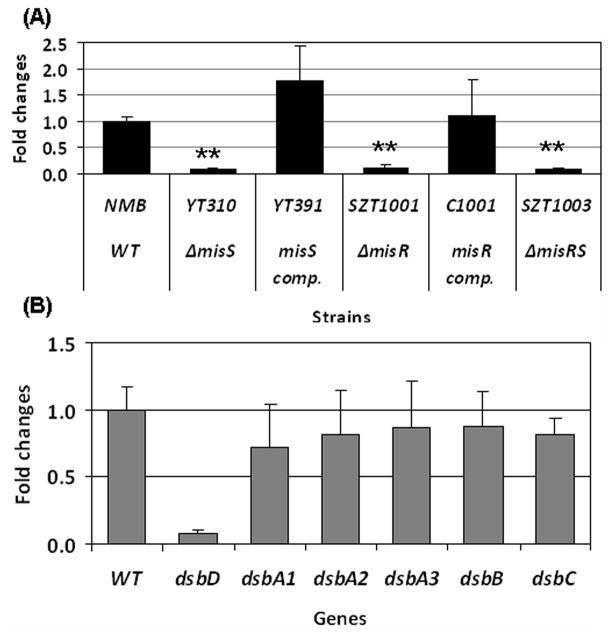
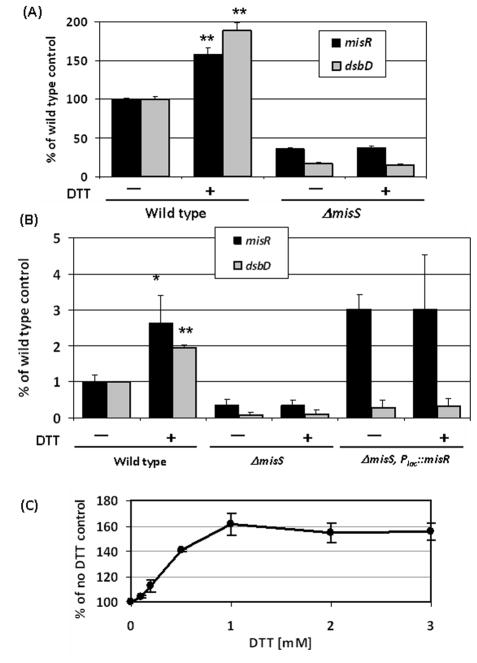
We have previously observed a significant down-regulation of dsbD in a misR mutant by microarray analysis (Tzeng et al., 2008) and this regulation was further characterized in this study. qRT-PCR was conducted to examine dsbD expression in the misR, misS and misRS mutants and the complemented misR and misS mutants. As shown in Fig. 1A, significant decreases in dsbD transcription (8–13 fold) were observed in all mutants, while complementation with an ectopically located second copy of misR or misS rescued the dsbD transcriptional defect. DsbD is one of several meningococcal Dsb proteins involved in the periplasmic disulfide formation that include DsbB, DsbC and three DsbAs. Although we did not detect transcriptional changes in these dsb genes by microarray, another microarray analysis (Newcombe et al., 2005) reported that expression of dsbA2, dsbA3, and dsbC was altered in a serogroup C misR mutant. Thus, we further assess whether MisR might influence expression of other dsb genes by qRT-PCR analyses. Transcription in the wild type strain and the misS mutant was compared and no significant differences in any other dsb genes were detected (Fig. 1B), thus the MisRS system specifically regulates dsbD expression.
Figure 1.
(A) qRT-PCR determinations of relative transcriptional changes of dsbD in the misR/S mutants and complemented mutants. The relative changes in transcriptional level were calculated by the 2−ΔΔCt method (Livak & Schmittgen, 2001)with the wild type strain as the calibrator. The expression level of the wild type strain was set as 1 for normalizing the relative transcription levels of mutants and complemented mutants. (B) qRT-PCR analyses of all dsb genes comparing the wild type strain and the misS mutant. The expression of individual dsb genes in the wild type is set as 1. Each qRT-PCR was examined in triplicate and was repeated with at least four independent RNA preparations. The asterisks indicate statistically significant difference between the wild type strain and the mutant as determined by the Student’s t test with a 2-tailed distribution (P < 0.01).
Mapping of the MisR regulatory region within the dsbD promoter
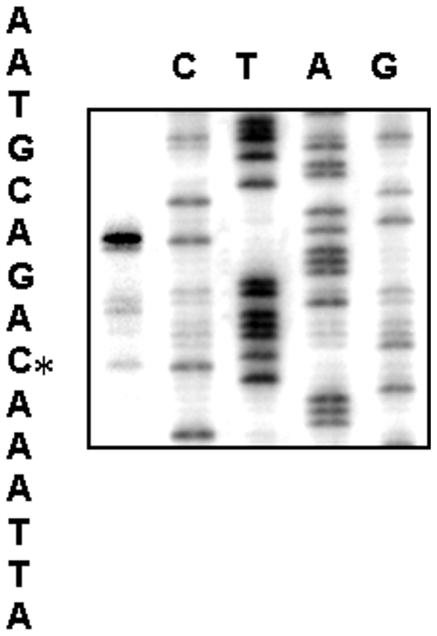
We first determined the transcriptional start site of dsbD by primer extension using total RNA isolated from the wild-type strain NMB and a reverse primer complementary to the coding strand of dsbD. A single major product was observed corresponding to the -29cytosine nucleotide (Fig. 2), indicating a single promoter. The same start site was verified with a second complementary primer (data not shown). Examination of the sequence upstream of the transcriptional start site revealed appropriate promoter elements of a -10 (TATAAT) and a -35 (ATCCCA) sequence separated by 17-bp (see Fig. 4B).
Figure 2.
Primer extension of dsbD. The YT144 primer was labeled and used to generate the sequencing ladder. Lanes C, T, A, G indicate the dideoxy sequencing reactions. The asterisk indicates the transcriptional start site.
Figure 4.
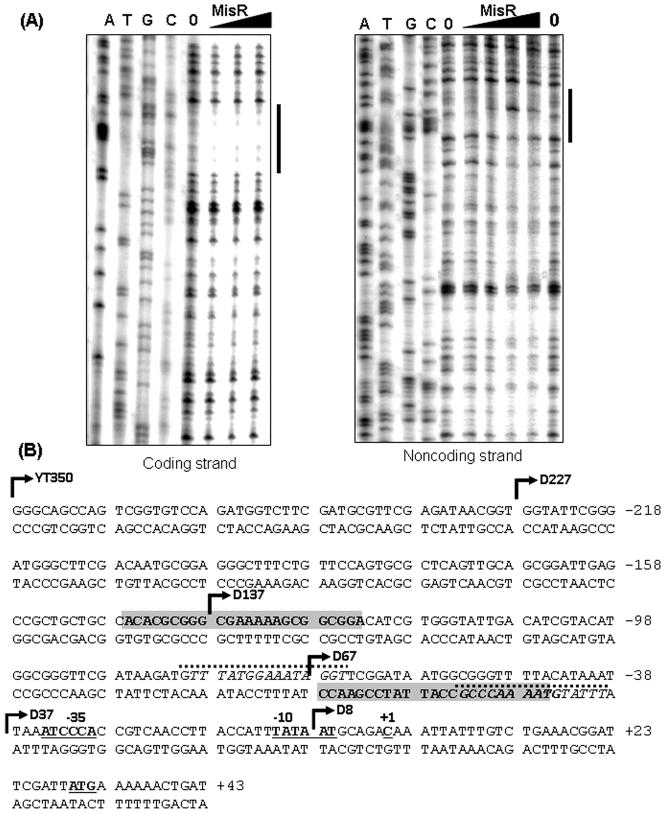
MisR binding sites located upstream of the dsbD promoter. (A) DNase I protection. The coding (left) or noncoding (right) strand of 32P-end labeled dsbD promoter fragment between −227 to +123 with respect to the dsbD transcriptional start site was incubated with increasing amounts of MisR for 20 minutes at 30°C and then subjected to DNase I digestion. The amounts of MisR used in the left panel are 0, 40, 80, and 120 pmol, while in the right panel are 0, 40, 80, 120, 200 and 0 pmol. The dideoxy chain-termination sequencing ladders were generated by extension of 32p-labeled pF2 primer for the coding strand or pR1 primer for the noncoding strand using the corresponding PCR product as template. Black bars indicate protected regions. (B) The dsbD promoter sequence. The protected regions are indicated as shaded boxes in both strands. The transcriptional start site +1, −10 and −35 promoter elements are bolded and underlined. The predicted MisR binding motifs are italicized and marked with dashed lines above the sequence. Bent arrows indicate the 5′ ends of various promoter fragments cloned in reporter strains.
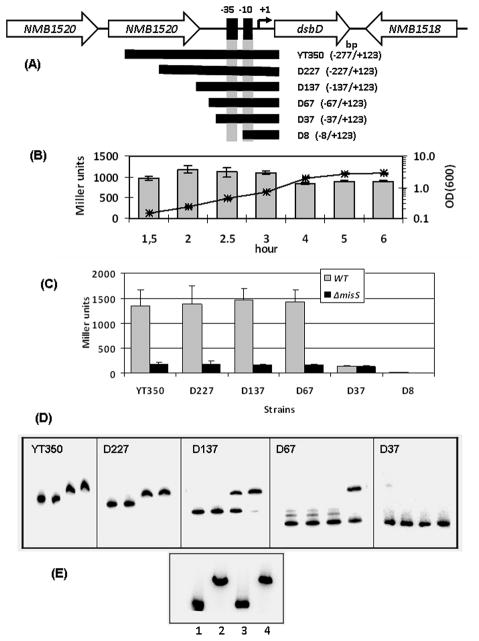
To delineate the dsbD promoter region required for transcriptional activation by MisR, a series of dsbD::lacZ transcriptional fusions with promoter lengths ranging from −277-bp to −8-bp upstream of the transcriptional start site were made (Fig. 3A). These fusions were inserted into a permissive chromosomal locus as a single copy. First, the activity of the YT350 strain with the longest promoter fragment was examined throughout the growth phase and Fig. 3B showed that the dsbD expression remained constant during the exponential growth phase and decreased slightly in the stationary phase. No β-galactosidase activity was detected in the D8 strain, thus confirming the location of the promoter(Fig. 3C). A dsbD promoter fragment that contains the nucleotide sequence between −67 and +123bp (strain D67) generated an activity similar to those with longer promoter sequences, indicating that the ~70-bp sequence upstream of the transcriptional start site was sufficient for full transcriptional activity. It was expected that in the absence of the sequence at which MisR exerts its regulation, transcription of dsbD will not be affected by inactivation of the MisRS system. Thus, the reporter constructs were introduced into the misS mutant for comparison. The activities of the D67 reporter and other longer promoter fusions were significantly reduced in the misS mutant. In contrast, the strain with only the −35 and −10 promoter elements (strain D37) yielded significantly reduced activity in the wild type background, and similar activities were observed between the wild type strain and the misS mutant. Thus, a minimal of ~ 30-bp sequence upstream of the dsbD promoter elements was required for MisR regulation as well as a full transcription activity.
Figure 3.
(A) Schematic of the dsbD locus. The cloned region with respect to the transcriptional start site (+1) is shown in parentheses. The schematic is not to scale. (B) The expression of dsbD was not growth phase dependent. The β-galactosidase activity of strain YT350 were monitored over the growth curve, which is shown as a line graph with a semi-log scale on the secondary axis. Triplicate measurements were performed for each time points and the average of two independent experiments is shown. Error bars indicate the standard deviation. (C) Activities of various dsbD::lacZ transcriptional fusions. β-Galactosidase activities of strains grown to mid-log phase are expressed as mean values ± standard deviations of at least four independent assays done in triplicate measurements. (D) The interaction of MisR with the dsbD promoter examined by EMSA. The PCR products of various dsbD promoter fragments were 32P-labeled by T4 kinase, mixed with increasing amounts of MisR for 20 minutes at 30°C and then subjected to gel electrophoresis. In each panel, the left lane contains DNA probe only and the remaining lanes contain 10, 20 and 40 pmol of MisR, respectively. (D) Competition EMSA. The 32P-labeled D227 dsbD probe (lane 1) was mixed with 40 pmol of MisR alone (lane 2) or with either specific (1 μg, lane 3) or nonspecific DNA (1 μg, lane 4), and analyzed as described above.
MisR directly interacted with the dsbD promoter
Although prior EMSA experiments have not detected direct binding of MisR to the dsbD promoter (Tzeng et al., 2008), in light of the strong regulatory effect of MisR on dsbD expression, we re-examined whether MisR directly interacts with the dsbD promoter. In contrast to our previous observation, we detected a specific binding of MisR protein to the dsbD promoter DNA using different MisR preparations (Fig. 3D). The binding of MisR to various promoter fragments used in the reporter constructs were evaluated. As shown in Fig. 3D, reduced binding were seen between the D227 and D137 fragments and between the D137 and D67 fragments. Consistent with the lacZ reporter data, no binding was detected with the D37 probe. The specificity of MisR binding was demonstrated with competition EMSA where only excess specific DNA, but not nonspecific DNA fragments, eliminated the shift (Fig. 3E). Thus, MisR directly interacted with the sequence upstream of the dsbD promoter, a commonly observed location for transcriptional activators.
To identify the precise location of MisR binding site(s), DNase I protection assays were performed on both strands of a 350-bp 32P-end-labeled dsbD probe (−227 to +123, the D227 fragment in Fig. 3), which displayed strong binding in EMSA. In the presence of increasing amounts of MisR, ~24-bp regions were protected from DNase I digestion on both the coding strand and the noncoding strand (Fig. 4A). The upstream DNase I protected region is present in the D227 fragment, truncated in the D137 fragment, and is absent in the D67 fragment, therefore consistent with the apparent gradual reduction in binding strength detected by EMSA (Fig. 3D). The downstream DNase I protected site fell within the dsbD promoter sequence deemed important in the reporter assays as the D67 fragment conferred a full MisR-regulated transcriptional activity (D67 vs. D37, Fig. 3C). Two putative MisR binding motifs were found to flank and partially overlap the downstream DNase I protected region (Fig. 4B). The motif at the 3′ end is most likely more important as it has a 10-nucleotide overlap with the protected sequence. A possible explanation for the absence of potential MisR binding motif in the upstream protected sequence is that the MisR binding consensus sequence (Tzeng et al., 2008) might be more degenerate than previously defined. Alternatively, a cooperative binding of MisR at the downstream site may result in an additional interaction with this upstream sequence. Taken together, these in vitro data supported a model that MisR directly activated dsbD expression.
Exposure to DTT enhanced dsbD expression via the induction of the MisRS system
DsbD functions as the redox hub of the Dsb system and is regulated by the environmental sensing MisRS two-component system, thus we tested whether redox changes can affect its expression. The dsbD::lacZ reporter strain (YT350) was treated with various redox agents and, interestingly, we discovered that exposure to 1 mM DTT for 30 minutes yielded ~2-fold induction of dsbD transcription(Fig. 5A). To determine whether the induction is dependent of the MisR regulation, we first tested whether DTT also induced misR expression. The MisRS system is autoregulated (Tzeng et al., 2006), thus its own expression will be up-regulated if the DTT signal is transmitted through the MisRS system. We found that the misR::lacZ expression was indeed induced in a similar manner (Fig. 5A). Next, we determined if the DTT induction required a functional MisS kinase. The dsbD and misR reporters in the ΔmisS background was treated with DTT and as shown in Fig. 5A, the induction was abolished by the misS mutation, thus supporting the involvement of the sensor kinase in transmitting the inducing signal. The induction of dsbD and misR by DTT and the dependence on MisS were further confirmed by qRT-PCR analyses (Fig. 5B). As the expression of misR was reduced in the misS mutant, we tested whether the lack of DTT response in the misS mutant was due to insufficient MisR protein. An IPTG-inducible misR construct was introduced into the misS mutant to generate strain PKT392. The expression of misR was induced by IPTG and then the cultures were similarly treated with DTT. As shown in Fig. 5B, in the presence of MisR over expression (~3-fold and 8-fold increases relative to the wild type strain and the misS parental strain, respectively) no DTT induction of dsbD expression was detected in strain PKT392, confirming that MisS was indeed required for transmitting the DTT signal. The fact that the dsbD expression in strain PKT392 was not significantly different from that of the misS strain despite over expression of MisR also indicated that MisR phosphorylation by MisS is critical in dsbD regulation. No growth inhibition was detected with the short DTT treatment, and the induction was dose-dependent (Fig. 5C) and time-dependent (data not shown). Subsequently, transcription of the autoregulated misR was monitored for specific signaling effect through the MisRS two-component system. No induction by oxidized DTT, N-acetyl cysteine, reduced and oxidized glutathione, tris(2-carboxyethyl) phosphine (TCEP), paraquat, copper, hydrogen peroxide (all at 1 mM) or heat shock (45°C, 30 minutes) was detected (107%, 115%, 118%, 103%, 114%, 113%, 107%, 97% and 99% relative to no treatment, respectively), suggesting that induction by DTT is most likely not a general stress response.
Figure 5.
Induction of dsbD and misR transcription by DTT. (A) The YT322 (misR) and YT350 (dsbD) reporter strains in the wild type and ΔmisS backgrounds were grown to mid-log phase and the cultures were divided into two: one treated with 1 mM DTT for 30 minutes and the other serves as non-induced control. The activities were normalized to those of the wild type backgrounds without DTT treatment, which were set as 100%. The data represent the means and standard deviation of at least four and two independent assays for the wild type and the misS backgrounds, respectively. The asterisks indicate statistically significant difference between the induced and non-induced samples as determined by the Student’s t test with a 2-tailed distribution (**, p < 0.001). (B) qRT-PCR analysis. The wild type strain NMB, the misS mutant YT310 and the PKY392 strain (YT310, Plac::misR) induced with 1 mM IPTG were treated with or without 1mM DTT for 30 minutes and total RNA isolated. The data are the mean and standard deviations of RT-PCR experiments done in triplicate using at least two independent RNA preparations. (*, p < 0.01; **, p < 0.001) (C) Dose-dependent induction by DTT. The YT322 strain was treated with various DTT concentrations for 30 minutes prior to activity assay. The sample without DTT treatment was set as 100%. The data are the means and standard deviations of two independent experiments done in triplicate.
DsbD is essential for N. meningitidis
In order to better understand the biological consequence of the MisRS regulation of dsbD in response to environmental changes and investigate whether DsbD plays a role in meningococcal virulence, we attempted to create a meningococcal dsbD mutant. Our data showed that dsbD is independently transcribed (Fig. 3A), thus a dsbD::Ω mutational construct was created. Despite numerous transformation attempts, we could not identify any correct meningococcal dsbD mutant (data not shown). Another dsbD deletion-insertion mutational construct utilizing the nonpolar aphA3 cassette was subsequently generated. Although many kanamycin resistant transformants were obtained, colony PCR analyses indicated that these clones carried a dsbD::aphA3 mutation as well as a wild type copy of dsbD. Transformation with either the purified PCR product or the linearized plasmid construct gave similar results. These results implicated that dsbD might be essential for meningococci. Thus, we generated a meningococcal strain with a second copy of dsbD at a permissive chromosomal lctP-aspC locus (Mehr & Seifert, 1998) under the control of an IPTG inducible promoter. The resulting strain was then transformed with the dsbD::aphA3 construct, and in contrast to previous attempts, transformants with a correct dsbD mutation can be readily recovered. The correct transformants can be obtained without IPTG induction as the Plac control was leaky and yielded a basal level of expression in N. meningitidis.
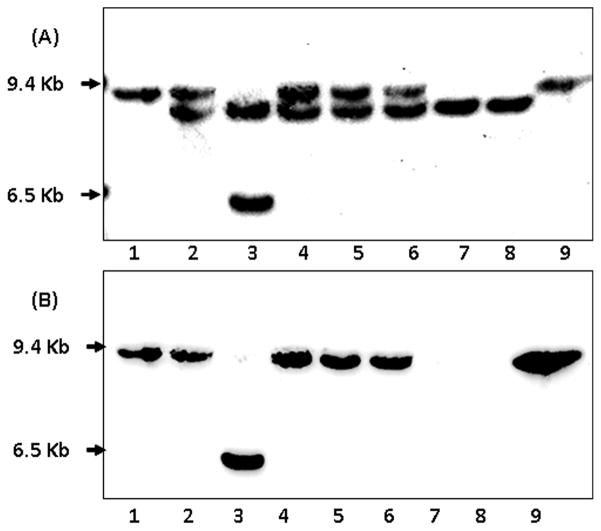
The results of PCR analyses were further confirmed by Southern blots. Detection with a probe that hybridizes with the remaining dsbD 3′ coding sequence (Fig. 6A) revealed a doublet in the dsbD* mutant that was recovered from the wild type strain (Fig. 6A, lane 2): the upper band corresponded to that of the wild type dsbD (lane 1), while the lower band was the mutated dsbD. In contrast, the strain with a dsbD mutation created in the complemented background (lane 3) only contained the lower band (the second smaller fragment is the complemented copy at the lctP-aspC locus). Further, a probe that hybridizes within the deleted region of the dsbD::aphA3 mutation detected the upper band in both the wild type and the dsbD* mutant (lanes 1 and 2 in Fig. 6B), but this signal was absent in the complemented dsbD mutant (lane 3). The probable duplication junctions and the extent of duplication were not determined. Therefore, the fact that the wild type dsbD locus can be disrupted only in the presence of a second functional copy of dsbD indicated that dsbD is essential for N. meningitidis.
Figure 6.
Characterization of dsbD mutants by Southern blots. SspI-digested chromosomal DNAs were resolved by 0.8% agarose gel. Lanes: 1) wild type strain NMB, 2) the incorrect mutant (dsbD*) obtained from the wild type strain, 3) the correct dsbD mutant created in the complemented strain, 4) dsbD* mutant of the dsbA1 strain, 5) dsbD* mutant of the dsbA2 strain, 6) dsbD* mutant of the dsbA2A3 strain, 7) dsbD mutant of the dsbA2A1 strain, 8) dsbD mutant of the dsbA2A1A3 strain. Removal of an 1.3-kb dsbD coding sequence and insertion of the 800-bp aphA3 cassette gave rise to a smaller fragment that is recognized by the probe made of the remaining 3′ dsbD sequence (A), but not by the probe hybridized to the deleted region (B). The dsbD* designation indicates incorrect dsbD clones that also carry a wild type copy of dsbD. An additional band in lane 3 of both panels represents the second copy of dsbD at the aspC-lctP locus.
A dsbD mutation can be established with compensatory dsbA mutations
DsbD together with the oxidoreductase DsbC constitute the reducing branch of the Dsb pathway and its main function is to transfer reducing power to the periplasm for correcting mis-formed disulfide bonds and for reducing apo-cytochrome c proteins prior to heme ligation. We postulated that if the oxidative Dsb pathway was inactivated, the requirement for a fully functional reductive branch might be lessened and consequently a dsbD mutation could be tolerated. To test this hypothesis, we created a strain with dsbA1, dsbA2 and dsbA3 triple mutations, and tested whether a clean dsbD mutation can be obtained. Interestingly, the correct dsbD mutation was indeed created in the dsbA1A2A3 triple null mutant (Fig. 6, lane 8). Subsequently, we tested combinations of double dsbA mutations and found that dsbA1A2, but not the dsbA2A3 and dsbA1A3 combinations, can sustain the dsbD mutation (Fig. 6, lanes 6 and 7 for dsbA2A3 and dsbA1A2, respectively; data not shown for dsbA1A3). Finally, an incorrect dsbD mutation was also detected in the single dsbA1, dsbA2 and dsbA3 mutants (Fig. 6, lanes 4 and 5 for dsbA1 and dsbA2, respectively, and data not shown). The correct dsbD mutation was stable with repeated passages. We have also attempted to generate the dsbD mutation in the wild type background by supplementing the selection media with various concentrations of DTT. How ever, no correct dsbD mutants could be recovered (Fig. S2). The fact that the dsbA1A2 double mutant produces the strongest phenotypes among the double mutational combinations and behaves similarly to the dsbA triple null mutant (Sinha et al., 2008, Tinsley et al., 2004) suggested that inactivation of DsbD can only be tolerated in situations with a minimal DsbA oxidative activity.
Characterization of the dsbD mutation in the dsbA1A2 background
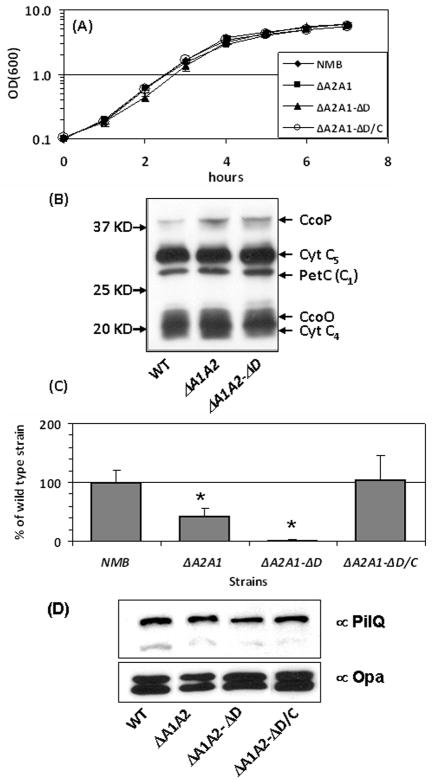
We examined the growth rates of the mutants in standard GC broth. Analogous to previous reports (Sinha et al., 2004, Tinsley et al., 2004), no obvious growth defect was detected for the dsbA1A2 mutant (Fig. 7A), while the triple dsbA null mutant exhibited reduced growth (data not shown). The growth rate of the dsbA1A2D mutant was also similar to that of the wild type strain (Fig. 7A), thus in subsequent characterization of DsbD contribution in various meningococcal phenotypes we compared the dsbA1A2D mutant to its parental dsbA1A2 strain. In addition, a complemented dsbD mutant with an IPTG inducible expression of dsbD at the nics locus (Mehr & Seifert, 1998) was also generated in the dsbA1A2 background for comparison.
Figure 7.
(A) Growth curves of the meningococcal strains in standard GC broth. (B) Heme staining of the wild type, dsbA2A1 and dsbA2A1D strains. The assignment of each band is based on the previous report (Li et al., 2010). (C) Natural competence of the wild-type NMB and its mutants. Bacteria were transformed with chromosomal DNA from a CmR derivative of NMB, and cfu counts were determined on selective and non-selective plates. Transformation frequencies were calculated by dividing the cfu numbers of selective plates by the cfu numbers of non-selective plates. The frequency of the mutant was normalized to that of the wild type, which was set as 100%. Data are the means ± standard deviations of two assays done in duplicate. (D) PilQ levels determined by Western blots. Equal amounts of whole cell lysates (8 μg) were resolved on 10% SDS-PAGE gels and transferred to PVDF membranes. The top half of the membrane was probed with PilQ antisera (Tonjum et al., 1998), while the lower half was probed with Opa antisera (Takahashi et al., 2008) for loading controls.
First, we assessed whether the cytochrome c maturation process was affected by the dsbD mutation, as c-type cytochromes are important in electron transfer chain and viability of meningococci (Deeudom et al., 2008, Li et al., 2010, Aspholm et al., 2010). The chemilumine scent detection of heme (Feissner et al., 2003) was used as a qualitative assessment of the overall cytochrome c profiles. Equal amounts of total proteins were loaded, resolved by SDS-PAGE in the absence of reducing agents and heat denaturing and then transferred to PVDF membranes for heme staining. As shown in Fig. 7B, wild type like cytochrome c profiles were detected in the dsbA1A2 and dsbA1A2D strains. Thus, the overall cytochrome c profiles appeared to be normal in both mutants, correlating with their growth phenotypes.
It has been previously reported that the meningococcal transformation efficiency was reduced by the dsbA1A2 mutation (Tinsley et al., 2004, Sinha et al., 2008) due to inefficient folding of the secret in PilQ (Sinha et al., 2008). PilQ is required for type IV pilin biogenesis and DNA binding, both necessary for natural transformation. We thus investigated whether the dsbD mutation will further influence meningococcal natural competence. Using the chromosomal DNA of strain NMB with a CmR marker at the nics locus as the transforming DNA, we found that the dsbA1A2D mutant exhibited ~10% of transformation frequency of its parental dsbA1A2 strain. Complementation with an IPTG inducible dsbD rescued the transformation defect (Fig. 7C). These results indicated that the folding defect of PilQ might be more profound in the absence of DsbD. However, the total PilQ protein levels of the mutants were not significantly different from that of the wild type strain when examined by Western blots (Fig. 7D). Thus, additional proteins involved in competence might be affected by the absence of DsbD.
The absence of dsbD enhanced the sensitivity of the dsbA1A2 mutant to oxidative stress
DsbC and DsbD of the isomerization pathway were shown to play a key role in protecting E. coli against oxidative copper stress (Hiniker et al., 2005). In addition, the methionine sulfoxide reductase MrsA/B, which repairs oxidative damage to proteins, acquires the reducing power from DsbD (Brot et al., 2006). It is likely that the increased numbers of periplasmic proteins carrying free thiol groups in the meningococcal dsbA1A2 mutant due to a slower oxidation process will render the strain more sensitive to oxidative stresses that cause incorrect disulfide bond formation and protein damage via nonspecific oxidation. We reasoned that this sensitivity will be further augmented in the absence of DsbD. Thus, we examined the sensitivity of the dsb mutants to hydrogen peroxide, paraquat and copper by disc diffusion assays. As shown in Table 2, relative to the wild type strain the dsbA1A2 mutant was more sensitive to paraquat and copper but not to hydrogen peroxide; while the dsbA1A2D mutant displayed enhanced sensitivities to all oxidants when compared to the parental dsbA1A2 strain. Complementation of dsbD reversed this sensitivity profile to levels consistent with the parental dsbA1A2 strain. Thus, we detected a decreased resistance to oxidative stress of the meningococcal dsbA1A2 strain and this sensitivity was further augmented by the dsbD mutation.
Table 2.
The dsbA and dsbD mutations increased sensitivity to oxidative stress
| Strain | Genotype | Zone of growth inhibition (mm) ± SDa with: | ||
|---|---|---|---|---|
| H2O2 (1%) | Paraquat (100 mM) | Copper (100 mM) | ||
| NMB | wild type | 35.2 ± 1.1 | 30.0 ± 1.4 | 20.4 ± 0.9 |
| PKT107 | dsbA1−dsbA2− | 36.6 ± 1.3 | 34.0 ± 2.5* | 31.2 ± 1.1** |
| PKT108 | dsbA1−dsbA2−dsbD− | 42.8 ± 1.1**(**) | 39.2 ± 3.0**(*) | 36.0 ± 0.0**(**) |
| PKT113 | dsbA1−dsbA2−dsbD−; Plac::dsbD | 37.2 ± 1.79 | 34.0 ± 2.5* | 30.4 ± 0.9** |
Data are the means ± standard deviations growth inhibition zones around at least four 6-mm filter discs obtained with the indicated concentration of reagents after overnight incubation. A Student’s t test showed significant differences when compared to the wild type strain (*, p < 0.05; **, p < 0.01), while the statistically significant differences when compared to the dsbA1A2 strain are shown in parentheses.
DISCUSSION
Many bacterial membrane and exported proteins are stabilized and attain their functional conformation by forming intramolecular disulfide bonds, which are catalyzed by the Dsb system in the periplasm. The activities of the Dsb system are necessary for many important pathogenic phenotypes including attachment, invasion and intracellular survival (Yu, 1998, Raivio, 2005, Bringer et al., 2007). In particular, DsbD plays a role in the translocation of reducing power from cytoplasm to several periplasmic thioredoxin-like proteins that include the disulfide isomerase DsbC, the cytochrome c reductase CcmG/CcsX, the peptide methionine sulfoxide reductase MsrA, a repair enzyme that contributes to the maintenance of functional adhesions (Brot et al., 2006, Wizemann et al., 1996), and additional periplasmic proteins shown to play a role in defense against oxidative stress and intracellular survival (Skaar et al., 2002, Brot et al., 2006, Achard et al., 2009). DsbD or its smaller homolog CcdA is present in all organisms that synthesize c-type cytochromes (Kranz et al., 2002) with the exception of Rickettsia, which is an obligate intracellular pathogen and likely uses host-derived reducing environment for its extracytoplasmic redox processes (Kranz et al., 2002).
Very little is known about how bacteria regulate the expression of Dsb proteins. The only regulation of the Dsb system illustrated to date is the control of dsbA. Expression of dsbA is regulated by the CpxRA two-component system in E. coli (Danese & Silhavy, 1997, Pogliano et al., 1997); while RtsA and H-NS regulate dsbA in Salmonella (Gallant et al., 2004, Ellermeier & Slauch, 2004). The response regulator CpxR has been shown to bind to the dsbA promoter and directly activate its expression (Pogliano et al., 1997). The MisRS two-component system is the first regulatory mechanism described for dsbD and is the first regulation identified for the meningococcal Dsb system. Our study has found that expression of dsbD, but not any of the other Dsb proteins, is significantly down regulated in the misR/S mutants (Fig. 1B), suggesting that MisR acts as a specific activator of dsbD transcription. Our current study further defined that MisR specifically binds to, and likely directly activates transcription from the dsbD promoter. A mutation of misS leads to a 5–10 fold decrease in dsbD expression. Hence, the MisRS system appears to contribute to the basal level of dsbD expression under normal growth conditions, suggesting that a low level of phosphorylated MisR may be present under unstimulated growth conditions. Why N. meningitidis evolves to specifically regulate DsbD of the reductive branch of the Dsb system is not clear. However, a coordinated control of three DsbAs in N. meningitidis presents a challenge. It seems likely that certain in vivo conditions may necessitate DsbD induction by the MisRS system to promote expedited proper disulfide folding as well as repair cell envelope/periplasmic proteins. For example, overexpression of DsbD upon the induction of the MisRS system by host derived environmental signals might combat stresses associated with certain host immune responses or microenvironments that resulted in protein damage within the extracytoplasmic compartments.
The regulatory mechanism(s) of the disulfide-folding process likely senses either cell envelope and/or periplasmic protein folding defects or senses the conditions that lead to folding defects. These signals must be transmitted across the inner membrane, a signal transduction mechanism that the two-component regulatory system is well suited for. We have found that exposure to the reducing agent DTT further induced expression of DsbD and its regulator MisR. DTT per se is not likely the actual signal, but rather its reducing effect on other physiological conditions or processes may signal the induction of the MisRS system. As DTT is freely diffusible through the inner membrane, it is also not clear the precise subcellular location for its effects. No induction was seen with a charged reducing agent, tris(2-carboxyethyl) phosphine (TCEP), which is a more powerful reducing agent and is commonly used to substitute DTT in many biochemical assays (Getz et al., 1999). TCEP is most likely unable to penetrate the inner membrane due to its charged nature, thus the induction of the MisRS system might require the reducing agent to diffuse through or act within the cytoplasmic membrane. Alternatively, TCEP has been found to be selective toward disulfide bonds, while DTT can react with Fe+3 or Ni+2 (Getz et al., 1999), implying that DTT might introduce a complicated effect on the cellular environment. Additional analyses are needed to clearly delineate the signal transduction mechanism initiated by DTT treatment that leads to the activation of the MisRS system. Further, it remains to be determined whether the perturbed redox balance of a particular subcellular environment or general or specific misfolding of proteins induces the global MisRS regulatory cascade.
In this study, we demonstrated that DsbD is essential for N. meningitidis. In the presence of an ectopically located dsbD, a correct mutation at the wild type dsbD locus can be achieved. On the other hand, numerous attempts to replace the dsbD gene by two independent mutational constructs in the wild type background were unsuccessful, and yielded clones carrying a duplicated wild type copy of dsbD elsewhere on the chromosome (Fig. 6). Similar duplication phenomenon has been described in N. gonorrhoeae. Attempts to inactivate the essential imp gene, which encoding an outer membrane protein important for lipooligosaccharide transport, in N. gonorrhoeae have resulted in clones carrying a wild type copy and a mutated copy of imp, representing merodiploids that were attributed to a rare recombination event (Bos et al., 2004, Tobiason & Seifert, 2010). Tobiason et al. have recently showed that both N. meningitidis and N. gonorrhoeae are polyploid and genetically haploid (Tobiason & Seifert, 2010). It is not clear whether the meningococcal polyploid genotype contributes to the duplication event.
Interestingly, a correct dsbD mutation can be attained in either the dsbA1A2 or dsbA1A2A3 genetic backgrounds, but not any other combinations of dsbA mutations. It has been shown that among the three double mutant combinations, the dsbA1A2 mutation yielded the most significant phenotypic changes that are similar to those of the triple dsbA null mutant (Tinsley et al., 2004, Sinha et al., 2008). Lafaye et al. showed that meningococcal DsbAs exhibit similar in vitro redox properties and are the most oxidizing thioredoxin-like enzymes known to date (Lafaye et al., 2009). The expression levels of dsbA genes normalized to 16s in the wild type strain NMB were assessed by qRT-PCR using 2−ΔΔCt comparison relative to dsbA1. The results indicated that expression levels among the dsbA genes are in the following order: dsbA1 ≥ dsbA2 >dsbA3 (1, 0.7 ± 0.3, 0.5 ± 0.1 for dsbA1, dsbA2 and dsbA3, respectively). Thus, in addition to the likely different substrate specificity of each DsbA, the amounts of DsbA1 and DsbA2 relative to DsbA3 may also contribute to the variations in phenotypes of the double mutants. A possible explanation for the compensatory mutation effect of dsbA1A2 and dsbD might be the redox requirement of cytochrome c maturation. Neisseria species produce several c-type cytochromes that participate in electron transfer chain (Deeudom et al., 2008) and the cytochrome c oxidase cbb3 is essential for aerobic growth (Aspholm et al., 2010). The survival of N. gonorrhoeae was also shown to depend upon the presence of either cytochrome c4 or c5 (Li et al., 2010). All c-type cytochromes are located in the periplasm and the maturation process involves the covalent attachment of heme to two cysteine residues of a CXXCH motif of the apo-protein. Two bacterial cytochrome c biogenesis pathways termed system I and II have been revealed (Kranz et al., 2002, Kranz et al., 1998), and N. meningitidis utilizes the System II pathway. The system II biogenesis pathway has been well characterized in Bordetella pertussis to require at least four specific assembly proteins, CcsA (ResC), CcsB (ResB), CcsX and DsbD (or its smaller homologue, CcdA) (Beckett et al., 2000). The CcsB and CcsA proteins form a cytochrome c synthetase complex that also functions in heme transport (Frawley & Kranz, 2009), while the thioredoxin like protein CcsX in cooperation with DsbD mediates the reduction of apo cytochrome c. A plausible model to explain the dsbA/dsbD dependence in meningococci is that the cysteine residues in the CXXCH motifs of apo c-type cytochrome proteins are oxidized by DsbAupon entering the periplasm and require re-reduction prior to heme ligation. This re-reduction involves DsbD as a donor of reducing power. Thus, the diminished reducing power within the periplasm caused by dsbD inactivation is compensated by the absence of both DsbA1 and DsbA2 and, therefore, allows for continued c-type cytochrome biosynthesis. Interestingly, Kranz et al. has reported failed attempts to make deletions in genes required for System II cytochrome c biogenesis in N. meningitidis, although the specific gene targeted was not described (Kranz et al., 2002). Analogous to our observation in N. meningitidis, the lack of DsbA in Bacillus subtilis or Rhodobacter capsulatus suppresses the cytochrome c deficiency of the ccdA (dsbD) mutants, indicating that the reductive branch becomes dispensable in the absence of the oxidative pathway (Erlendsson & Hederstedt, 2002, Deshmukh et al., 2003). In addition, the defect in c-type cytochrome biosynthesis of dsbD mutants in E. coli can be restored by the cytoplasmic membrane-impermeable thiol, mercaptoethane sulfonic acid, supplemented in the growth medium (Sambongi & Ferguson, 1994), again suggesting that a balance of oxidative and reductive reactions is required for cytochrome c maturation. However, an exogenous supplementation of DTT could not rescue the dsbD defect in N. meningitidis.
Alternatively, the more error prone oxidation by other periplasmic oxidants that are oxidized by DsbAs could potentially damage essential enzymatic functions within periplasm. In a study designed to identify periplasmic substrates of DsbG, Depuydt et al. showed that the single cysteines of several proteins belonging to the family of L, D-transpeptidases, which catalyze the crosslinking of peptidoglycan for cell wall synthesis, are oxidized to a sulfenic acid (Cys-SOH) by oxidants present in the periplasm and are protected by the function of DsbG and DsbC (Depuydt et al., 2009). DsbC and DsbG (absent in meningococci) that are kept reduced by DsbD were shown to control the level of cysteine sulfenylation in the periplasm and provides reducing equivalents to rescue oxidatively damaged proteins (Depuydt et al., 2009). Thus, the electron flux mediated by DsbD provides the reducing power required for not only the correction of incorrect disulfides but also the rescue of sulfenylated orphan cysteines (Depuydt et al., 2009)in critical periplasmic enzymes. Consequently, together with the role of DsbD in cytochrome c maturation discussed above, only a less oxidative periplasmic environment lacking DsbAs can sustain a dsbD null mutation in N. meningitidis.
Due to the necessity of dsbA1 and dsbA2 mutations in maintaining the dsbD mutation, we have examined the contribution of DsbD to several phenotypes with respect to that of the parental dsbA1A2 background. In general, the dsbD mutation further enhanced the defect of the dsbA1A2 mutant. This was seen in the transformation efficiency (Fig. 7C). The transformation deficiency of the dsbA1A2 mutant has been attributed to folding defects of PilQ and PilE that reduce the PilQ protein level to cause deficiency in DNA binding and uptake but allow the formation of non-functional pilus fibers on cell surface (Tinsley et al., 2004, Sinha et al., 2008). Similar to these earlier studies, we did not detect differences in levels of the pilin subunits (PilE) between the dsbA1A2D mutant and the wild type strains by immunoblots (data not shown); while in contrast to the earlier report (Sinha et al., 2008), no clear reduction in PilQ protein levels can be detected in the dsbA1A2 and dsbA1A2D mutants. Our results suggested that inactivation of dsbA1A2 and dsbD in the meningococcal strain NMB did not appear to severely impair the disulfide folding and assembly of PilQ multimers but likely affected other competence-related proteins with multiple cysteine residues. For example, the DprA protein that is essential for natural competence (Smeets et al., 2000, Takata et al., 2005) has four cysteines, thus may requires a functional DsbD for proper expression.
Mutation in dsbA1A2 increased the meningococcal sensitivity to oxidative stresses elicited by paraquat, and copper, but not hydrogen peroxide, whereas the dsbD mutation further enhanced the sensitivity of N. meningitidis to all three stresses (Table 2). Hydrogen peroxide is known to oxidize critical cysteine or methionine residues in folded proteins, but it likely elicits only modest effect on DsbA-catalyzed oxidative protein folding. Most of the copper-mediated hydroxyl radical formation occurs in the periplasm (Macomber et al., 2007). In E. coli, the dsbA mutant is not sensitive to oxidative copper stress comparing to wild type, but the dsbA mutation does increase copper sensitivity of a dsbD− strain (Hiniker et al., 2005). Mutation in meningococcal dsbA1A2, however, increased the sensitivity to oxidative copper stress. It is possible that due to the need of three DsbA proteins, N. meningitidis relies more on DsbA for proper disulfide folding than does E. coli, and thus displays a stronger copper phenotype when DsbAs are inactivated. The further augmentation of oxidative stress sensitivity of the meningococcal dsbA mutant by the dsbD mutation may due to the lack of reducing power from DsbD to the methionine sulfoxide reductase MrsAB, which is involved in survival against oxidative stress (Brot et al., 2006, Skaar et al., 2002).
DsbD has not been shown to be essential for any other bacteria, likely because 1) the presence of other types of cytochrome oxidases allows the lack of c-type cytochrome biosynthesis or 2) other mechanisms or proteins capable of delivering reducing power to the periplasm can compensate for the absence of DsbD. For example, a heterodimeric ABC transport system, CydDC, which exports glutathione and cysteine from cytoplasm into the periplasm, represents a redundant pathway of DsbD and has been shown to be required for assembly of the cytochrome bd-type terminal oxidase in E. coli (Pittman et al., 2005). As overexpression of cydDC restores defects of a dsbD mutant, it appears that in E. coli the CydDC exporter can compensate the inadequate periplasmic reducing capability of a dsbD mutant for cytochrome assembly (Pittman et al., 2005). However, no clear cydDC homolog can be found in N. meningitidis. Taken together, our data suggest that the meningococcal Dsb system plays an important role in combating periplasmic oxidative stress and that the MisRS system is critical in controlling the function of the Dsb system via the regulation of dsbD expression.
Experimental procedures
Plasmids, strains and media
The strains and plasmids used in this study are listed in Table 1. Neisseria meningitidis strain NMB (CDC 8201085) is a serogroup B meningococcal strain originally isolated in 1982 from the cerebrospinal fluid of a patient with meningococcal meningitis in Pennsylvania. Meningococcal strains were grown with 5% CO2 at 37°C on GC base (GCB; Difco) agar containing supplements of 0.4% glucose and 0.68 mM Fe(NO3)3, or GC broth with the same supplements and 0.043% NaHCO3. Brain heart infusion (BHI) medium with 1.25% fetal bovine serum was used when kanamycin selection was required. Escherichia coli DH5α or Top 10 strains were routinely grown in Luria Bertani broth for cloning and propagation of plasmids. N. meningitidis was transformed by the procedure of Janik et al. (Janik et al., 1976). E. coli strains were transformed by chemical competence (Top 10) or by electroporation (DH5α) with a GenePulser (Bio-Rad) according to the manufacturer’s protocol. Antibiotic concentrations (μg/ml) for N. meningitidis were: kanamycin 80, chloramphenicol, 5 (Neil & Apicella, 2009), tetracycline, 5, spectinomycin, 60 and erythromycin 3; and for E. coli strains were: ampicillin 100, kanamycin 50, chloramphenicol 34, spectinomycin 100, and erythromycin 300.
Table 1.
Strains and plasmids used in this study
| strain | genotype | reference |
|---|---|---|
| N. meningitidis | ||
| NMB | B:2b:P1.2, 5:L2(CDC8201085) | (Stephens et al., 1991) |
| YT310 | NMB/misS::aphA-3 | (Tzeng et al., 2006) |
| YT391 | NMB310 complemented with Plac::misS at the lctP- aspC locus | (Sannigrahi et al., 2009) |
| SZT1001/YT0336 | NMB/misR::aphA-3 | (Tzeng et al., 2008) |
| C1001 | NMB/misR::aphA-3 complemented with Ptrc::misR in the iga locus | (Tzeng et al., 2008) |
| SZT1003 | NMB/misRS::aphA-3 | (Tzeng et al., 2008) |
| PKT101 | dsbD::aphA3 | This study |
| PKT102 | dsbD::aphA3, Plac::dsbD | This study |
| PKT103 | dsbA1::Ω(Sp) | This study |
| PKT104 | dsbA1::Ω(Sp), dsbD::aphA3 | This study |
| PKT105 | dsbA2::tetM | This study |
| PKT106 | dsbA2:: tetM, dsbD::aphA3 | This study |
| PKT107 | dsbA1::Ω(Sp), dsbA2:: tetM | This study |
| PKT108 | dsbA1::Ω(Sp), dsbA2:: tetM, dsbD::aphA3 | This study |
| PKT109 | dsbA2:: tetM, dsbA3::ermC | This study |
| PKT110 | dsbA2:: tetM, dsbA3:: ermC, dsbD::aphA3 | This study |
| PKT111 | dsbA1::Ω(Sp), dsbA2:: tetM, dsbA3:: ermC | This study |
| PKT112 | dsbA1::Ω(Sp), dsbA2:: tetM, dsbA3:: ermC, dsbD::aphA3 | This study |
| PKT113 | dsbA1::Ω(Sp), dsbA2:: tetM, dsbD::aphA3, Plac::dsbD | This study |
| PKT392 | Plac::misR in YT310 | This study |
| YT350 | dsbD::lacZ (−277/+123) | (Tzeng et al., 2008) |
| YT350s | dsbD (−277/+123)::lacZ in YT310 | (Tzeng et al., 2008) |
| YT322 | misR::lacZ in NMB | (Tzeng et al., 2006) |
| YT322s | misR::lacZ in YT310 | (Tzeng et al., 2006) |
| D227 | dsbD::lacZ (−227/+123)::lacZ | This study |
| D227s | dsbD (−227/+123)::lacZ in YT310 | This study |
| D137 | dsbD (−137/+123)::lacZ | This study |
| D137s | dsbD (−137/+123)::lacZ in YT310 | This study |
| D67 | dsbD (−67/+123)::lacZ | This study |
| D67s | dsbD (−67/+123)::lacZ in YT310 | This study |
| D37 | dsbD (−37/+123)::lacZ | This study |
| D37s | dsbD (−37/+123)::lacZ in YT310 | This study |
| D8 | dsbD (−8/+123)::lacZ | This study |
| E. coli | ||
| DH5α | Cloning strain | New England Biolab |
| TOP10 | Cloning strain | Invitrogen |
| Plasmid | ||
| pUC18k | Source of aphA-3 cassette | (Menard et al., 1993) |
| pHP45Ω | Source of Ω(Sp) cassette | (Prentki & Krisch, 1984) |
| pCR2.1 | TOPO cloning vector | Invitrogen |
| pSmartLCKan | Blunt-end cloning vector | Lucigen |
| pYT328 | Cloning vector for chromosomal lacZ fusion | (Tzeng et al., 2006) |
| pGCC4 | Complementation vector for integration into lctP- aspC locus | (Mehr & Seifert, 1998) |
| pJKD2639 | dsbA1::Ω(Sp) | This study |
| pJKD2641 | dsbA2::tetM | This study |
| pJKD2643 | dsbA3:: ermC | This study |
| pPK101 | dsbD::aphA3 | This study |
| pPK102 | Plac::dsbD in pGCC4 | This study |
| pYT382 | dsbD::Ω(Sp) | This study |
| pYT392 | Plac::misR in pGCC4 | This study |
The ΔdsbD constructs and the dsbD complementation plasmid
A nonpolar aphA3 insertional mutation with a 1296-bp deletion of dsbD (1803 bp) was generated by overlapping PCR. A 562-bp PCR product containing the upstream and 5′ dsbD coding sequence (94-bp) was generated using primer pair 5Dkan-F and 5Dkan-R. Another 779-bp PCR product containing the 416-bp 3′ sequence of dsbD was amplified using primer pair 3Dkan-F and 3Dkan-R. The aphA3 cassette from pUC18K (Menard et al., 1993) was amplified with primers aphA3-cla-f2 and aphA3-cla-r2 and digested with SmaI. The first overlapping PCR was performed with the aphA3 and 3′ dsbD fragments, and the resulting product was purified, digested with SmaI, and used for the second round of overlapping PCR with the 5′ fragment. The final PCR product with the expected size was purified and cloned into pCR2.1. Removal of the dsbD sequence and the presence of a correctly oriented aphA-3 cassette in the resulting pPK101 plasmid were confirmed by sequencing analysis. To generate the meningococcal dsbD mutant, pPK101 was linearized with ScaI to disrupt the AmpR gene and the digestion mixture was used to transform meningococcal strains. Alternatively, the PCR product was used directly for transformation of meningococci. Colonies were selected on BHI agar plates with kanamycin.
A dsbD::Ω(Sp) construct was also made. A 2745-bp DNA containing the entire dsbD and the flanking sequence was obtained with primers ΔdsbD-F and ΔdsbD-R and cloned into pCR2.1 by TOPO cloning method (Invitrogen). The resulting plasmid was digested with HincII to remove ~ 1.3 kb insert, gel purified and then ligated with the Ω(Sp) fragment released from pHP45Ω (Prentki & Krisch, 1984) with SmaI digestion. Deletion of the dsbD sequence and insertion of Ω in the resulting pYT382 plasmid was confirmed by sequencing analysis.
For complementation, the dsbD coding sequence was PCR amplified with dsbD-PacI and dsbD-PmeI primers from strain NMB. The PCR product was digested with PacI and PmeI and then cloned into pGCC4 (Mehr & Seifert, 1998) cut with the same enzymes to yield pPK102. The presence of correct insert was confirmed by PCR and sequencing analyses.
The dsbA mutants
dsbA1
A 2.5 Kb fragment encompassing the dsbA1 gene was amplified using DAP275 and DAP263 primers and cloned into the HincII site of pHSG576 by blunt end ligation. The resulting plasmid was digested with BssHII, blunt-ended and ligated with the Ω cassette released from pHP45Ω to yield pJKD2639. N. meningitidis strain NMB was transformed with pJKD2639 and SpecR colonies selected. The correct transformants were confirmed by PCR with the chromosomal specific primer DAP418 and a Ω cassette specific primer DAP140.
dsbA2
The dsbA2 locus of strain NMB was amplified by PCR using primers DAP292 and DAP201. The 3.5 kb PCR product was cloned into the HincII site of pHSG576. The resulting plasmid was cut with the unique NotI site within the dsbA2 coding sequence, ligated with the EcoRV fragment of tetM obtained from pJKD2410 to produce pJKD2641. The pJKD2641 plasmid was used to transform strain NMB and TetR transformants screened with primers DAP285 and DAP420, which anneal outside of the cloned region. An increase of 4 Kb in PCR product size compared to that obtained from strain NMB confirmed the allelic exchange.
dsbA3
Similarly, the dsbA3 fragment (1.2 Kb using primers DAP268 and DAP269) was cloned into the HincII site of pHSG576 and a HincII-BssHII fragment was deleted prior to the insertion of the ermC cassette. The ErmR colonies were screened for correct ermC orientation relative to dsbA3, and the resulting pJKD2643 plasmid was then used for meningococcal transformation. Transformants were examined with primers DAP267 and DAP265 flanking the cloned fragment and clones with a 0.8 Kb increase in PCR product size were saved.
The dsbD::lacZ transcriptional fusion and β-galactosidase assays
Promoter fragments were obtained using the following primer pairs: dsbD-f2/dsbD-r1 (350-bp), dsbD-f3/dsbD-r1 (260-bp), dsbD-f4/dsbD-r1 (190-bp), dsbD-f6/dsbD-r1 (160-bp) and dsbD-f5/dsbD-r1 (131-bp). Each promoter fragment with flanking EcoRI sites was PCR amplified with chromosomal DNA of strain NMB as template using Phusion polymerase (New England Biolab). After EcoRI digestion, the fragments were purified and cloned into the EcoRI site of pYT328 (Tzeng et al., 2006). The resulting plasmids were linearized with NcoI and used to transform meningococcal strain NMB. Correct transformants were identified by colony PCR as described and named D227, D137, D67, D37, and D8, respectively. The reporters in the misS mutant background were generated by transformation of strain YT310, and designated similarly to those of the wild type strain with an S extension. β-Galactosidase activity was assayed by the Miller method (Miller, 1972) performed in triplicates. The reporter strains were grown in GC broth with supplements at 37°C with 200 rpm aeration to mid-log phase (OD600 ~ 0.5).
qRT-PCR
Cultures were grown in standard GC broth and cells were collected at mid-log phase. Two volume of RNA protect Bacteria Reagent was added into cultures and RNA was isolated using RNeasy mini kit (Qiagen) following the manufacturer’s recommendations with on-column DNase digestion. PCR amplification of the RNA samples using primers confirmed no genomic DNA contamination. cDNA was obtained by reverse transcription of total RNA (1 μg) using GeneAmp RNA PCR core kit (Applied Biosystems) and reactions without the reverse transcriptase were used as a negative control. The transcription of genes of interest was measured using the SYBR green detection method (SYBR Green Supermix, BioRad) and the reaction conditions have been described previously (Tzeng et al., 2004). The 16s rRNA was used as an internal control in each experiment for normalization. The primers designed with Primer 3 software (http://fokker.wi.mit.edu/primer3/; (Rozen & Skaletsky, 2000)) were listed in Table S1 and have been confirmed to yield similar amplification efficiencies and thus were suitable for the 2−ΔΔCt method (Livak & Schmittgen, 2001)to determine the relative transcriptional changes between the mutant strains and the wild type strain. The values of the wild type strain were used as the calibrator for comparison. RT negative control reactions were also analyzed to measure whether there was contaminating chromosomal DNA and melting curve analyses were performed following each qRT-PCR experiment to ensure that each reaction contained only a single specific product. Each qRT-PCR was performed in triplicates. Student’s t test with a two-tailed hypothesis was used to determine the significant difference (p<0.01) between two variables in this study.
Electrophoretic mobility shift assay (EMSA) and DNase I protection assay
Both EMSA and DNase I protection assays were performed following previously reported procedures (Tzeng et al., 2006). The promoter fragments used in EMSA were obtained by PCR amplification and end-labeled using T4 kinase, while the probes for DNase I protection assay was prepared by PCR using 32P-labeled primers, dsbD-pf2 and dsbD-pr1. Competition with excess specific (unlabeled probes) and nonspecific competitors, a 237-bp internal coding sequence of misS obtained by PCR amplification using primers YT150 and YT141 was performed to assess the specificity of the interaction.
Primer extension
Total RNA (30 μg) was used in primer extension reactions with 32P-labeled primers (YT144 and dsbD-PE1) following previously described procedure (Sannigrahi et al., 2009). The corresponding DNA sequencing reactions were carried out using the same labeled oligonucleotides and PCR fragments of the promoter regions with Thermo Sequenase dye primer manual cycle sequencing kit (USB). The extension products and the sequencing ladders were resolved on an 8% sequencing gel.
Chromosomal DNA isolation and Southern blots
Meningococcal chromosomal DNA was prepared according to the method of Nath (Nath, 1990). The PCR DIG probe synthesis and detection kit (Roche) was used to perform DNA hybridization. The digoxigenin labeled probe for detecting the deleted dsbD sequence was generated by with dsbD-For and dsbD-Rev, while the probe for the remaining dsbD sequence was made with 3Dkan-F and dsbD-PmeI. Chromosomal DNA was digested with SspI overnight and resolved on a 0.8 % agarose gel. DNA was transferred to a nylon membrane and hybridization and development of the blots were performed following the manufacturer’s protocol.
Heme staining
The previously described procedure (Feissner et al., 2003, Feissner et al., 2005) was followed. Plate grown cultures was collected and suspended in PBS. One ml of cell suspensions at an OD600 of 1.5 was pelleted and the supernatant removed. Pellets frozen at −80°C for a minimum of 15 min to aid in cell disruption were thawed on ice and resuspended in 100 μl of B-PER extraction reagent (Pierce) by pipetting and vortexing. The supernatant obtained after centrifugation at 15,000 × g for 5 minutes was saved and protein concentration determined by BCA assay using BSA as a standard (Pierce). Equal amounts of proteins were mixed with DTT-free loading buffer and loaded directly without heat denaturing. Samples were separated by 12% SDS-PAGE and transferred to nitrocellulose membrane. Membranes were washed twice in PBS and SuperSignal West Pico chemiluminescent substrates (Pierce) prepared according to manufacturer’s instruction were applied to the membranes for 5 minutes, followed by exposure to X-ray film.
Transformation efficiency
Plate-grown meningococcal strains were suspended in GC broth supplemented with 5 mM MgCl2. One μg chromosomal DNA was added to aliquots (100 μl) of cell suspension at an OD550 of 0.1 and then incubated for 1 hr at 37°C. Pre-warmed GC broth with complete supplements (500 μl) and DNase I (2 units) was added and the incubation continued for another hour. Serial dilutions were plated onto both selective and non-selective plates and the colony forming units (cfu) determined after overnight growth. The efficiencies were calculated as the ratio of cfu from the selection plate to the cfu of non-selective plates. The transforming DNA was chromosomal DNA prepared from strain NMB carrying a chloramphenicol cassette at the aspC-lctP locus.
Western blot
The meningococcal strains were grown on GC plates overnight and resuspended in PBS. Whole cell lysates were prepared by sonication and equal amounts of proteins (8 μg) were resolved by 10% SDS-PAGE, and transferred to nitrocellulose membranes. The antisera against PilQ (Tonjum et al., 1998) and Opa (Takahashi et al., 2008) were used at 1:2000 dilutions and anti-rabbit IgG-HRP conjugate secondary antibody was used at 1:15,000 dilution.
Supplementary Material
Acknowledgments
This work was supported by grants R01 AI061031 to Y. T. from the National Institutes of Health. We thank Dr. Sarah Satola for providing Southern hybridization reagents. We are grateful to Dr. H. Takahashi for the generous gift of Opa and PilE antisera and to Dr. Ashild Vik for the PilQ antisera.
References
- Achard ME, Hamilton AJ, Dankowski T, Heras B, Schembri MS, Edwards JL, Jennings MP, McEwan AG. A periplasmic thioredoxin-like protein plays a role in defense against oxidative stress in Neisseria gonorrhoeae. Infect Immun. 2009;77:4934–4939. doi: 10.1128/IAI.00714-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspholm M, Aas FE, Harrison OB, Quinn D, Vik A, Viburiene R, Tonjum T, Moir J, Maiden MC, Koomey M. Structural alterations in a component of cytochrome c oxidase and molecular evolution of pathogenic Neisseria in humans. PLoS Pathog. 2010:6. doi: 10.1371/journal.ppat.1001055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett CS, Loughman JA, Karberg KA, Donato GM, Goldman WE, Kranz RG. Four genes are required for the system II cytochrome c biogenesis pathway in Bordetella pertussis, a unique bacterial model. Mol Microbiol. 2000;38:465–481. doi: 10.1046/j.1365-2958.2000.02174.x. [DOI] [PubMed] [Google Scholar]
- Bilukha OO, Rosenstein N. Prevention and control of meningococcal disease. Recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR Recomm Rep. 2005;54:1–21. [PubMed] [Google Scholar]
- Bos MP, Tefsen B, Geurtsen J, Tommassen J. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc Natl Acad Sci U S A. 2004;101:9417–9422. doi: 10.1073/pnas.0402340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringer MA, Rolhion N, Glasser AL, Darfeuille-Michaud A. The oxidoreductase DsbA plays a key role in the ability of the Crohn’s disease-associated adherent-invasive Escherichia coli strain LF82 to resist macrophage killing. J Bacteriol. 2007;189:4860–4871. doi: 10.1128/JB.00233-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brot N, Collet JF, Johnson LC, Jonsson TJ, Weissbach H, Lowther WT. The Thioredoxin Domain of Neisseria gonorrhoeae PilB Can Use Electrons from DsbD to Reduce Downstream Methionine Sulfoxide Reductases. J Biol Chem. 2006;281:32668–32675. doi: 10.1074/jbc.M604971200. [DOI] [PubMed] [Google Scholar]
- Collet JF, Bardwell JC. Oxidative protein folding in bacteria. Mol Microbiol. 2002;44:1–8. doi: 10.1046/j.1365-2958.2002.02851.x. [DOI] [PubMed] [Google Scholar]
- Danese PN, Silhavy TJ. The sigma(E) and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 1997;11:1183–1193. doi: 10.1101/gad.11.9.1183. [DOI] [PubMed] [Google Scholar]
- Deeudom M, Koomey M, Moir JW. Roles of c-type cytochromes in respiration in Neisseria meningitidis. Microbiology. 2008;154:2857–2864. doi: 10.1099/mic.0.2008/020339-0. [DOI] [PubMed] [Google Scholar]
- Depuydt M, Leonard SE, Vertommen D, Denoncin K, Morsomme P, Wahni K, Messens J, Carroll KS, Collet JF. A periplasmic reducing system protects single cysteine residues from oxidation. Science. 2009;326:1109–1111. doi: 10.1126/science.1179557. [DOI] [PubMed] [Google Scholar]
- Deshmukh M, Turkarslan S, Astor D, Valkova-Valchanova M, Daldal F. The dithiol:disulfide oxidoreductases DsbA and DsbB of Rhodobacter capsulatus are not directly involved in cytochrome c biogenesis, but their inactivation restores the cytochrome c biogenesis defect of CcdA-null mutants. J Bacteriol. 2003;185:3361–3372. doi: 10.1128/JB.185.11.3361-3372.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier CD, Slauch JM. RtsA coordinately regulates DsbA and the Salmonella pathogenicity island 1 type III secretion system. J Bacteriol. 2004;186:68–79. doi: 10.1128/JB.186.1.68-79.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlendsson LS, Hederstedt L. Mutations in the thiol-disulfide oxidoreductases BdbC and BdbD can suppress cytochrome c deficiency of CcdA-defective Bacillus subtilis cells. J Bacteriol. 2002;184:1423–1429. doi: 10.1128/JB.184.5.1423-1429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feissner R, Xiang Y, Kranz RG. Chemiluminescent-based methods to detect subpicomole levels of c-type cytochromes. Anal Biochem. 2003;315:90–94. doi: 10.1016/s0003-2697(02)00658-9. [DOI] [PubMed] [Google Scholar]
- Feissner RE, Beckett CS, Loughman JA, Kranz RG. Mutations in cytochrome assembly and periplasmic redox pathways in Bordetella pertussis. J Bacteriol. 2005;187:3941–3949. doi: 10.1128/JB.187.12.3941-3949.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frawley ER, Kranz RG. CcsBA is a cytochrome c synthetase that also functions in heme transport. Proc Natl Acad Sci U S A. 2009;106:10201–10206. doi: 10.1073/pnas.0903132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant CV, Ponnampalam T, Spencer H, Hinton JC, Martin NL. H-NS represses Salmonella enterica serovar Typhimurium dsbA expression during exponential growth. J Bacteriol. 2004;186:910–918. doi: 10.1128/JB.186.4.910-918.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getz EB, Xiao M, Chakrabarty T, Cooke R, Selvin PR. A comparison between the sulfhydryl reductants tris(2-carboxyethyl)phosphine and dithiothreitol for use in protein biochemistry. Anal Biochem. 1999;273:73–80. doi: 10.1006/abio.1999.4203. [DOI] [PubMed] [Google Scholar]
- Hiniker A, Collet JF, Bardwell JC. Copper stress causes an in vivo requirement for the Escherichia coli disulfide isomerase DsbC. J Biol Chem. 2005;280:33785–33791. doi: 10.1074/jbc.M505742200. [DOI] [PubMed] [Google Scholar]
- Hoch JA. Two-component and phosphorelay signal transduction. Curr Opin Microbiol. 2000;3:165–170. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- Janik A, Juni E, Heym GA. Genetic transformation as a tool for detection of Neisseria gonorrhoeae. J Clin Microbiol. 1976;4:71–81. doi: 10.1128/jcm.4.1.71-81.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CR, Newcombe J, Thorne S, Borde HA, Eales-Reynolds LJ, Gorringe AR, Funnell SG, McFadden JJ. Generation and characterization of a PhoP homologue mutant of Neisseria meningitidis. Mol Microbiol. 2001;39:1345–1355. doi: 10.1111/j.1365-2958.2001.02324.x. [DOI] [PubMed] [Google Scholar]
- Kadokura H, Katzen F, Beckwith J. Protein Disulfide Bond Formation in Prokaryotes. Annu Rev Biochem. 2003 doi: 10.1146/annurev.biochem.72.121801.161459. [DOI] [PubMed] [Google Scholar]
- Kranz R, Lill R, Goldman B, Bonnard G, Merchant S. Molecular mechanisms of cytochrome c biogenesis: three distinct systems. Mol Microbiol. 1998;29:383–396. doi: 10.1046/j.1365-2958.1998.00869.x. [DOI] [PubMed] [Google Scholar]
- Kranz RG, Beckett CS, Goldman BS. Genomic analyses of bacterial respiratory and cytochrome c assembly systems: Bordetella as a model for the system II cytochrome c biogenesis pathway. Res Microbiol. 2002;153:1–6. doi: 10.1016/s0923-2508(01)01278-5. [DOI] [PubMed] [Google Scholar]
- Lafaye C, Iwema T, Carpentier P, Jullian-Binard C, Kroll JS, Collet JF, Serre L. Biochemical and structural study of the homologues of the thiol-disulfide oxidoreductase DsbA in Neisseria meningitidis. J Mol Biol. 2009;392:952–966. doi: 10.1016/j.jmb.2009.07.056. [DOI] [PubMed] [Google Scholar]
- Li Y, Hopper A, Overton T, Squire DJ, Cole J, Tovell N. Organization of the electron transfer chain to oxygen in the obligate human pathogen Neisseria gonorrhoeae: roles for cytochromes c4 and c5, but not cytochrome c2, in oxygen reduction. J Bacteriol. 2010;192:2395–2406. doi: 10.1128/JB.00002-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Macomber L, Rensing C, Imlay JA. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J Bacteriol. 2007;189:1616–1626. doi: 10.1128/JB.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehr IJ, Seifert HS. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol Microbiol. 1998;30:697–710. doi: 10.1046/j.1365-2958.1998.01089.x. [DOI] [PubMed] [Google Scholar]
- Menard R, Sansonetti PJ, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1972. [Google Scholar]
- Nath K. A rapid DNA isolation procedure from petri dish grown clinical bacterial isolates. Nucleic Acids Res. 1990;18:6462. doi: 10.1093/nar/18.21.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil RB, Apicella MA. Role of HrpA in biofilm formation of Neisseria meningitidis and regulation of the hrpBAS transcripts. Infect Immun. 2009;77:2285–2293. doi: 10.1128/IAI.01502-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcombe J, Eales-Reynolds LJ, Wootton L, Gorringe AR, Funnell SG, Taylor SC, McFadden JJ. Infection with an avirulent phoP mutant of Neisseria meningitidis confers broad cross-reactive immunity. Infect Immun. 2004;72:338–344. doi: 10.1128/IAI.72.1.338-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcombe J, Jeynes JC, Mendoza E, Hinds J, Marsden GL, Stabler RA, Marti M, McFadden JJ. Phenotypic and transcriptional characterization of the meningococcal PhoPQ system, a magnesium-sensing two-component regulatory system that controls genes involved in remodeling the meningococcal cell surface. J Bacteriol. 2005;187:4967–4975. doi: 10.1128/JB.187.14.4967-4975.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima T, Aiba H, Masuda Y, Kanaya S, Sugiura M, Wanner BL, Mori H, Mizuno T. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol Microbiol. 2002;46:281–291. doi: 10.1046/j.1365-2958.2002.03170.x. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Achtman M, James KD, Bentley SD, Churcher C, Klee SR, Morelli G, Basham D, Brown D, Chillingworth T, Davies RM, Davis P, Devlin K, Feltwell T, Hamlin N, Holroyd S, Jagels K, Leather S, Moule S, Mungall K, Quail MA, Rajandream MA, Rutherford KM, Simmonds M, Skelton J, Whitehead S, Spratt BG, Barrell BG. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- Pittman MS, Robinson HC, Poole RK. A bacterial glutathione transporter (Escherichia coli CydDC) exports reductant to the periplasm. J Biol Chem. 2005;280:32254–32261. doi: 10.1074/jbc.M503075200. [DOI] [PubMed] [Google Scholar]
- Pogliano J, Lynch AS, Belin D, Lin EC, Beckwith J. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 1997;11:1169–1182. doi: 10.1101/gad.11.9.1169. [DOI] [PubMed] [Google Scholar]
- Prentki P, Krisch HM. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Raivio TL. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol Microbiol. 2005;56:1119–1128. doi: 10.1111/j.1365-2958.2005.04625.x. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Sambongi Y, Ferguson SJ. Specific thiol compounds complement deficiency in c-type cytochrome biogenesis in Escherichia coli carrying a mutation in a membrane-bound disulphide isomerase-like protein. FEBS Lett. 1994;353:235–238. doi: 10.1016/0014-5793(94)01053-6. [DOI] [PubMed] [Google Scholar]
- Sannigrahi S, Zhang X, Tzeng YL. Regulation of the type I protein secretion system by the MisR/MisS two-component system in Neisseria meningitidis. Microbiology. 2009;155:1588–1601. doi: 10.1099/mic.0.023945-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchat A, Robinson K, Wenger JD, Harrison LH, Farley M, Reingold AL, Lefkowitz L, Perkins BA. Bacterial meningitis in the United States in 1995. Active Surveillance Team. N Engl J Med. 1997;337:970–976. doi: 10.1056/NEJM199710023371404. [DOI] [PubMed] [Google Scholar]
- Sinha S, Ambur OH, Langford PR, Tonjum T, Kroll JS. Reduced DNA binding and uptake in the absence of DsbA1 and DsbA2 of Neisseria meningitidis due to inefficient folding of the outer-membrane secretin PilQ. Microbiology. 2008;154:217–225. doi: 10.1099/mic.0.2007/010496-0. [DOI] [PubMed] [Google Scholar]
- Sinha S, Langford PR, Kroll JS. Functional diversity of three different DsbA proteins from Neisseria meningitidis. Microbiology. 2004;150:2993–3000. doi: 10.1099/mic.0.27216-0. [DOI] [PubMed] [Google Scholar]
- Skaar EP, Tobiason DM, Quick J, Judd RC, Weissbach H, Etienne F, Brot N, Seifert HS. The outer membrane localization of the Neisseria gonorrhoeae MsrA/B is involved in survival against reactive oxygen species. Proc Natl Acad Sci. 2002;99:10108–10113. doi: 10.1073/pnas.152334799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeets LC, Bijlsma JJ, Kuipers EJ, Vandenbroucke-Grauls CM, Kusters JG. The dprA gene is required for natural transformation of Helicobacter pylori. FEMS Immunol Med Microbiol. 2000;27:99–102. doi: 10.1111/j.1574-695X.2000.tb01418.x. [DOI] [PubMed] [Google Scholar]
- Stephens DS. Conquering the meningococcus. FEMS Microbiol Rev. 2007;31:3–14. doi: 10.1111/j.1574-6976.2006.00051.x. [DOI] [PubMed] [Google Scholar]
- Stephens DS, Greenwood B, Brandtzaeg P. Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet. 2007;369:2196–2210. doi: 10.1016/S0140-6736(07)61016-2. [DOI] [PubMed] [Google Scholar]
- Stephens DS, Swartley JS, Kathariou S, Morse SA. Insertion of Tn916 in Neisseria meningitidis resulting in loss of group B capsular polysaccharide. Infect Immun. 1991;59:4097–4102. doi: 10.1128/iai.59.11.4097-4102.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens DS, Zimmer SM. Pathogenesis, therapy, and prevention of meningococcal sepsis. Curr Infect Dis Rep. 2002;4:377–386. doi: 10.1007/s11908-002-0004-4. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Carlson RW, Muszynski A, Choudhury B, Kim KS, Stephens DS, Watanabe H. Modification of lipooligosaccharide with phosphoethanolamine by LptA in Neisseria meningitidis enhances meningococcal adhesion to human endothelial and epithelial cells. Infect Immun. 2008;76:5777–5789. doi: 10.1128/IAI.00676-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata T, Ando T, Israel DA, Wassenaar TM, Blaser MJ. Role of dprA in transformation of Campylobacter jejuni. FEMS Microbiol Lett. 2005;252:161–168. doi: 10.1016/j.femsle.2005.08.052. [DOI] [PubMed] [Google Scholar]
- Tettelin H, Saunders NJ, Heidelberg J, Jeffries AC, Nelson KE, Eisen JA, Ketchum KA, Hood DW, Peden JF, Dodson RJ, Nelson WC, Gwinn ML, DeBoy R, Peterson JD, Hickey EK, Haft DH, Salzberg SL, White O, Fleischmann RD, Dougherty BA, Mason T, Ciecko A, Parksey DS, Blair E, Cittone H, Clark EB, Cotton MD, Utterback TR, Khouri H, Qin H, Vamathevan J, Gill J, Scarlato V, Masignani V, Pizza M, Grandi G, Sun L, Smith HO, Fraser CM, Moxon ER, Rappuoli R, Venter JC. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- Thony-Meyer L. Cytochrome c maturation: a complex pathway for a simple task? Biochem Soc Trans. 2002;30:633–638. doi: 10.1042/bst0300633. [DOI] [PubMed] [Google Scholar]
- Tinsley CR, Nassif X. Analysis of the genetic differences between Neisseria meningitidis and Neisseria gonorrhoeae: two closely related bacteria expressing two different pathogenicities. Proc Natl Acad Sci U S A. 1996;93:11109–11114. doi: 10.1073/pnas.93.20.11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinsley CR, Voulhoux R, Beretti JL, Tommassen J, Nassif X. Three homologues, including two membrane-bound proteins, of the disulfide oxidoreductase DsbA in Neisseria meningitidis: effects on bacterial growth and biogenesis of functional type IV pili. J Biol Chem. 2004;279:27078–27087. doi: 10.1074/jbc.M313404200. [DOI] [PubMed] [Google Scholar]
- Tobiason DM, Seifert HS. Genomic content of Neisseria species. J Bacteriol. 2010;192:2160–2168. doi: 10.1128/JB.01593-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonjum T, Caugant DA, Dunham SA, Koomey M. Structure and function of repetitive sequence elements associated with a highly polymorphic domain of the Neisseria meningitidis PilQ protein. Mol Microbiol. 1998;29:111–124. doi: 10.1046/j.1365-2958.1998.00910.x. [DOI] [PubMed] [Google Scholar]
- Tzeng Y-L, Stephens DS. Epidemiology and pathogenesis of Neisseria meningitidis. Microbes Infect. 2000;6:687–700. doi: 10.1016/s1286-4579(00)00356-7. [DOI] [PubMed] [Google Scholar]
- Tzeng YL, Datta A, Ambrose KD, Davies JK, Carlson RW, Stephens DS, Kahler CM. The MisR/MisS two-component regulatory system influences inner core structure and immunotype of lipooligosaccharide in Neisseria meningitidis. J Biol Chem. 2004;279:35053–35062. doi: 10.1074/jbc.M401433200. [DOI] [PubMed] [Google Scholar]
- Tzeng YL, Kahler CM, Zhang X, Stephens DS. MisR/MisS two-component regulon in Neisseria meningitidis. Infect Immun. 2008;76:704–716. doi: 10.1128/IAI.01007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng YL, Zhou X, Bao S, Zhao S, Noble C, Stephens DS. Autoregulation of the MisR/MisS two-component signal transduction system in Neisseria meningitidis. J Bacteriol. 2006;188:5055–5065. doi: 10.1128/JB.00264-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney CG, Farley MM, Hadler J, Harrison LH, Bennett NM, Lynfield R, Reingold A, Cieslak PR, Pilishvili T, Jackson D, Facklam RR, Jorgensen JH, Schuchat A. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N Engl J Med. 2003;348:1737–1746. doi: 10.1056/NEJMoa022823. [DOI] [PubMed] [Google Scholar]
- Wizemann TM, Moskovitz J, Pearce BJ, Cundell D, Arvidson CG, So M, Weissbach H, Brot N, Masure HR. Peptide methionine sulfoxide reductase contributes to the maintenance of adhesins in three major pathogens. Proc Natl Acad Sci U S A. 1996;93:7985–7990. doi: 10.1073/pnas.93.15.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Hirao K, Oshima T, Aiba H, Utsumi R, Ishihama A. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J Biol Chem. 2005;280:1448–1456. doi: 10.1074/jbc.M410104200. [DOI] [PubMed] [Google Scholar]
- Yu J. Inactivation of DsbA, but not DsbC and DsbD, affects the intracellular survival and virulence of Shigella flexneri. Infect Immun. 1998;66:3909–3917. doi: 10.1128/iai.66.8.3909-3917.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Montanez GE, Kumar P, Sannigrahi S, Tzeng YL. Regulatory role of the MisR/S two-component system in hemoglobin utilization in Neisseria meningitidis. Infect Immun. 2010;78:1109–1122. doi: 10.1128/IAI.00363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.