Abstract
Despite the high prevalence and poor outcome of patients with metastatic lung cancer, the mechanisms of tumour progression and metastasis remain largely uncharacterized. We modelled human lung adenocarcinoma, which frequently harbours activating point mutations in KRAS1 and inactivation of the p53-pathway2, using conditional alleles in mice3–5. Lentiviral-mediated somatic activation of oncogenic Kras and deletion of p53 in the lung epithelial cells of KrasLSL-G12D/+;p53flox/flox mice initiates lung adenocarcinoma development4. Although tumours are initiated synchronously by defined genetic alterations, only a subset become malignant, suggesting that disease progression requires additional alterations. Identification of the lentiviral integration sites allowed us to distinguish metastatic from non-metastatic tumours and determine the gene expression alterations that distinguish these tumour types. Cross-species analysis identified the NK-2 related homeobox transcription factor Nkx2-1 (Ttf-1/Titf1) as a candidate suppressor of malignant progression. In this mouse model, Nkx2-1-negativity is pathognomonic of high-grade poorly differentiated tumours. Gain-and loss-of-function experiments in cells derived from metastatic and non-metastatic tumours demonstrated that Nkx2-1 controls tumour differentiation and limits metastatic potential in vivo. Interrogation of Nkx2-1 regulated genes, analysis of tumours at defined developmental stages, and functional complementation experiments indicate that Nkx2-1 constrains tumours in part by repressing the embryonically-restricted chromatin regulator Hmga2. While focal amplification of NKX2-1 in a fraction of human lung adenocarcinomas has focused attention on its oncogenic function6–9, our data specifically link Nkx2-1 downregulation to loss of differentiation, enhanced tumour seeding ability, and increased metastatic proclivity. Thus, the oncogenic and suppressive functions of Nkx2-1 in the same tumour type substantiate its role as a dual function lineage factor.
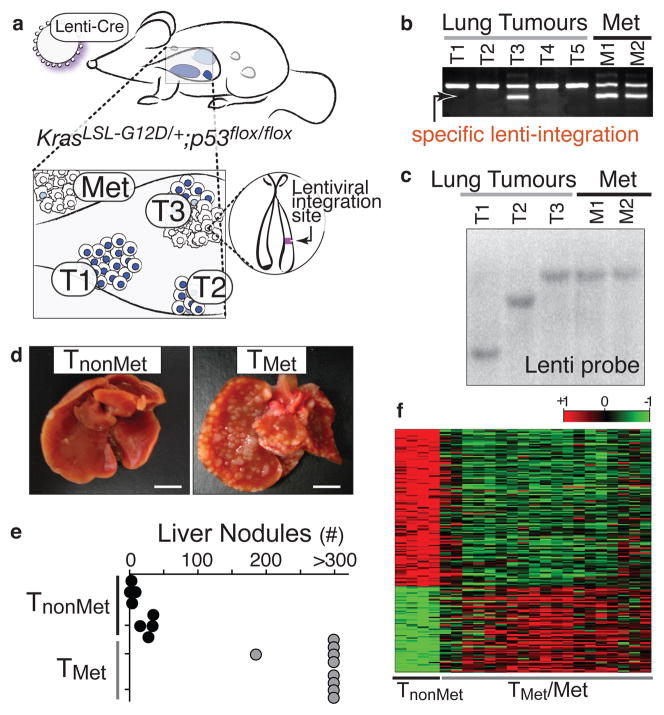
We developed lentiviral vectors that express Cre-recombinase (Lenti-Cre)10 and determined the dose that results in 5 to 20 lung tumours per KrasLSL-G12D/+;p53flox/flox mouse after intratracheal administration. These mice lived 8–14 months after tumour initiation and developed macroscopic metastases to the draining lymph nodes, pleura, kidneys, heart, adrenal glands, and liver (Supplementary Fig. 1). Because lentiviruses integrate stably into the genome, the integration site was a unique molecular identifier that unambiguously linked primary tumours to their related metastases (Fig. 1a). We used linker-mediated PCR (LM-PCR) to determine the genomic sequence directly 3′ of the integrated lentiviral genome followed by a specific PCR for the lentiviral integration site (Fig. 1b). To have samples of sufficient quantity and purity for our analyses, we derived cell lines from primary tumours and metastases. Cell lines were pure tumour cells as determined by recombination of the p53floxed alleles (data not shown). The clonal relationship of these cell lines was established using LM-PCR or Southern blot analysis for the lentiviral genome (Fig. 1c and data not shown). We termed cell lines derived from verified metastatic primary lung tumours TMet.
Figure 1. A lentiviral vector-induced mouse model of lung adenocarcinoma identifies gene expression alterations during tumour progression.
a, Infection of KrasLSL-G12D/+;p53flox/flox mice with Cre-expressing lentiviral vectors initiates lung adenocarcinoma. b, Linker-mediated PCR cloning of the lentiviral integration site in metastases (Met) allows specific PCR amplification of that lentiviral-integration (lower band) to identify which primary tumour gave rise to the metastases. Top band is a control product. c, Southern blot on cell lines for the integrated lentiviral genome. d, Representative images of livers after intrasplenic transplantation of TnonMet or TMet cells. Scale bar = 0.5cm. e, Quantification of liver nodules after intrasplenic injection of two TnonMet and TMet cell lines. f, Gene expression alterations (log2) between TnonMet and TMet/Met samples.
Gene expression profiling was performed on cell lines from twenty-three lung tumours and metastases (nine metastases, seven TMet primary tumours, and seven potentially non-metastatic primary tumours). Using unsupervised consensus clustering11, we identified four cell lines from likely non-metastatic tumour samples that had highly concordant gene expression and were separate from all TMet and metastasis (Met) samples (Supplementary Fig. 2). Therefore, we surmised that these could represent non-metastatic primary tumours and classified them as TnonMet. These TnonMet cell lines consistently formed fewer tumour nodules in the liver after intrasplenic injection despite equivalent proliferation rates in cell culture (Fig. 1d–e and Supplementary Fig. 2).
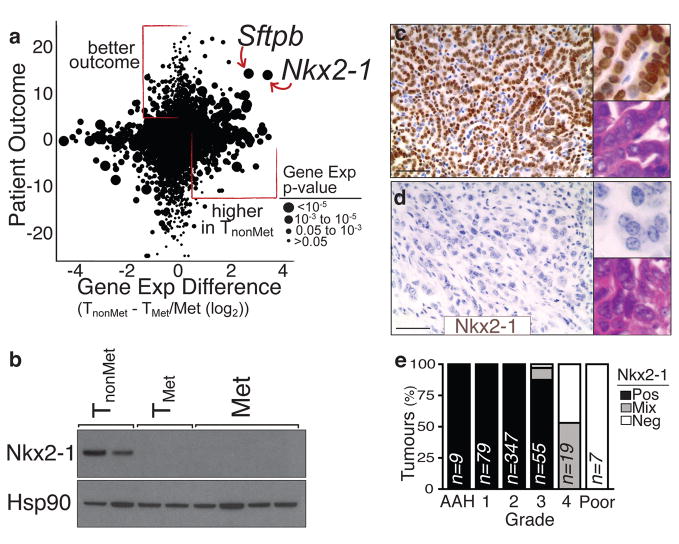
Significant gene expression alterations distinguished TnonMet from TMet and Met-derived cell lines (Fig. 1f and Supplementary Table 1), many of which were validated by qRT-PCR, flow cytometry, and western blotting (data not shown). A gene expression signature generated by comparing TnonMet to TMet/Met samples predicted patient outcome in human lung adenocarcinoma gene expression datasets12,13, suggesting the possibility of evolutionarily-conserved molecular mechanisms of tumour progression (Supplementary Fig. 2). Thus, we integrated mouse and human data by comparing the differences in expression between TnonMet and TMet/Met samples with the association of human gene expression and patient survival (Fig. 2a). Two genes were particularly notable from this analysis: the NK-related homeobox transcription factor Nkx2-1 and the Nkx2-1 target gene surfactant protein B (Sftpb; Fig. 2a). Nkx2-1 regulates lung development and is expressed in Type II pneumocytes and bronchiolar cells in the adult14–16. Nkx2-1 expression was >10-fold higher in TnonMet samples, and higher NKX2-1 expression in human tumours correlated with longer survival. Of note, NKX2-1 is focally amplified in ~10% of human lung adenocarcinoma, with functional data supporting oncogenic activity6–9,17. Conversely, most immunohistochemical analyses of NKX2-1 in this disease suggest an association between NKX2-1-negative tumours and poor patient outcome17,18. Thus, we focused on validating and characterizing the function of this transcription factor in suppressing tumour progression and metastasis.
Figure 2. Reduced Nkx2-1 in advanced lung adenocarcinoma correlates with a less differentiated state.
a, Cross-species analysis of human lung adenocarcinoma12 patient outcome (likelihood ratio with the sign from correlation value) versus differential gene expression in murine TnonMet cells. b, Nkx2-1 protein is absent from TMet and Met-derived cell lines. c–d, Nkx2-1 expression is high in well differentiated adenomas and early murine adenocarcinoma (top) but is downregulated in moderately to poorly differentiated advanced carcinomas (bottom). Scale bar = 50μm. Upper inlay Nkx2-1 staining. Lower inlay H+E staining. e, Quantification of Nkx2-1 expression in murine lung tumours relative to tumour grade from most differentiated (atypical adenomous hyperplasia (AAH)) to least differentiated (Poor)).
We confirmed reduced Nkx2-1 mRNA and protein in TMet and Met cell lines without evidence of focal genomic loss of this region (Fig. 2b, Supplemental Fig. 4, and data not shown). Nkx2-1 was consistently downregulated in high-grade poorly differentiated tumours from our mouse model (Fig. 2c–e and Supplementary Fig. 3). Nkx2-1 expression was also reduced in advanced KrasG12D-driven lung adenocarcinomas with p53R270H or p53R172H point mutations4,19. Using our LM-PCR assay, we identified three primary lung tumours as metastatic based on the presence of metastases with the same lentiviral integration site (Fig. 1b and data not shown). These tumours each contained poorly-differentiated areas that were Nkx2-1neg (Supplementary Fig. 6). Interestingly, Nkx2-1 expression was low/absent in almost all lymph node and distant macrometastases, though some micrometastases were Nkx2-1pos or Nkx2-1mixed (Supplementary Fig. 3). Whether certain micrometastases were seeded by Nkx2-1pos cells or reverted to an Nkx2-1pos phenotype due to cues from their new environment is unknown.
In human lung adenocarcinoma12,13 the expression of NKX2-1 correlated with a mouse TnonMet gene expression signature (Supplementary Fig. 3). Additionally, the TnonMet signature was anti-correlated with an embryonic stem cell signature, supporting the notion that TMet/Met cells have transitioned to a less differentiated and more stem-like state20 (Supplementary Fig. 3).
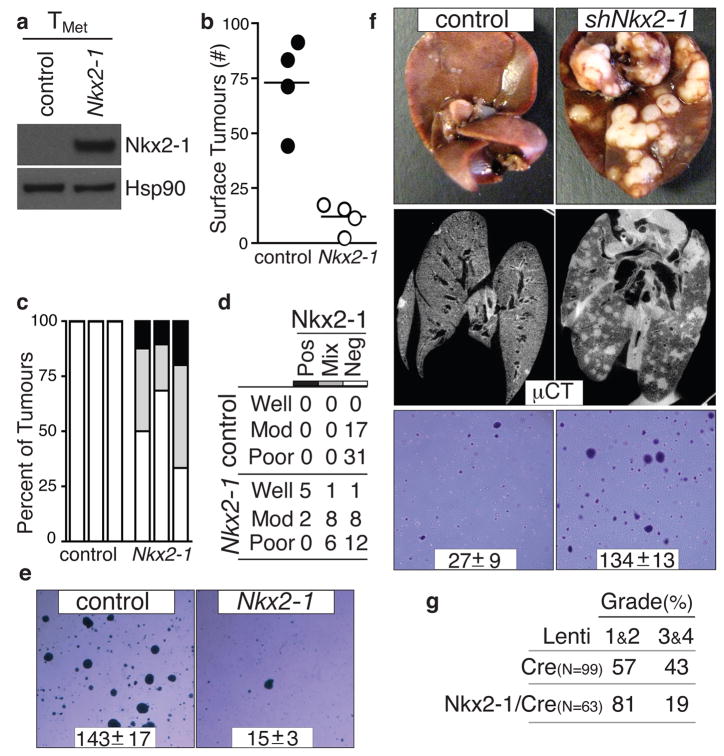
The correlative mouse and human data were consistent with Nkx2-1 being either a marker or a functional regulator of tumour progression. Nkx2-1 expression in a TMet cell line greatly suppressed tumour formation after intravenous transplantation (Fig. 3a, 3b, and Supplementary Fig. 5). Moreover, of the tumours that formed after injection of TMet-Nkx2-1 cells, many were either Nkx2-1neg or Nkx2-1mixed (Fig. 3c). In general, tumours that continued to express Nkx2-1 were well-differentiated, while Nkx2-1neg tumours often displayed solid architecture or areas of poorly-differentiated cells (Fig. 3d and Supplementary Fig. 5). Intrasplenic transplantation unveiled a similar diminution of tumour formation by TMet–Nkx2-1 cells (Supplementary Fig. 5). Nkx2-1 expression did not alter proliferation or cell death in cell culture, or affect established tumour proliferation in vivo (Supplementary Fig. 5 and data not shown), but dramatically reduced the ability of these cells to grow in anchorage-independent conditions and initiate tumours after subcutaneous transplantation (Fig. 3e and Supplementary Fig. 5)
Figure 3. Nkx2-1 controls lung adenocarcinoma differentiation and restricts metastatic ability.
a, Nkx2-1 protein expression in TMet cells. b, Nkx2-1 expression reduces lung nodule formation after intravenous transplantation. p < 0.002. c, Quantification of Nkx2-1 in lung nodules after TMet or TMet-Nkx2-1 transplantation. n=3/group. d, Association of Nkx2-1 expression with differentiation state after TMet or TMet-Nkx2-1 transplantation. Fisher’s exact test on the association of differentiation state with Nkx2-1 p < 0.002. e, Nkx2-1 expression reduces anchorage-independent growth of TMet cells. Representative images and colony number (mean +/− SD of quadruplicate wells, p < 0.0001). f, Nkx2-1 knockdown increases liver nodules after intrasplenic injection (top) and lung nodules after intravenous transplantation (middle) of TnonMet cells. Representative of 7 mice/group. shNkx2-1 enhanced anchorage-independent growth of TnonMet cells (bottom). Representative images and colony number (mean +/− SD of triplicate wells, p < 0.0001). g. Induction of tumours in KrasLSL-G12D/+;p53flox/flox mice with Nkx2-1/Cre lentivirus reduces the development of advanced tumours (grades 3&4). Numbers indicate percent of tumours in each group.
To further elucidate the function of Nkx2-1, we knocked-down Nkx2-1 in TnonMet cell lines (Supplementary Fig. 7). Nkx2-1 knockdown allowed the formation of more liver nodules after intrasplenic injection and more lung nodules after intravenous transplantation (Fig. 3f). Nkx2-1 knockdown did not alter proliferation or cell death in cell culture (Supplementary Fig. 7) but enhanced the cells’ ability to form colonies under anchorage-independent conditions and tumours after subcutaneous transplantation (Fig. 3f and Supplementary Fig. 7). Reexpression of an shRNA-insensitive Nkx2-1 cDNA (Nkx2-1*) reverted the phenotypic alterations elicited by shNkx2-1 confirming that the effects of the shNkx2-1 were specifically due to Nkx2-1 knockdown (Supplementary Fig. 7). Finally, we induced tumours in KrasLSL-G12D/+;p53flox/flox mice with either Lenti-Cre or a lentiviral vector expressing both Nkx2-1 and Cre (Lenti-Nkx2-1/Cre). Expression of exogenous Nkx2-1 limited tumour progression resulting in fewer tumours of advanced histopathological grades (Fig. 3g).
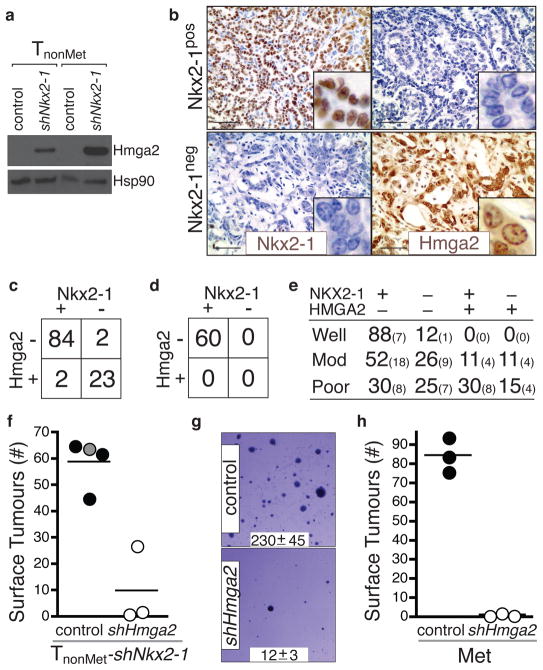
To discover Nkx2-1 regulated genes, we compared gene expression in TnonMet and TnonMet–shNkx2-1 cells. Overlapping this gene list with the genes expressed at different levels in TnonMet versus TMet/Met cells uncovered high priority candidate genes (Supplementary Fig. 8). We elected to focus on Hmga2 given its role in altering global gene expression through the regulation of chromatin structure and its association with embryonic and adult stem cell states21–24 as well as malignant tumours of diverse origins25–29. Hmga2 was derepressed by Nkx2-1 knockdown in TnonMet cells and regions of KrasG12D/+;p53Δ/Δ tumours that lacked Nkx2-1 expression were almost universally Hmga2pos (Fig. 4a–c). Importantly, Nkx2-1neg areas of known metastatic primary tumours and metastases were also Hmga2pos (Supplementary Fig. 9 and data not shown). Additionally, Hmga2 was downregulated in TMet cells after expression of Nkx2-1 cDNA and in TnonMet-shNkx2-1 cells after reexpression of Nkx2-1* (data not shown).
Figure 4. Nkx2-1 regulates the expression of Hmga2 in advanced lung adenocarcinoma.
a, Nkx2-1 knockdown derepresses Hmga2 in TnonMet cell lines. b, Hmga2 and Nkx2-1 are reciprocally expressed in KrasG12D/+;p53Δ/Δ murine lung adenocarcinomas. Scale bar = 50μm. Inlaid images show cellular features and protein localization. c, Hmga2 and Nkx2-1 expression in advanced KrasG12D/+;p53Δ/Δ murine lung adenocarcinomas. Fisher’s exact test, p-value < 10−11. d, Early KrasG12D/+;p53Δ/Δ tumours are Nkx2-1posHmga2neg. e, NKX2-1 and HMGA2 expression in human lung adenocarcinomas. Large numbers are percentages. Small numbers are absolute numbers. f, Hmga2 knockdown reduces the tumourigenic potential of TnonMet-shNkx2-1 cells after intravenous transplantation. Control samples include the parental TnonMet-shNkx2-1 cells (grey circle) and cells infected by a control retrovirus (black circles). p < 0.003. g, shHmga2 reduces anchorage-independent growth of a metastasis-derived cell line (Met). Representative images and colony number (mean +/− SD of quadruplicate wells, p < 0.0001). h, shHmga2 reduces the tumour-seeding potential of a Met cell line after intravenous transplantation. p <0.0001.
Although Hmga2 can be regulated by the Let7 family of miRNAs21,25,30, Let7 levels, Lin28 expression, and Let7 activity were equivalent in TnonMet, TMet, and Met cell lines and were unaltered in TnonMet-shNkx2-1 cells (Supplementary Fig. 10 and data not shown). Hmga2 promoter activity was derepressed in TnonMet-shNkx2-1 cells and repressed in TMet-Nkx2-1 cells, indicating that expression of Hmga2 in lung adenocarcinoma cells is regulated at least in part through differential promoter activity (Supplementary Fig. 10).
We hypothesized that lung adenocarcinomas progress from an Nkx2-1posHmga-2neg to an Nkx2-1negHmga-2pos state. However, metastatic and non-metastatic tumours could be fundamentally distinct at the time of initiation. Hmga2 is highly expressed in embryonic lung but not in any normal adult lung cells, and early after initiation, KrasG12D/+;p53Δ/Δ tumours were uniformly Nkx2-1posHmga-2neg (Fig. 4d and Supplementary Fig. 11). KrasG12D/+;p53-proficient tumours, which maintain their differentiated phenotype and never metastasize even late after tumour initiation5, were almost universally Nkx2-1posHmga-2neg (Supplementary Fig. 11). Poorly differentiated areas of KrasG12D/+;p53Δ/Δ tumours with reduced Nkx2-1 expression were never found as in situ lesions and were almost always associated with lower grade Nkx2-1-expressing areas (Supplementary Fig. 6). Finally, we induced KrasG12D/+;p53Δ/Δ tumours with a pool of lentiviral vectors that contain nucleotide barcodes. Amplification and sequencing of the lentivirus-encoded barcodes from adjacent low-grade Nkx2-1posHmga2neg and high-grade Nkx2-1negHmga2pos areas showed that these areas were clonally related (Supplemental Fig. 12). While alternate mechanisms leading to the generation of clonally-related but phenotypically-distinct tumour cell populations are possible, including the expansion of rare Nkx2-1neg cells that pre-exist within the tumour, we believe that our data strongly suggest that lung adenocarcinomas undergo a transition from an Nkx2-1posHmga-2neg state to a more aggressive Nkx2-1negHmga-2pos state. Our data additionally indicate that an Nkx2-1-dependent gene expression program is a key regulator of this transition.
We next analyzed the expression of NKX2-1 and HMGA2 in human adenocarcinoma. Although the expression patterns were diverse, two important conclusions could be made. First, tumours of NKX2-1posHMGA2neg and NKX2-1negHMGA2pos phenotypes exist within the spectrum of human lung adenocarcinoma (Fig. 4e and Supplementary Fig. 10). Second, there was a trend towards well-differentiated tumours being NKX2-1pos/HMGA2neg whereas moderately and poorly differentiated tumours were more often represented by other combinations of these proteins. Most notably, the moderately and poorly differentiated groups contained NKX2-1neg/HMGA-2pos tumours (Fig. 4e). These results underscore the diversity within this single human tumour type and suggest that our genetically defined model likely represents, at the molecular level, a subset of these tumours.
Next we knocked-down Hmga2 in TnonMet-shNkx2-1 cells and found that their metastasis seeding potential was greatly reduced after transplantation (Fig. 4f and Supplementary Fig. 13). Additionally, Hmga2 knockdown in a metastasis-derived cell line reduced its anchorage-independent growth and tumour seeding ability after transplantation (Fig 4g–h and Supplementary Fig. 13). A future challenge will be to understand the molecular mechanism by which Hmga2 controls lung adenocarcinoma metastatic potential. The expansion of Nkx2-1negHmga-2pos regions within primary lung tumours suggests the acquisition of phenotypes that are advantageous to the primary tumours and also increase the probability of metastatic spread.
That NKX2-1 can have both oncogenic and tumour suppressive functions in lung cancer presumably illustrating context-dependent functions within individual tumours of the same type. Lung adenocarcinomas may differ in their cell of origin, mutation spectrum, or gene expression leading to distinct requirements for continued NKX2-1 expression and different capacity to tolerate or benefit from NKX2-1 downregulation. Our studies uncovered the molecular and cellular basis for the association of NKX2-1 expression with good patient outcome17,18 and HMGA2 expression with poor patient outcome26,27. Our results emphasize the power of genetically-engineered mouse models of advanced disease, used in conjunction with human studies, to elucidate mechanisms that control cancer progression and metastatic spread. Through this approach we identified one molecular mechanism by which a highly prevalent tumour type can progresses to its malignant state.
Methods Summary
Mice, tumour initiation, and derivation of cell lines
KrasLSL-G12D, p53flox, p53LSL-R270H, and p53LSL-R172H mice have been described3,5,19. Tumours were initiated by intratracheal infection of mice with a lentiviral vector expressing Cre-recombinase10. The MIT Institutional Animal Care and Use Committee approved all animal studies and procedures. Cell lines were created by enzymatic and mechanical dissociation of individual lung tumours and metastases harvested from mice 8-14 months after tumour initiation.
LM-PCR, Southern blotting, and gene expression analysis
LM-PCR was performed with forward primers specific for the lentiviral LTR. Southern blotting used a Cre probe and standard methods. RNA was extracted using Trizol, analyzed for RNA integrity, and prepared with Affymetrix GeneChip® WT Sense Target Labelling and Control Reagents kits, followed by hybridization to Affymetrix GeneChip® Mouse Exon 1.0 ST Arrays.
Protein and RNA analysis
Western blotting used standard methods and antibodies to Nkx2-1 (Epitomics, Inc), Hmga2 (BioCheck, Inc), and Hsp90 (BD Transduction Laboratories) as a loading control. Immunohistochemistry was performed on formalin-fixed, paraffin-embedded 4μm sections using the ABC Vectastain kit (Vector Laboratories) with antibodies described above. Sections were developed with DAB and counterstained with hematoxylin.
Gene expression and knockdown
Nkx2-1 was stably knocked-down with a pLKO-based lentiviral vector (OpenBiosystems/TRC). MSCV-Puro retroviral vectors were used for stable expression of Nkx2-1 and Nkx2-1* (created with 4 silent mutations using QuikChange® Lightning Site-Directed Mutagenesis (Stratagene). Hmga2 was stably knocked-down with an MSCV-Hygro retroviral vector.
Transplantation experiments
For intravenous transplantation 105 cells resuspended in 200μl PBS were injected into the lateral tail vein. For intrasplenic transplantation 105 cells resuspended in 50μl PBS were injected. In all graphs each circle represents an individual mouse and the bar represents the mean. Statistical significance was determined using the Student’s t-test.
Supplementary Material
Acknowledgments
The authors thank Alison Dooley, Nadya Dimitrova, Trudy Oliver, and Margaret Ebert for providing reagents; Manlin Luo (Biology/Koch Institute BioMicro Core) for array processing; Madhu Kumar for experimental assistance; Tracy Staton, David McFadden, Alice Shaw, and the entire Jacks laboratory for comments. M.M.W was a Merck Fellow of the Damon Runyon Cancer Research Foundation and was funded by a Genentech Postdoctoral Fellowship. R.G.W.V. is supported by a Fellowship from the Dutch Cancer Society KWF. E.L.S is supported by a training grant (T32-HL007627). D.M.F is a Leukemia and Lymphoma Postdoctoral Fellow. This work was supported by National Institutes of Health grants U01-CA84306 (to T.J) and K99-CA151968 (to M.M.W.), the Howard Hughes Medical Institute, the Ludwig Center for Molecular Oncology at MIT, and in part by the Cancer Center Support (core) grant P30-CA14051 from the National Cancer Institute. T.J. is the David H. Koch Professor of Biology and a Daniel K. Ludwig Scholar.
Footnotes
Author Contributions M.M.W. and T.J. designed the study; M.M.W., T.L.D., C.K-K., and E.L.S performed experiments; R.G.W.V., C.A.W., D.D.H., S.H., and D.Y.C. conducted bioinformatic analyses; E.L.S. and R.T.B. provided pathology assistance; S.Y. and D.C. provided technical assistance; M.J.D. provided reagents; D.M.F. and M.M. gave conceptual advice; M.M.W and T.J. wrote the paper with comments from all authors.
Author Information Gene expression data was deposited in Gene Expression Omnibus (GSE26874). Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Rodenhuis S, et al. Incidence and possible clinical significance of K-ras oncogene activation in adenocarcinoma of the human lung. Cancer Res. 1988;48:5738–5741. [PubMed] [Google Scholar]
- 2.Takahashi T, et al. p53: a frequent target for genetic abnormalities in lung cancer. Science. 1989;246:491–494. doi: 10.1126/science.2554494. [DOI] [PubMed] [Google Scholar]
- 3.Jonkers J, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 4.Jackson EL, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 5.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weir BA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka H, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007;67:6007–6011. doi: 10.1158/0008-5472.CAN-06-4774. [DOI] [PubMed] [Google Scholar]
- 8.Kendall J, et al. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci U S A. 2007;104:16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwei KA, et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008;27:3635–3640. doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monti S, Tamayo P, Mesirov J, Golub T. Consensus Clustering: A Resampling-Based Method for Class Discovery and Visualization of Gene Expression Microarray Data. Machine Learning. 2003;52:91–118. [Google Scholar]
- 12.Shedden K, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen DX, et al. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009;138:51–62. doi: 10.1016/j.cell.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura S, et al. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- 15.Krude H, et al. Choreoathetosis, hypothyroidism, and pulmonary alterations due to human NKX2-1 haploinsufficiency. J Clin Invest. 2002;109:475–480. doi: 10.1172/JCI14341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeda Y, Dave V, Whitsett JA. Transcriptional control of lung morphogenesis. Physiol Rev. 2007;87:219–244. doi: 10.1152/physrev.00028.2006. [DOI] [PubMed] [Google Scholar]
- 17.Barletta JA, et al. Clinical Significance of TTF-1 Protein Expression and TTF-1 Gene Amplification in Lung Adenocarcinoma. J Cell Mol Med. 2008 doi: 10.1111/j.1582-4934.2008.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berghmans T, et al. Thyroid transcription factor 1--a new prognostic factor in lung cancer: a meta-analysis. Ann Oncol. 2006;17:1673–1676. doi: 10.1093/annonc/mdl287. [DOI] [PubMed] [Google Scholar]
- 19.Olive KP, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu F, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 23.Li O, Vasudevan D, Davey CA, Droge P. High-level expression of DNA architectural factor HMGA2 and its association with nucleosomes in human embryonic stem cells. Genesis. 2006;44:523–529. doi: 10.1002/dvg.20242. [DOI] [PubMed] [Google Scholar]
- 24.Rommel B, et al. HMGI-C, a member of the high mobility group family of proteins, is expressed in hematopoietic stem cells and in leukemic cells. Leuk Lymphoma. 1997;26:603–607. doi: 10.3109/10428199709050896. [DOI] [PubMed] [Google Scholar]
- 25.Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 26.Meyer B, et al. HMGA2 overexpression in non-small cell lung cancer. Mol Carcinog. 2007;46:503–511. doi: 10.1002/mc.20235. [DOI] [PubMed] [Google Scholar]
- 27.Hristov AC, et al. HMGA2 protein expression correlates with lymph node metastasis and increased tumor grade in pancreatic ductal adenocarcinoma. Mod Pathol. 2009;22:43–49. doi: 10.1038/modpathol.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogalla P, et al. Expression of HMGI-C, a member of the high mobility group protein family, in a subset of breast cancers: relationship to histologic grade. Mol Carcinog. 1997;19:153–156. doi: 10.1002/(sici)1098-2744(199707)19:3<153::aid-mc2>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 29.Di Cello F, et al. HMGA2 participates in transformation in human lung cancer. Mol Cancer Res. 2008;6:743–750. doi: 10.1158/1541-7786.MCR-07-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.