Abstract
All isolates of serotype M1 of group A streptococci possess a gene for streptococcal inhibitor of complement (SIC) in the mga regulon, which harbors genes for other virulence factors, such as M and M-like proteins, C5a peptidase, and a regulator. In serotype M57 the gene for a protein that is closely related to SIC (crs57) is located outside the mga regulon. We mapped the location of the crs57 gene in six strains of emm57 (gene encoding the M57 protein) sequence types to an intergenic region between the ABC transporter gene (SPy0778) and the gene for a small ribosomal protein (rpsU). The noncoding sequences on both sides of crs57 exhibited high degrees of identity to the corresponding regions of sic from M1 strains. This included one of the inverted repeat sequences of IS1562 but not the insertion element itself. These observations suggest that crs57 was recently acquired by serotype M57 or its progenitor via horizontal acquisition from serotype M1. The six emm57 sequence type isolates analyzed in this study belong to two distinct molecular types (vir types VT8 and VT101). Although the crs57 sequences from VT8 strains had very few substitution mutations, the VT101 crs57 sequence had a large number of such mutations. The CRS57 proteins from these strains are secretory products and have the ability to bind to complement proteins. All these proteins contain several tryptophan-rich repeats designated DWS motifs and internal repeat sequences. In all of these structural and biochemical characteristics CRS57 resembles SIC from M1 strains. Hence, CRS57 has a functional role similar to that of SIC in an M1 strain.
Group A streptococcus (GAS) (Streptococcus pyogenes) is a human-specific pathogen that is responsible for a wide range of diseases, including immune-mediated postinfective sequelae, such as acute rheumatic fever and post-streptococcus glomerulonephritis. This pathogen has evolved diverse mechanisms to overcome host defenses against infection. The surface-exposed major protective antigen, M protein, binds to factor H and thereby may inhibit deposition of opsonin C3b on the GAS surface (9). M protein also binds to a 570-kDa human plasma protein designated C4b-binding protein that inhibits complement activation via the classical pathway (3, 15). All S. pyogenes strains express a specific protease that cleaves chemotactic complement C5a protein (4) to an inactive molecule, retarding phagocyte recruitment at the site of infection. Additionally, isolates of S. pyogenes with M1-type specificity express a secretory protein designated SIC (streptococcal inhibitor of complement function) that inhibits complement-mediated cell lysis (1). While the biological significance of SIC as an inhibitor of complement function is not clear, the presence of the gene encoding it in all serotype M1 strains (1), the extreme divergence (20), and the rapid emergence of variants (14) indicate that this protein has an important biological role. In fact, recent studies of SIC showed that it also inhibits the functions of innate immune proteins, such as secretory leukocyte proteinase inhibitor and lysozyme (5).
Biochemical studies have suggested that SIC interferes with the function of the membrane attack complex by possibly binding to one or more protein components associated with the complex (1). Fernie-King et al. (6) showed that M1 SIC binds to the C6 and C7 complement proteins, preventing their incorporation into the membrane attack complex. Despite considerable sequence diversity, all SIC variants from M1 strains have the complement-inhibiting activity (14).
Molecular studies (12) have shown that like serotype M1 strains, all serotype M57 strains possess a gene which encodes a protein closely related to the M1 SIC, designated CRS (closely related to SIC). However, while the gene encoding SIC is part of the M1 mga regulon, which comprises genes encoding M or M-like proteins (emm or emmL), C5a peptidase (scpA), and a regulator (mga), the crs57 gene (the gene encoding CRS57 in M57 strains) is located outside the mga regulon in M57 strains. While most studies have been carried out with SIC variants of M1 strains, little is known about CRS57. In this study, we determined the exact location of crs57 in the M57 genome, examined the diversity of CRS57 from six M57 isolates belonging two distinct molecular types (vir types), and tested the ability of the molecules to bind to the complement proteins. CRS57 is highly conserved in the major vir type (VT8) of M57 strains, whereas in the minor vir type (VT101) the protein is more diverse than VT8 CRS57. We show here that the CRS57 proteins are excretory products and that they have the ability to bind to the C6 and C7 complement proteins. Taken together, our results are consistent with single lateral acquisition of the sic gene from an emm1 strain by M57 or its progenitor.
MATERIALS AND METHODS
Strains, culture, and genomic DNA
Table 1 summarizes information about the S. pyogenes strains used in this study. S. pyogenes serotype M1 and 57 reference strains (strains 2031 and 2077, respectively) were obtained from Public Health Laboratory Services, Prague, Czech Republic. All other strains were isolates obtained in the Northern Territory (NT). The GAS isolates were typed by vir typing (Fig. 1). Vir typing is based on restriction fragment length polymorphism of the mga regulon (8, 11, 12). Growth of GAS and genomic DNA extraction were carried out as described previously (10).
TABLE 1.
S. pyogenes strains used in this study
| Strain | M57 vir type | emm sequence type | Source | Presence of sic genea |
|---|---|---|---|---|
| 2077 | VT8 | emm57 | Prague | Present |
| NS1140 | VT101 | emm57 | NT | Present |
| NS38 | VT8 | emm57 | NT | Present |
| NS48 | VT8 | emm57 | NT | Present |
| BSA16 | VT8 | emm57 | NT | Present |
| BSA5 | VT8 | emm57 | NT | Present |
| 2031 | emm1 | Prague | Present | |
| NS844 | emm1 | NT | Present | |
| NS27 | NDb | NT | Absent |
The presence of the sic gene was determined by PCR by using CRS-specific primers (12).
ND, not determined.
FIG. 1.
Vir typing of emm57 strains was performed as described previously (11). The amplified products of the mga regulon were digested with HaeIII (A) or HinfI (B). A marker (1-kb ladder; New England Biolabs) and strain 2031 (an M1 strain) were included (lanes M and 7, respectively). Lanes 1 to 6, isolates 2077 (a reference strain from Prague), NS1140, BSA16, BSA5, NS38, and NS48, respectively.
Sequencing of crs57
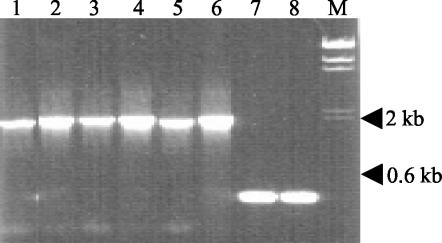
Based on a partial sequence for crs57 (accession numbers AF060764 and AF060765) (12), a specific reverse primer (primer crs57R; GAGACAAACCAACTCCAGACCGG) corresponding to this gene was designed. To determine the sequence upstream of crs57, a bubble PCR was employed (17). Genomic DNA was digested with Sau3A1, and to 15 μg of the digest, 10 μg of preannealed linker oligonucleotides (GCACGTCTGACGATCTCAGTACAGACTGGAGTCACAGCTGC and GATCGCAGCTGTGACTTAGTCACTCCAATGATCTGTCAGACGTGC) containing a Sau3A1 overhang and an internal mismatch (bubble) was ligated. Three units of T4 DNA ligase in 10× buffer containing ATP (Amersham) was used. The ligation product was then used as a template in a PCR with Taq DNA polymerase (Amersham) by using the bubble-specific primer (GACGATCTCAGTACAGACTG) and the crs57R primer. The cycling conditions were 95°C for 30 s for denaturation, 53°C for 30s for annealing, and 68°C for 90 s for extension. The bubble PCR amplified a product corresponding to a region between the crs57R target site and an upstream Sau3A1 site. Sequencing of the PCR product from an emm57 strain revealed 112 bp which exhibited 96% homology to the 3′ end of an ABC transporter (Spy0778) immediately adjoining a gene encoding a small ribosomal protein subunit (rpsU) (data not shown). Based on these results and the S. pyogenes genomic sequence data (accession number AE004092), we designed primers ABCF2 and rpsUR1 (GATCTGGCTTTAGCACCCTTTAGC and GATCTGGCTTTAGCACCCTTTAGĆ, respectively) spanning the region between Spy0778 and rpsU. A PCR product that was about 2 kb long was obtained from all six emm57 strains, while PCR with M1 and sic-negative strains resulted in a product that was approximately 400 bp long (Fig. 2). The complete sequence of the PCR product from the emm57 strains was determined.
FIG. 2.
PCR amplification of the SPy0778-rpsU intergenic region in isolates of the emm57 sequence type (lanes 1 to 6), in an isolate of the emm1 sequence type (lane 7), and in NS27 (lane 8). Lanes 1 to 6, amplification products from 2077, NS1140, BSA16, BSA5, NS38, and NS48, respectively. Lane M contained HindIII-digested lambda phage DNA as a size marker.
Production of recombinant CRS57
The CRS57 gene was amplified by using oligonucleotides CTACTAGGAGCTACACAACC and CGTTGCTGATGGTGTATATGG. Recombinant CRS57 constructs were then generated in either the pQE30 expression system (Qiagen) or the pBAD/TOPO TA (Thiofusion) expression system (Invitrogen). Sticky-end cloning of sic into pQE30 was performed at the BamHI and PstI (New England Biolabs) restriction enzymes sites. DNA manipulation, transformation, expression, and purification were performed according to the manufacturer's recommendations. The pBAD TA constructs in TOP10 cells (Invitrogen) were screened by PCR. Sequence analysis (by using an ABI 3700 DNA sequencer and the BigDye 3 terminator mixture) was performed to confirm positive clones. Plasmid preparations of positive clones were transformed in Escherichia coli BL21 for expression. One-liter cultures of BL21 containing the recombinant plasmid were grown in Luria-Bertani broth with 100 μg of ampicillin per ml until the optical density at 600 nm reached 0.5. The best yields with the pBAD system were obtained upon induction with 0.2% arabinose for 2 h.
To purify recombinant His-tagged CRS57, cell pellets from induced cultures were sonicated to lyse the cells and centrifuged to remove the insoluble cellular debris. The expressed recombinant protein was isolated from the resultant cleared lysate by using a Superflow Ni-nitrilotriacetic acid column (Qiagen) and the Biologic HR system (fast protein liquid chromatography; Bio-Rad). The recombinant SIC was eluted with an imidazole (ICN Biomedical) gradient (20 to 300 mM) and dialyzed against phosphate-buffered saline (PBS). Fractionation by polyacrylamide gel electrophoresis (PAGE) (Gradipore; Miniprotean II; Bio-Rad) followed by immunoblotting with anti-SIC (this study) and anti-His (Invitrogen) antibodies was used to identify the purified proteins.
Antibodies
CRS-specific rabbit antiserum (IMVS, Gilles Plains, South Australia, Australia) was obtained by immunizing rabbits with recombinant M1 SIC (pQE30; Qiagen) from reference strain 2031.
Detection of crs57 in culture supernatants
Overnight cultures (10 ml) of S. pyogenes 2077, NS38, BSA16, BSA5, NS844, and NS27 in Todd-Hewitt broth (Oxoid) were centrifuged at 12,000 × g for 10 min, and 1 ml of each culture supernatant was transferred to a new tube. The culture supernatants were concentrated by using trichloroacetic acid (final concentration, 10%) at −20°C for approximately 20 min to induce precipitation. To retrieve the precipitated proteins, each mixture was centrifuged at 16,000 × g for 20 min. The supernatant was discarded, and the pellet was resuspended in 100 μl of 0.1 M NaOH. SIC was detected by separation of the sample by PAGE (Gradipore; Miniprotean II; Bio-Rad), followed by Western blotting and detection with anti-SIC antibody.
Binding of CRS57 to complement
Assays for binding of CRS57 to the complement proteins C6 and C7 were performed by using an enzyme-linked immunosorbent assay. Ninety-six-well plates (TITERTEK) were coated with recombinant proteins (50 μg/ml; CRS57 or control proteins) in PBS at 4°C overnight. After blocking with 5% skim milk in PBS, complement from human sera (diluted 1/100 in PBS) or commercial C6 and C7 (diluted 1/1,000 in PBS; Sigma) were added and incubated for 1 h at 37°C in a 100-μl (final volume) mixture. The wells were then washed three times with PBS containing 0.5% Tween. Horseradish peroxidase-conjugated secondary antibodies to the complement components C6 and C7 (diluted 1:1,000; ICN Biomedical) were used to determine the extent of binding. The reaction mixture was developed with 4-chloro-1-naphthol (Sigma), and the absorbance at 450 nm was determined with a Bio-Rad benchmark microplate reader.
Nucleotide sequence accession numbers
Nucleotide sequences of the PCR products obtained from the six independent isolates of the emm57 type have been deposited in the GenBank database under accession numbers AY229856, AY229857, AY229858, AY229859, AY229860, and AF060764.
RESULTS
Divergence of crs57 sequence
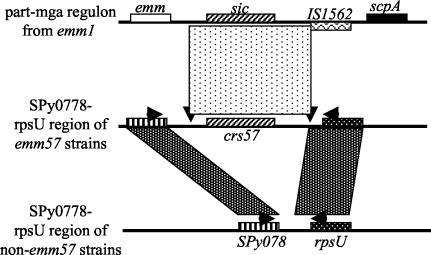
PCR performed with primers flanking the 3′ end of the ABC transporter gene (SPy0778) and the 5′ end of rpsU suggested that in all emm57 isolates the two genes are farther apart than they are in serotype M1strains or 39 other strains tested. Data for the six isolates of the emm57 sequence type and strains of representative non-emm57 sequence types are shown in Fig. 2. The six isolates of the emm57 sequence type belong to two distinct vir types, VT8 and VT101. The results of a comparison of the SPy0778-rpsU regions in emm1 and emm57 strains are presented schematically in Fig. 3. In all six emm57 isolates tested, the region between the SPy0778 and rpsU genes contained crs57. Remnants of the emm1 mga regulon were present immediately upstream and downstream of the crs57 gene. In emm1, there is an IS1562 sequence in reverse orientation between the sic gene and the scpA gene. The IS1562 sequence was not found within the SPy0778-rpsU region in the emm57 strains. However, the intervening sequence between the sic gene and the IS1562 in M1, including a copy of the terminal inverted repeat of the insertion element, was conserved in the SPy0778-rpsU region in all of the emm57 strains and occurred in the same position in relation to the crs57 gene (Fig. 3). These findings suggest that a crs57 gene was recently acquired by M57 or its progenitor, possibly by horizontal transfer from M1.
FIG. 3.
Comparison of part of the mga locus in M1 and the SPy0778-rpsU region in M57 and other strains. The shaded areas in the SPy0778-rpsU region represent homologous regions. The mottled area between the regions surrounding the sic gene in M1 and the regions surrounding the crs57 gene in M57 shows a high level of similarity. This area also includes one of the inverted repeats of IS1562. The downward arrowheads beneath the mottled rectangle suggest that there was horizontal acquisition of this region from M1. The horizontal arrows show the relative positions of the ABC-F2 and rpsU-R1 primers used to amplify SPy0778-rpsU regions (the sequences are given in the text). The figure is not to scale.
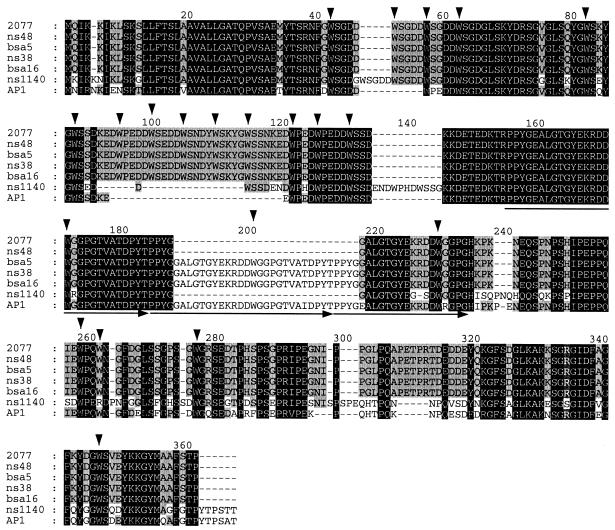
The emm57 strains used in this study include one strain from Prague (a reference strain) and five strains from the NT that belong to two distinct vir types (Fig. 1). Interestingly, the vir type profile of strain 2077 is identical to the VT8 profile. The derived CRS57 sequences of all of the VT8 isolates are highly homologous except for a few insertions or deletions of residues (including short and long repeats) (Fig. 4). A comparison of the corresponding DNA sequences also showed that there is a silent mutation in addition to the mutations that account for the amino acid changes. By contrast, the crs57 gene in NS1140, which is a VT101 strain, had several replacement mutations. Interestingly however, examination of the C-proximal half of the NS1140 CRS57 revealed a greater resemblance to the corresponding region of SIC from strain AP1 than to the corresponding region of VT8 CRS57. A comparison of the sic sequence from strain AP1 (accession number X92968) and the VT8 crs57 sequence showed that there was a relatively high number of replacement mutations in the C-proximal third of the molecule (26 replacements in the C-proximal third and 12 replacements in the rest of the protein). Of particular interest is the conservation of tryptophan-containing short repeats. The frequencies of the five amino acids surrounding the tryptophan residues were determined (Fig. 5). The most conserved amino acid residues in the first three of the five positions are (D/G)-W-(S/G). We therefore refer to this as the DWS motif. The M57 isolates tested in this study each had 15 to 21 DWS motifs.
FIG. 4.
Alignment of CRS57 from strains 2077, NS48, BSA5, NS38, BSA16, and NS1140 with SIC from M1 strain AP1. The three backgrounds indicate levels of conservation. The residues conserved in all seven sequences are indicated by a black background, and the residues conserved in five or six of the seven sequences are indicated by a grey background. The downward arrowheads indicate tryptophan residues conserved in at least four sequences. The horizontal arrows indicate repeat sequences.
FIG. 5.
Frequency of residues within the tryptophan-containing short repeat domains. To generate the frequency histogram, the BSA5-derived amino acid sequence data were used. There were 21 tryptophan residues, 19 of which were in the short conserved motifs. The letters within the bars indicate the amino acid residues. The sections of the bars labeled “others” could include up to four different residues.
CRS57 is an excretory product
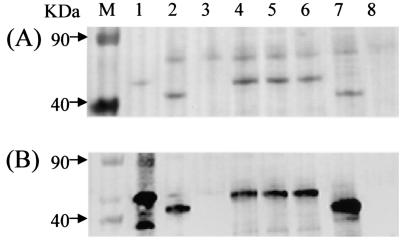
The results described above showed that the crs57 gene is at the end of the ABC transporter immediately upstream of the rpsU gene in all of the emm57 strains. Since all of the crs57 sequences analyzed have a signal sequence, it is reasonable to expect that, like the SIC protein, the CRS57 protein in these strains is an excretory product. We confirmed this by analyzing culture supernatants and whole-cell extracts from overnight cultures of all six emm57 isolates by PAGE and Western blotting (Fig. 6; data for NS1140 not shown). Anti-SIC antibodies reacted with 40- to 50-kDa bands in culture supernatants of strain 2031 (a positive control strain) and the emm57 strains, as reported previously (6). The observed size differences between the CRS57 proteins of different strains in Fig. 6 is consistent with derived crs57 sequence data.
FIG. 6.
Expression of CRS57 by M57 strains. Supernatants of overnight cultures of GAS strains were concentrated by precipitation in 10% cold trichloroacetic acid for 1 h. Each pellet was dissolved in 100 mM NaOH. The samples (40 μl) were electrophoresed in 4 to 12% (Gradipore) acrylamide under denaturing conditions. (A) Ponceau-stained membrane. (B) Western blot of the same gel reacted with polyclonal rabbit serum raised against SIC from M1 strain 2031. Lane M contained a marker. Lane 1, recombinant SIC from M1; lanes 2 to 7, culture supernatants from 2077, NS38, BAS16, BSA5, NS844 (an NT emm1 strain), and NS27 (a SIC-negative strain), respectively.
Demonstration of binding of CRS57 to complement proteins C6 and C7
Binding of SIC from an M1 strain to intermediate terminal complement complexes was recently demonstrated by Fernie-King et al. (6). To show that recombinant CRS57 proteins bind to C6 and C7 complement proteins in serum, we purified the fusion protein containing thioredoxin (pBAD thio cloning system). In our hands the expression of CRS57 in the pQE system gave low yields, the recombinant proteins accumulated in inclusion bodies, and the proteins were insoluble and often degraded upon purification. The thioredoxin fusion improved the yield, solubility, and stability of the recombinant proteins considerably. Thioredoxin by itself did not bind to the complement proteins (Fig. 7). Plates coated with the recombinant proteins were reacted with human serum, and binding of the C6 and C7 complement proteins was detected by using secondary antibodies. The results demonstrated that the complement component C6 and C7 proteins were bound to CRS57 (Fig. 7A). To confirm this, we also tested binding using purified C6 and C7. In these experiments the recombinant CRS57 coated onto plates was reacted with the C6 and C7 proteins (Fig. 7B). The results confirmed that there is an interaction between the complement proteins and CRS57.
FIG. 7.
Binding of CRS57 to the complement C6 (solid bars) and C7 (open bars) proteins as determined by an enzyme-linked immunosorbent assay. (A) Binding to the complement proteins in the sera. (B) Binding to the purified C6 and C7 components. CRS1 and CRS57 were recombinant CRS proteins from strains 2031 and BSA5, respectively. The controls included recombinant His-tagged thioredoxin (thio), bovine serum albumin (BSA), and PBS (no coat). The data are averages for triplicate experiments for each protein, and the error bars indicate standard deviations. OD540, optical density at 540 nm.
DISCUSSION
We reported previously (12) that in type 57 isolates the gene for CRS57 is outside the mga regulon, which contains emm, emm-like, scpA, and mga genes. In addition to confirming this, in this study we showed that in all six type 57 isolates tested the gene is located between the ABC transporter (SPy0778) and the rpsU gene. The organization of the Spy0778-rpsU region is highly conserved in the non-emm57 GAS strains tested and in three genomic sequences (M1, M3, and M18) (2, 7, 18). The CRS57 sequence of NS1140 exhibited considerable diversity compared to the other CRS57 sequences analyzed. Despite the difference, the junction points on either side of crs57 in all six emm57 strains are highly conserved. These observations suggest that horizontal acquisition of the crs gene must have been a one-off process and may have been a recent evolutionary event. Recently, Ma et al. (16) found that sic was distributed in a greater number of strains when they examined pharyngitis isolates from Japanese children. We believe that if horizontal acquisition was the mode of spread of sic among GAS strains, as these authors indicate, the source may been serotype M1 because this serotype has an intact IS1562.
Since CRS57 proteins have all of the structural characteristics of the SIC from an M1 strain, it is reasonable to expect that CRS57 proteins also have similar biological properties. We tested this hypothesis by determining whether the CRS57 proteins, like SIC from M1 strains, bind to the C6 and C7 complement proteins. Our results show that the CRS57 proteins interact with the complement proteins. Hence, it may be inferred that CRS57 has the same properties as SIC from M1 with respect to the interaction with the complement proteins.
Recent serological observations in the NT indigenous population, in which post-streptococcus glomerulonephritis is highly endemic, revealed that 57% of the population has antibodies to SIC (19). However, isolation of type 1 or close relatives of this type from this population has been rare in the last 10 years (unpublished observations). By contrast, the rate of isolation of emm57 strains is high. Thus, at least the newly acquired antibodies to SIC in our study population are most likely due to emm57 strains. If so, this further supports our hypothesis that CRS57 is expressed and is antigenic during natural infection. Thus, if antibodies confer selection pressure for variants of sic from M1 strains (as proposed by Hoe et al. [14]), the same pressure could also operate to select variants of crs57 given that SIC and CRS57 have common biological and biochemical properties. Whereas a large number of mutations have been observed in the crs57 gene from VT101, the frequency of point mutations in the remaining five epidemiologically unrelated emm57 isolates was low; only short insertions or deletions, particularly of the repeat sequences, accounted for most of the mutations.
The mga regulon harbors antigenically highly variable genes. This regulon may be a mutational hot spot in the GAS genome. Since in M57 the crs57 gene is outside this region, it may be less prone to mutations than sic from M1.
All CRS proteins have short conserved DWS repeats. The tryptophan content of some CRS molecules could be as high as 6%. The role of the conserved tryptophan-containing motif is not known. In some proteins tryptophan-rich motifs may have a role in membrane binding. For instance, streptolysin O, a thiol-activated cytolysin, has a tryptophan-rich domain in the C-terminal region which is essential for membrane binding (21). Since SIC is known to enter host cells efficiently (13), it is possible that the DWS motif is responsible for promoting an interaction between SIC and the host cell membrane. Further work to test this hypothesis is in progress.
Acknowledgments
This work was supported by the Australian National Health and Medical Research Council.
We thank Jon Hartas for technical help with the initial cloning experiments.
Editor: V. J. DiRita
REFERENCES
- 1.Akesson, P., A. G. Sjoholm, and L. Bjorck. 1996. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J. Biol. Chem. 271:1081-1088. [DOI] [PubMed] [Google Scholar]
- 2.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berggard, K., E. Johnsson, E. Morfeldt, J. Persson, M. Stalhammar-Carlemalm, and G. Lindahl. 2001. Binding of human C4BP to the hypervariable region of M protein: a molecular mechanism of phagocytosis resistance in Streptococcus pyogenes. Mol. Microbiol. 42:539-551. [DOI] [PubMed] [Google Scholar]
- 4.Cleary, P. P., U. Prahbu, J. B. Dale, D. E. Wexler, and J. Handley. 1992. Streptococcal C5a peptidase is a highly specific endopeptidase. Infect. Immun. 60:5219-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernie-King, B. A., D. J. Seilly, A. Davies, and P. J. Lachmann. 2002. Streptococcal inhibitor of complement inhibits two additional components of the mucosal innate immune system: secretory leukocyte proteinase inhibitor and lysozyme. Infect. Immun. 70:4908-4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernie-King, B. A., D. J. Seilly, C. Willers, R. Wurzner, A. Davies, and P. J. Lachmann. 2001. Streptococcal inhibitor of complement (SIC) inhibits the membrane attack complex by preventing uptake of C567 onto cell membranes. Immunology 103:390-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardiner, D., J. Hartas, B. Currie, J. D. Mathews, D. J. Kemp, and K. S. Sriprakash. 1995. Vir typing: a long-PCR typing method for group A streptococci. PCR Methods Applic. 4:288-293. [DOI] [PubMed] [Google Scholar]
- 9.Giannakis, E., D. A. Male, R. J. Ormsby, C. Mold, T. S. Jokiranta, S. Ranganathan, and D. L. Gordon. 2001. Multiple ligand binding sites on domain seven of human complement factor H. Int. Immunopharmacol. 1:433-443. [DOI] [PubMed] [Google Scholar]
- 10.Gillen, C. M., R. J. Towers, D. J. McMillan, A. Delvecchio, K. S. Sriprakash, B. Currie, B. Kreikemeyer, G. S. Chhatwal, and M. J. Walker. 2002. Immunological response mounted by aboriginal Australians living in the Northern Territory of Australia against Streptococcus pyogenes serum opacity factor. Microbiology 148:169-178. [DOI] [PubMed] [Google Scholar]
- 11.Hartas, J., M. Hibble, and K. S. Sriprakash. 1998. Simplification of a locus-specific DNA typing method (Vir typing) for Streptococcus pyogenes. J. Clin. Microbiol. 36:1428-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartas, J., and K. S. Sriprakash. 1999. Streptococcus pyogenes strains containing emm12 and emm55 possess a novel gene coding for distantly related SIC protein. Microb. Pathog. 26:25-33. [DOI] [PubMed] [Google Scholar]
- 13.Hoe, N. P., R. M. Ireland, F. R. DeLeo, B. B. Gowen, D. W. Dorward, J. M. Voyich, M. Liu, E. H. Burns, Jr., D. M. Culnan, A. Bretscher, and J. M. Musser. 2002. Insight into the molecular basis of pathogen abundance: group A Streptococcus inhibitor of complement inhibits bacterial adherence and internalization into human cells. Proc. Natl. Acad. Sci. USA 99:7646-7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoe, N. P., K. Nakashima, S. Lukomski, D. Grigsby, M. Liu, P. Kordari, S. J. Dou, X. Pan, J. Vuopio-Varkila, S. Salmelinna, A. McGeer, D. E. Low, B. Schwartz, A. Schuchat, S. Naidich, D. De Lorenzo, Y. X. Fu, and J. M. Musser. 1999. Rapid selection of complement-inhibiting protein variants in group A Streptococcus epidemic waves. Nat. Med. 5:924-929. [DOI] [PubMed] [Google Scholar]
- 15.Johnsson, E., A. Thern, B. Dahlback, L. O. Heden, M. Wikstrom, and G. Lindahl. 1996. A highly variable region in members of the streptococcal M protein family binds the human complement regulator C4BP. J. Immunol. 157:3021-3029. [PubMed] [Google Scholar]
- 16.Ma, X., H. Kikuta, N. Ishiguro, M. Yoshioka, T. Ebihara, T. Murai, I. Kobayashi, and K. Kobayashi. 2002. Association of the prtF1 gene (encoding fibronectin-binding protein F1) and the sic gene (encoding the streptococcal inhibitor of complement) with emm types of group A streptococci isolated from Japanese children with pharyngitis. J. Clin. Microbiol. 40:3835-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riley, J., R. Butler, D. Ogilvie, R. Finniear, D. Jenner, S. Powell, R. Anand, J. C. Smith, and A. F. Markham. 1990. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 18:2887-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sriprakash, K. S., J. Hartas, and A. White. 2002. Antibodies to streptococcal inhibitor of complement function and M peptides in a post-streptococcal glomerulonephritis endemic region of Australia. J. Med. Microbiol. 51:589-594. [DOI] [PubMed] [Google Scholar]
- 20.Stockbauer, K. E., D. Grigsby, X. Pan, Y. X. Fu, L. M. Mejia, A. Cravioto, and J. M. Musser. 1998. Hypervariability generated by natural selection in an extracellular complement-inhibiting protein of serotype M1 strains of group A Streptococcus. Proc. Natl. Acad. Sci. USA 95:3128-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weis, S., and M. Palmer. 2001. Streptolysin O: the C-terminal, tryptophan-rich domain carries functional sites for both membrane binding and self-interaction but not for stable oligomerization. Biochim. Biophys. Acta 1510:292-299. [DOI] [PubMed] [Google Scholar]