Abstract
Nine different near-atomic resolution structures of icosahedral viruses, determined by electron cryo-microscopy and published between early 2008 and late 2010, fulfill predictions made 15 years ago that single-particle cryo-EM techniques could visualize molecular detail at 3 – 4 Å resolution. This review summarizes technical developments, both in instrumentation and in computation, that have led to the new structures, which advance our understanding of virus assembly and cell entry.
Introduction
Most cellular activities are outcomes of interactions among many components, including proteins, nucleic acids and lipids. Electron cryo-microscopy of isolated macromolecular complexes ("single particle cryo-EM") can now visualize icosahedral viruses at near-atomic resolution (Table 1, [1–9]), and it should soon achieve similar resolution with less symmetrical particles. In a seminal review 15 years ago, Henderson predicted the success of single-particle techniques in visualizing detail at 3 – 4 Å resolution [10], and Glaeser extended the analysis a few years later [11]. A major step towards this goal was visualization of the hepatitis B icosahedral capsid at sub-nanometer resolution [12,13], allowing for the first time the identification of a protein fold using single-particle cryo-EM. Reconstructions at sub-nanometer resolution of particles with lower symmetry followed, and it is now possible to obtain reconstructions of ribosomes at about 5 Å resolution [14].
Table 1.
Chronological list of icosahedral virus structures imaged at near-atomic resolution. "Averaged subunits" is the number of molecules (or molecular units) averaged to give the stated resolution. References to image processing software: Signature[70], CTFTILT/CTFFIND3[41], Frealign[45], Uppsala[71,72], EMAN[52], IMIRS[51], MPSA[73].
| Virus | Cryo- gen |
Cryo stage | Detector | Image processing |
Averaged subunits |
MW [MDa] |
Resolution [Å] |
|---|---|---|---|---|---|---|---|
| Rotavirus DLP [1] | LN2 | FEI F30, Gatan side entry | Film | Signature, CTFTILT, Frealign, Uppsala | 6.6 × 106 | 70 | 3.8 (FSC=0.143) |
| ε15 phage [2] | LHe | JEOL JEM3000SFF | Film | EMAN | 1.2 × 106 | 22 | 4.5 (FSC=0.5) |
| CPV [3] | LN2 | FEI Polara | 4k × 4k CCD | IMIRS | 7.7 × 105 | 50 | 3.8 (FSC=0.5) |
| Rotavirus 7RP [4] | LN2 | FEI F30, Gatan side entry | Film | Signature, CTFTILT, Frealign, Uppsala | 2.7 × 106 | 95 | 4.2 (FSC=0.143) |
| Bovine papillomavirus [5] | LN2 | FEI F30, Gatan side entry | Film | Signature, CTFTILT, Frealign, Uppsala | 1.4 × 106 | 26 | 3.6 (FSC=0.143) |
| Aquareovirus ISVP [6] | LN2 | FEI Titan Krios | Film | CTFFIND3, Frealign, IMIRS | 1.1 × 107 | 110 | 3.3 (FSC=0.143) |
| Cyanophage P-SSP7 [7] | LN2 | JEOL JEM3200FSC | Film | ETHAN, EMAN, MPSA | 2.2 × 106 | 17 | 4.6 (FSC=0.5) |
| Adenovirus [8] | LN2 | FEI Titan Krios | Film | CTFFIND3, IMIRS, EMAN | 1.9 × 106 | 150 | 3.6 (FSC=0.143) |
| Rotavirus TLP [9] | LN2 | FEI F30, Gatan side entry | Film | Signature, CTFTILT, Frealign, Uppsala | 2.5 × 105 | 110 | 4.3 (FSC=0.143) |
The leading role of icosahedral viruses in achieving near-atomic resolution is due primarily to: (i) their high symmetry, which effectively increases the size of the data set 60-fold, or more if quasi-equivalent subunits can be averaged; (ii) their large molecular mass, producing strong image contrast and hence more accurate alignments and reconstructions; and (iii) their rigidity and uniformity, ensuring near-perfect superposition of structural features in three-dimensional (3D) reconstructions. In this review, we consider technological improvements made over the last decade that have enabled the recent successes, explain some of the remaining challenges and discuss some of the resulting functional conclusions about the organization of non-enveloped virions.
Problems and progress
Radiation damage and beam-induced sample movement
Inelastic scattering of high-energy electrons by the sample leads to radiolysis of macromolecules and of the embedding medium, usually vitrified water. The extent of radiation damage depends on the electron dose and sample temperature and beam energy, thus limiting the useful dose for imaging to 5 – 10 e−/Å2 at liquid nitrogen temperature when using 100 kV electrons [10,15]. The doses used for the near-atomic resolution reconstructions listed in Table 1 range between 15 and 30 e−/Å2 which is a bit higher due to the smaller scattering cross-sections at the higher voltage used (300 kV). The dose limitation imposes a lower molecular mass limit of 20 to 50 kDa for molecules that can be detected and aligned in an image [10,11], well below the typical mass of an icosahedral virus (see Table 1). The products of radiolysis require more space than the original molecules, leading to buildup of internal pressure in the sample and ultimately to breakdown (“bubbling”) of the sample matrix [16]. The pressure increase and bubbling cause movement in the exposed area of the sample and blurring in the image, thereby attenuating or eliminating the signal at higher resolution [15]. A recent study suggests that decreasing the rate of radiolysis by using an electron beam with lower intensity can reduce pressure buildup and bubbling [17]. Inelastic scattering also leads to ejection of electrons from the sample and buildup of positive charge that can act as a weak “lens”, again shifting the image and causing blurring [15].
While the dose limitation represents a fundamental limit of the technique, sample movement and charging can be minimized by using more rigid and conductive sample support films (for example, TiSi [18], doped silicon carbide [19]). The performance of these new support films at near-atomic resolution remains to be evaluated. Virus sample is usually prepared using perforated support films in which the vitrified sample is imaged in ice over the holes. It is generally recognized that charging can be reduced by using an objective aperture [15] and by including a small amount of the support film in the illuminated sample area at the edge of a hole, which acts as a drain for the evolving charge. Machined grids (Quantifoil™ [20], C-Flat™ [21]) provide reproducible hole sizes and allow symmetrical imaging conditions, in which a centered beam includes the rim of the circular hole. Machined grids also help produce a more uniform ice thickness, which can then be optimized by adjusting the vitrification procedure. Circular holes and optimized ice thickness have led to higher average image quality and thus to higher resolution in 3D reconstructions of icosahedral viruses and other samples. The spot-scan technique [22] in which only a small sample area is exposed at a time can also reduce beam-induced sample movement in single-particle EM but it is predominantly used in two-dimensional crystallography.
Cooling the sample at liquid helium temperature has been discussed as a way to prolong the molecular integrity of the sample under the high-energy electron beam [23,24] and may generate superior results for 2D crystals. Most recent high-resolution virus structures were calculated from data recorded at liquid nitrogen temperature, however (see Table 1), as increased charging and reduced image contrast at liquid helium temperature annul the advantage gained by the increased dose tolerance of single particles [25,26].
Instrumentation
New sample stages for cryo-microscopy have been introduced during the last ten years to improve cooling and mechanical stability. The new stages include top-entry designs (for example, JEOL JEM3000SFF) as well as specially designed side-entry stages (for example, FEI Polara and FEI Titan Krios), all of which use detachable sample cartridges that afford better vacuum, insulation from the environment and temperature stability. The FEI Titan Krios also features a symmetrical lens design to allow operation with a constant total lens current to keep the heat generation constant. Furthermore, most modern microscopes allow remote operation, to eliminate instabilities caused by the presence of a human operator. The FEI Titan Krios offers additional protection against environmental changes with a box housing the microscope. While the new stage designs and environmental protection are clear improvements over older designs, several of the recent near-atomic resolution structures were obtained with traditional side-entry stages and electron microscopes that were available a decade ago (see Table 1). The older designs are therefore still a competitive and cost-effective alternative to the much more expensive latest-generation instruments.
The data for all reconstructions listed in Table 1, except CPV, were collected on photographic film. Film has a higher detective quantum efficiency (DQE) and larger number of pixels per frame compared with the scintillator-based electronic detectors in general use today [27]. A new generation of solid state detectors is emerging, based on direct detection of the high-energy electrons [27,28]. Future improvements to these detectors will include back-thinning and a single-electron counting mode, in which incident electron positions can be detected with sub-pixel accuracy. These detectors will represent a substantial improvement over photographic film with respect to DQE, speed and effective number of pixels per frame. They will therefore have an important role in high-resolution cryo-EM of macromolecular assemblies, either by reducing the number of images required to reach a certain resolution or by extending the attainable resolution.
A fundamental improvement in image quality is also anticipated with the introduction of phase plates [29–32], which boost low-resolution contrast. This enhancement will be especially important for work on particles of lower molecular mass but may still help in the accurate alignment of large virus particles (see below).
Determination of single particle parameters
The single-particle technique gains its greatest advantage over x-ray crystallography by not requiring crystals. While molecules in a crystal have fixed orientations, imposed by crystal contacts, the orientation and x,y position of each single particle in an image must be determined separately. Errors in these parameters, in particular in the Euler angles specifying the view of a particle, probably pose one of the most severe limitations in attainable resolution for single-particle work [33]. The angular alignment error of rotavirus DLPs was estimated to be about 0.2° which attenuates the signal at the periphery of this 700-Å particle by roughly 70% [1]. The accuracy with which alignment parameters can be determined depends primarily on the signal strength associated with each particle, especially at low resolution where the benefit of a phase plate is expected to be greatest. The alignment accuracy can be improved by introducing appropriate resolution-dependent weighting of the data [34] and parameter restraints [4]. The use of symmetry-adapted functions can also help in determining more accurate particle angles and shifts [35,36]. Another promising approach is the use of maximum likelihood functions in estimating 3D reconstructions [37], as this approach should be more noise-tolerant and should increase the radius of convergence in a refinement [38]. The significantly higher computational cost of this approach has so far prevented its application to reconstructions at near-atomic resolution.
Particle heterogeneity
Even the most accurate particle alignment cannot overcome limitations introduced by molecular flexibility and disorder within a particle. This restriction applies also to icosahedral viruses, which are generally extremely regular and rigid assemblies, when internal disorder limits the attainable resolution, just as in x-ray crystallography. Depending on the nature of the disorder, introducing additional parameters or image classes can separate particles into more homogeneous subsets, each of which can (in principle) yield a higher resolution reconstruction. The development of methods dealing with particle heterogeneity represents one of the most important current research topics in the field. The separation of damaged or distorted particles and particle images affected by beam-induced movement and other imperfections from intact particles and high-quality images can be made more easily for large assemblies such as icosahedral viruses than for smaller particles due to their strong image contrast [1].
Signal restoration from images
Cryo-EM images are affected by small variations in magnification [39] and corruption by the contrast transfer function (CTF) of the microscope. While the former did not require special accommodation in image processing procedures to reach near-atomic resolution, the latter must be corrected to retrieve the high-resolution signal. The oscillating behavior of the CTF depends on the image defocus and produces so-called Thon rings [40] in the image power spectrum. The Thon ring pattern can be used to determine the CTF by fitting a calculated ring pattern. Inaccuracies in the determined defocus were estimated between 100 Å and 1000 Å, depending on the presence of carbon support film in the images [41]. These inaccuracies lead to phase errors in the corrected data and affect the signal at high resolution more strongly than at low resolution. For example, an average defocus error of 300 Å will lead to about 60% signal loss in a reconstruction at 4 Å resolution [1]. Because virus particles produce strong image contrast, it is possible to refine the defocus per particle using restraints, leading to a modest resolution improvement [4].
Additional phase error comes from beam tilt [42,43]. Careful alignment of the microscope can eliminate beam tilt, but unless the C3 lens of the microscope is freely adjustable, as in the FEI Titan Krios and JEOL JEM3200FSC, it is hard to obtain a strictly parallel beam. A small amount of variable tilt will therefore remain (axial coma [42]). In this case, the beam tilt will be a function of the position within the image, and a computational procedure will be needed to retrieve corrected phases. Beam tilt is routinely determined and corrected for in 2D crystallography by minimizing the phase difference between an image and a reference [44]. Although beam tilt correction has been implemented in the software package Frealign [45] which was used for some of the reconstructions listed in Table 1, it has not been attempted for any of the published near-atomic resolution virus structures because its measurement is less accurate for single particles than for 2D crystal due to the weaker signal. As beam tilt does not change significantly from image to image, it should be possible to measure it for an entire data set of large single particles such as viruses, using an appropriate reference to maximize the average correlation coefficient with all aligned particles in the data set.
Ewald sphere curvature (limited depth of focus) also introduces phase errors, which are expected to reach about 60° at 3.3 Å for a 700 Å particle imaged at 300 kV [46]. A correction procedure for the Ewald sphere curvature [47] was applied in the reconstruction of BPV [5], leading to a small improvement in the map. This result suggests that Ewald sphere curvature is not limiting at the resolution levels reported in Table 1. The icosahedral particle symmetry implies that many of the asymmetric units within the particle will be imaged close to the average defocus determined for the entire particle; for these asymmetric units the phase error will be small and Ewald sphere curvature will not be a major resolution-limiting factor.
Contrast transfer is also affected by the limited coherence of the electron beam. To avoid resolution loss, focal pairs can be recorded with a close-to-focus image containing strong high-resolution signal, and a far-from-focus image containing strong low-resolution signal needed for reliable particle alignment (for example, see [3,13]). The brightness of modern field emission guns generates highly coherent electron beams, while keeping exposure times within one to two seconds. Therefore, focal pairs for large icosahedral particles are no longer required, and the procedure was not used in any of the recent cryo-EM studies, except for the work on CPV (see Table 1).
3D reconstruction
The performance of computer hardware, including capacity of in-core memory and speed of program execution, has restricted achievable resolution at all stages since the first icosahedral reconstructions in 1970 from 6 particles of negatively stained TBSV (to 28 Å resolution) and two of human wart virus (to 60 Å) [48]. The resolution of early virus reconstructions from cryo-EM was limited by the pixel size. For example, the HepB capsid reconstructions published in 1997 [12,13] used a pixel size of about 2.5 Å, imposing a limit of 6 – 7 Å on the reconstruction, when the typical signal loss due to interpolation is taken into account. The reconstructions listed in Table 1 were all done with pixel sizes of 1.3 Å or smaller. Limiting computing resources also affected the pioneering reconstruction algorithms developed for icosahedral viruses, which used symmetry-adapted Fourier-Bessel synthesis to reduce the determination of the 3D Fourier transform of the virus to a set of one-dimensional problems that could be solved using standard matrix inversion techniques [49]. This simplification limited the accuracy of the least-squares solution found for each matrix inversion. Contemporary computing hardware has enabled the development of more powerful algorithms that calculate least-squares solutions of the inversion problem using spherical harmonics [35,36,50]. These new algorithms have been implemented in the software packages RIco [36,50] and IMIRS [35,51], used for some of the reconstructions listed in Table 1. Other reconstructions in Table 1 were calculated using EMAN [52] and Frealign [45], which employ a more general algorithm commonly referred to as direct Fourier inversion [53].
The resolution of a 3D reconstruction is less objectively determined than in x-ray crystallography, where "resolution" has come to mean the highest spatial frequency contributing in a meaningful way to the map or to the refinement. A resolution cutoff is important, however, not only for reporting a reasonable assessment of detail in 3D-reconstruction density maps, but also for avoiding oversharpening when amplitude scaling is used to restore high-resolution features. Scaling is generally accomplished by using reference data from x-ray solution scattering profiles [54] or by determining an effective B-factor and figure-of-merit [33].
Different resolution criteria are in use, but each can be somewhat unreliable. The values listed in Table 1 are those provided in the respective publications. In all cases, the Fourier Shell Correlation (FSC [55]) was calculated, but in some the resolution was determined at FSC = 0.5 and in others, at FSC = 0.143. The former cutoff was introduced ad hoc [12], while the latter was proposed to adjust the FSC resolution criterion to relate more closely to figure-of-merit criteria used in x-ray crystallography [33]. In each case, using the other resolution criterion will lead to an unrealistic estimate as judged by the detail seen in the maps. For example, a resolution of 4.5 Å was reported for the reconstruction of bacteriophage epsilon15 [2] at FSC = 0.5. The FSC curve published for this structure shows that FSC = 0.143 at a resolution of about 3.3 Å, an estimate inconsistent with the lack of clear density for many of the side chains. Conversely, a resolution of 3.3 Å was reported for the VP1 protein of aquareovirus [6] at FSC = 0.143; the estimate would have been only 3.8 Å at FSC = 0.5, incompatible with the distinctive density for many VP1 side chains.
A possible source of these inconsistencies lies in the alignment algorithms. The use of a correlation coefficient can lead to amplification of noise in 3D reconstructions and a strong bias towards higher values in the FSC, depending on the weighting of the data and the resolution limits used for the alignment [34,56]. In reconstructions affected by a bias in the FSC, it may then be necessary to compensate by reporting the resolution at FSC = 0.5. The software package Frealign reduces noise bias by using a weighted correlation coefficient [34] and parameter restraints [4], and FSC=0.143 appears to provide a reliable estimate for the resulting maps. In several cases listed in Table 1, the resolution of the reconstruction extends beyond the visibility of Thon rings in power spectra calculated from image fields of the viruses [2,3,6,8], showing that useful signal can be obtained beyond detectable Thon rings, contrary to earlier suggestions [57].
Near-atomic resolution virus structures
Double-stranded DNA (dsDNA) bacteriophage
The head-protein subunits of a large class of dsDNA bacteriophages have a folded structure first recognized in crystallographic analyses of the HK97 phage head and its assembly precursors [58]. Unlike the largely α-helical HepB capsid protein, which could be traced correctly at about 7 Å resolution [12,13], the phage-head subunit is largely β-sheet, for which strands are resolved only if the resolution extends beyond 5 Å. One of the first group of higher-resolution cryo-EM reconstructions showed that the bacteriophage epsilon15 head subunit is a member of the HK97 class [2]. This conclusion derived from a chain trace based on automatic assignment of secondary structural elements, and it may therefore not have exact sequence assignments in some parts of the fit. Local averaging within the T-7 icosahedral asymmetric unit, as in the work on bovine papillomavirus (BPV: see below [5]), and direct visual model building, as in most of the other efforts reviewed here, might enable a number of adjustments. A subsequent study by the same group of the T7-like cyanophage, P-SSP7, at 4.6 Å resolution showed, as expected, conservation of the same subunit design [7].
Double-stranded RNA (dsRNA) viruses
These viruses have segmented RNA genomes, packaged into a shell formed by 120 subunits of an "inner shell protein" (ISP). The ISPs are homologous in all cases described here [59]. The "inner capsid particle" (ICP) based on this shell also contains an RNA-dependent RNA polymerase, a capping activity, and in some cases an additional set of stabilizing subunits. The capping enzymes (guanylyl transferase and methylases) are part of a single polypeptide chain, which may be on the outer surface of the ICP ("turreted" ICPs) or within its shell ("non-turreted" ICPs).
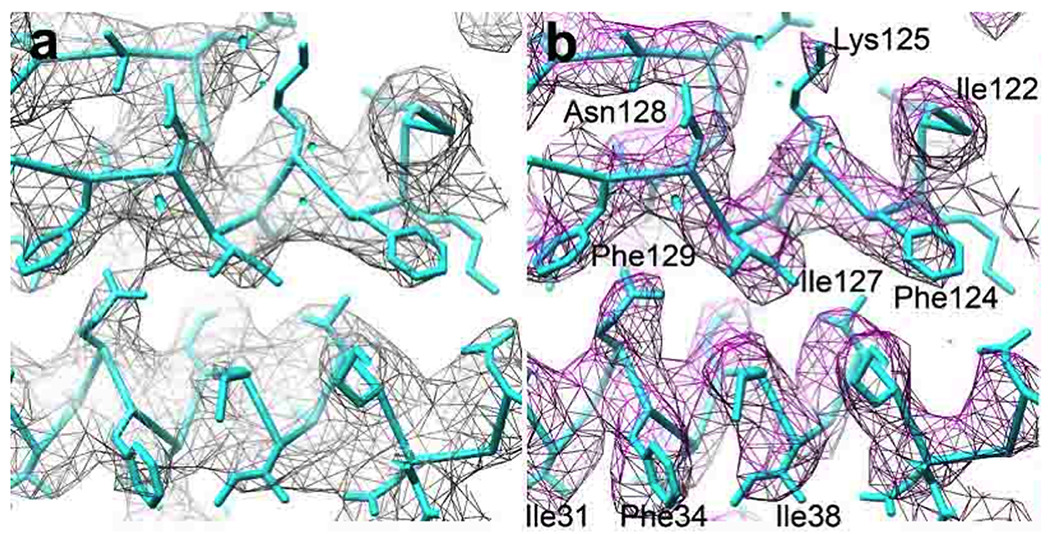
The rotavirus ICP is known as a "double layered particle" (DLP), because a complete layer of a protein known as VP6 surrounds the ISP layer, forming a T=13 icosahedral lattice. DLP structures have been determined both by cryo-EM at 3.8 Å nominal resolution [1] and by x-ray crystallography at about the same resolution [59]. Fig. 1 shows a comparison of the density maps, filtered to 4.2Å resolution. The cryo-EM density is roughly equivalent to the 2Fo-Fc x-ray map, even though the latter has been calculated with phases from a refined model.
Fig. 1.
Comparison of x-ray crystallographic and EM density maps of the rotavirus DLP [1]. (a) X-ray crystallographic 2Fo-Fc density map, filtered to 4.2 Å resolution, with refined atomic model superimposed [59]. The residue numbering refers to the VP6 polypeptide chain. (b) Same area as in (a) but showing the cryo-EM map. Modified from [1].
One or both of the outer-layer proteins (VP7 and VP4) can be added back to the DLP -- a step known as recoating [60]. A cryo-EM structure of the DLP recoated with VP7 (4.2 Å resolution) shows that a VP7 trimer caps each VP6 trimer in the T=13 intermediate layer, through contacts mediated largely by a 25-residue, N-terminal arm, which is not ordered in VP7 crystals [4]. The polypeptide chain of the N-terminal arm of VP7 was traced unambiguously, with a glycan at position 65 as a particularly prominent landmark. In the complete rotavirus triple-layered particle (TLP), nearly half of the VP4 polypeptide chain was traced de novo in a 4.2 Å resolution cryo-EM map [9]. By excising density from the final three-dimensional reconstruction and inserting it into a suitable P1 unit cell, "structure factors" were obtained and used as targets in standard x-ray crystallographic refinement procedures, with strong constraints to compensate for limited resolution.
The layer of 120 ISP subunits in turreted reovirus ICPs is stabilized by a set of "clamp" subunits [61]. Cytoplasmic polyhedrosis virus (CPV) does not require an additional outer layer to penetrate. A model, published in 2008, was the first example of a complete de novo chain trace from a cryo-EM reconstruction with clear evidence of amino-acid residue positions from side-chain density [3]. Homology of the ISP with the corresponding proteins of orthoreoviruses and orbiviruses, for which crystal structures had been determined previously [61,62], added confidence to the interpretation.
Surrounding the turreted ICP of orthoreoviruses is an outer protein layer based on a T=13 icosahedral lattice, through which the ICP turrets project at each fivefold [63]. The assembly unit is a threefold symmetric, outer-layer heterohexamer [64]. Proteolytic removal of one of the two subunits (a "chaperone") primes the other (a "penetration protein") to mediate ICP penetration into a host cell. Autolytic cleavage of the penetration protein liberates a pore-forming, myristoylated, N-terminal peptide. Reconstruction of an aquareovirus particle, protease treated to remove the outer-layer chaperone, generating a so-called "infectious subviral particle" (ISVP), confirmed earlier conclusions about the autolytic cleavage derived from a crystal structure of the othroreovirus penetration-protein:chaperone heterohexamer [64] and a ~6 Å resolution reconstruction of the virion [65]. The aquareovirus ISVP density map was of sufficient clarity to allow building of a full atomic model, which also showed the myristoyl groups inserted into long, hydrophobic pockets.
dsDNA viruses of eukaryotes
The papillomavirus capsid is a shell of 72 pentamers (protein L1), which surrounds a closed circular DNA genome coiled into about 25 nucleosomes. BPV has been, until recently, the only full virion that could be prepared in quantities sufficient even for cryo-EM. A reconstruction of BPV at about 3.8Å resolution [5] has permitted fitting and refinement of a complete model for the L1 shell, using a crystal structure of the HPV16 L1 pentamer as a homology template for the framework of the subunit [66]. Projecting loops and N- and C-terminal arms were build de novo. The framework and loops have identical structures on all subunits, allowing further local averaging of the initial icosahedral reconstruction. This sixfold "non-capsid symmetry" (ncs) averaging greatly facilitated model building. An earlier effort to fit a much lower resolution (~ 13 Å) BPV reconstruction had misconnected one part of the C-terminal arm [67]. The full BPV atomic model was refined against calculated structure factors, as described above for parts of the rotavirus TLP. Weights for the refinement (Hendrickson-Lattman coefficients) were assigned in shells, based on the Fourier shell correlation [5]. This calculation is effectively a real-space refinement carried out indirectly in reciprocal space. That is, there is no iterative improvement of the unmeasured parameters of the reconstruction (Euler angles, etc., for each image), unlike the phase improvement that accompanies cycles of refinement and rebuilding in x-ray crystallography. Whether some combination of model density with the reconstruction used to build the model could be used as a reference for further improvement of image parameters, leading to a better map for extending or adjusting the model, is an important open question.
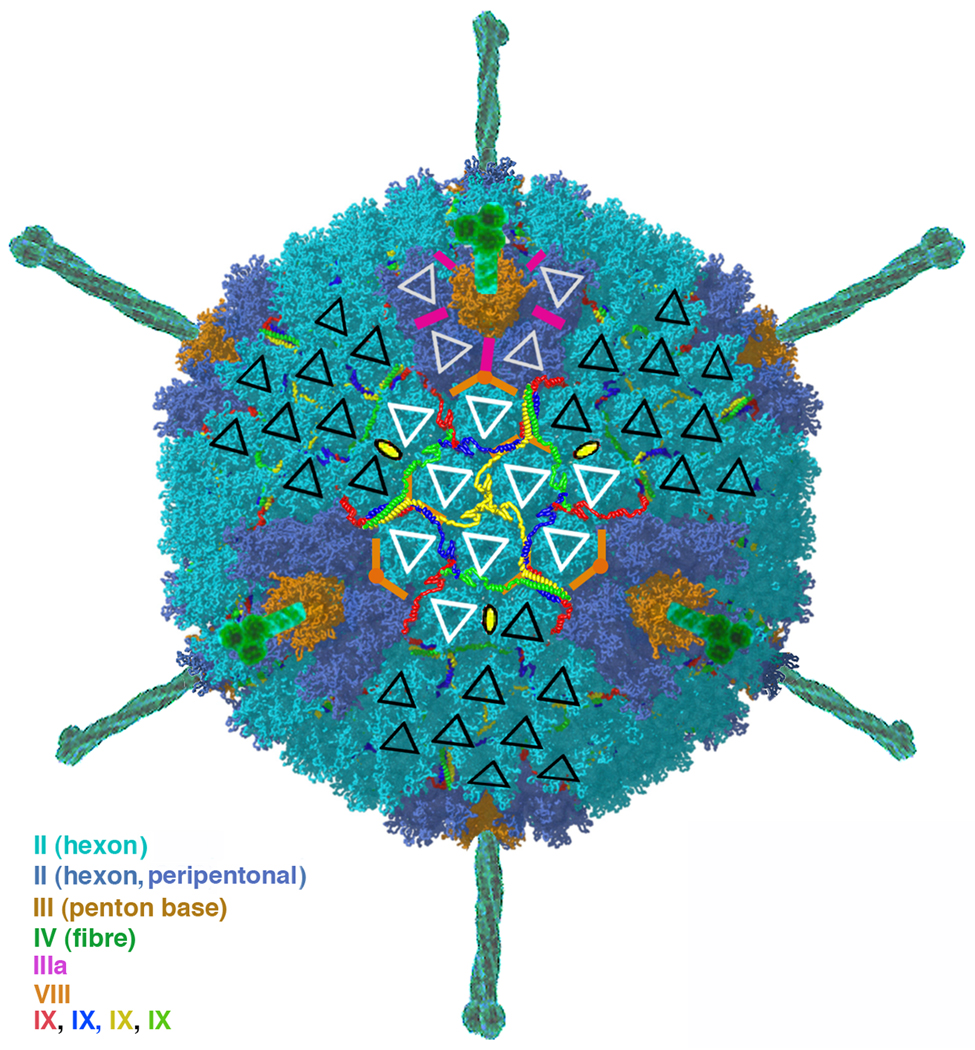
Adenoviruses have a striking icosahedral shape and a complex molecular design. A recently published 3D reconstruction of adenovirus type 5 [8] is a tour de force of cryo-EM, as it allows assignment of density (with amino-acid residue resolution in most places) not only to the polypeptide chains of the principal outer-shell subunits (the so-called "hexon" and "penton"), but also to the set of "minor" proteins that cement the capsid together (Fig. 2). A recurring architectural feature of most non-enveloped virus particles is the interlacing of extended "arms" (N-terminal, C-terminal, or both), which hold the principal subunits together in a way that can adapt to various symmetry environments [68]. The size of the adenovirus genome allows it to encode a set of separate cement proteins that are, in effect, detached arms. These proteins switch the characteristics of the subunit interface at which they are found, leading to accurate assembly of a complex shell.
Fig. 2.
3D reconstruction of adenovirus type 5 [8]. The overall diameter of the particle (not including the spikes) is about 900 Å. Superposed on a surface rendering are triangles highlighting the hexon subunits. Polypeptide chains of one of the arm-like "cement" proteins (protein IX), shown as colored "worms", winds between hexons on the external surface of the particle. The other cement proteins are on the interior of the hexon shell; their locations are symbolized by magenta (protein IIIa) and orange (protein VIII) bars. Modified from [69].
Conclusions
The first image reconstructions of icosahedral viruses, derived from images of negatively stained particles, were published 40 years ago [48]. Reconstructions of icosahedral viruses at near-atomic resolution published during the past two years show that in favorable cases, atomic models can be derived from cryo-EM images [1–9], yielding valuable new information about virus assembly. Advances in EM instrumentation, in computational algorithms, and in available computing power have all contributed. The large size and high symmetry of icosahedral viruses have made them particularly favorable objects for analysis by high-resolution cryo-EM. On-going developments in both instrumentation and computation should extend the range of structures that can be studied in this way to smaller and less symmetrical particles and to image fields containing particles in more than one conformational state.
Acknowledgments
The authors are grateful to David DeRosier and Alexis Rohou for comments and careful reading of the manuscript. Supported by NIH Grant P01 GM-62580. The authors are Investigators in the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1. Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci U S A. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. Reports a 3.8 Å resolution map of the rotavirus double-layered particle, validated in molecular detail by comparison with a crystal structure of the same particle.
- 2. Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. Reports a 4.5 Å resolution map of bacteriophage ε15. Two proteins make up a heterodimeric protomer in the T=7 icosahedral surface lattice. The secondary structure and approximate chain trace of one of the two proteins establishes its homology with the coat protein of bacteriophage HK97.
- 3. Yu X, Jin L, Zhou ZH. 3.88 A structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. Reports a 3.9 Å resolution map of cytoplasmic polyhedrosis virus, with a de novo polypeptide chain tracing of the component proteins.
- 4. Chen JZ, Settembre EC, Aoki ST, Zhang X, Bellamy AR, Dormitzer PR, Harrison SC, Grigorieff N. Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM. Proc Natl Acad Sci U S A. 2009;106:10644–10648. doi: 10.1073/pnas.0904024106. Reports a structure at 4.2 Å resolution of a VP7-recoated, rotavirus double-layered particle (DLP) and shows how trimers of VP7, one of the two viral outer-layer proteins, fit onto trimers of VP6, the DLP structural component arrayed in a T=13 l icosahedral lattice.
- 5. Wolf M, Garcea RL, Grigorieff N, Harrison SC. Subunit interactions in bovine papillomavirus. Proc Natl Acad Sci U S A. 2010;107:6298–6303. doi: 10.1073/pnas.0914604107. A 3.6 Å resolution map of a bovine papillomavirus particle yields a complete atomic model of the 72-pentamer icosahedral shell, allowing refinement of coordinates (R=37.7%, 15-3.6 Å). The structure corrects and extends an earlier model for key interactions in papillomavirus particle assembly.
- 6. Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH. 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010;141:472–482. doi: 10.1016/j.cell.2010.03.041. The structure of the infectious subviral particle of aquareovirus, determined de novo from a 3.6 Å resolution map, confirms and extends earlier analyses of the myristoyl switch that has a key role in penetration.
- 7. Liu X, Zhang Q, Murata K, Baker ML, Sullivan MB, Fu C, Dougherty MT, Schmid MF, Osburne MS, Chisholm SW, et al. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat Struct Mol Biol. 2010;17:830–836. doi: 10.1038/nsmb.1823. Icosahedral and asymmetric reconstructions of bacteriophage P-SSP7 (4.6 Å and 9 Å resolution, respectively) show that the folded structure of the principal head protein corresponds to the HK97 family and indicate how the portal proteins insert at one of the vertices.
- 8. Liu H, Jin L, Koh SB, Atanasov I, Schein S, Wu L, Zhou ZH. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science. 2010;329:1038–1043. doi: 10.1126/science.1187433. Reports the structure of adenovirus type 5 at 3.6 Å resolution and shows how the "cement proteins" tie together the principal structural proteins (hexon and penton base) into the icosahedral capsid. This is the largest and most complex of the structures described in this review.
- 9. Settembre EC, Chen JZ, Dormitzer PR, Grigorieff N, Harrison SC. Atomic model of an infectious rotavirus particle. Embo J. 2010 doi: 10.1038/emboj.2010.322. in press. Reports a 4.3 Å resolution map of the rotavirus triple-layered particle (the infectious virion). Averaging within the icosahedral asymmetric unit ("non-capsid symmetry averaging") enhances the effective resolution. A substantial part of the VP4 protein has been traced de novo and combined with crystallographically derived structures for other parts of the particle to yield a nearly complete atomic model.
- 10.Henderson R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q Rev Biophys. 1995;28:171–193. doi: 10.1017/s003358350000305x. [DOI] [PubMed] [Google Scholar]
- 11.Glaeser RM. Review: electron crystallography: present excitement, a nod to the past, anticipating the future. J Struct Biol. 1999;128:3–14. doi: 10.1006/jsbi.1999.4172. [DOI] [PubMed] [Google Scholar]
- 12.Böttcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 13.Conway JF, Cheng N, Zlotnick A, Wingfield PT, Stahl SJ, Steven AC. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature. 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- 14.Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. Localization of eukaryotespecific ribosomal proteins in a 5.5-A cryo-EM map of the 80S eukaryotic ribosome. Proc Natl Acad Sci U S A. 2010;107:19754–19759. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henderson R. Image contrast in high-resolution electron microscopy of biological macromolecules: TMV in ice. Ultramicroscopy. 1992;46:1–18. doi: 10.1016/0304-3991(92)90003-3. [DOI] [PubMed] [Google Scholar]
- 16.Glaeser RM. Retrospective: radiation damage and its associated "information limitations". J Struct Biol. 2008;163:271–276. doi: 10.1016/j.jsb.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Chen JZ, Sachse C, Xu C, Mielke T, Spahn CM, Grigorieff N. A dose-rate effect in single-particle electron microscopy. J Struct Biol. 2008;161:92–100. doi: 10.1016/j.jsb.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhinow D, Kuhlbrandt W. Electron cryo-microscopy of biological specimens on conductive titanium-silicon metal glass films. Ultramicroscopy. 2008;108:698–705. doi: 10.1016/j.ultramic.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Yoshioka C, Carragher B, Potter CS. Cryomesh: a new substrate for cryo-electron microscopy. Microsc Microanal. 2010;16:43–53. doi: 10.1017/S1431927609991310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ermantraut E, Wohlfart K, Tichelaar W. Perforated support foils with pre-deÞned hole size, shape and arrangement. Ultramicroscopy. 1998;74:75–81. [Google Scholar]
- 21.Quispe J, Damiano J, Mick SE, Nackashi DP, Fellmann D, Ajero TG, Carragher B, Potter CS. An improved holey carbon film for cryo-electron microscopy. Microsc Microanal. 2007;13:365–371. doi: 10.1017/S1431927607070791. [DOI] [PubMed] [Google Scholar]
- 22.Bullough P, Henderson R. Use of spot-scan procedure for recording low-dose micrographs of beam-sensitive specimens. Ultramicroscopy. 1987;21:223–230. [Google Scholar]
- 23.Fujiyoshi Y. The structural study of membrane proteins by electron crystallography. Adv Biophys. 1998;35:25–80. doi: 10.1016/s0065-227x(98)90004-1. [DOI] [PubMed] [Google Scholar]
- 24.Stark H, Zemlin F, Boettcher C. Electron radiation damage to protein crystals of bacteriorhodopsin at different temperatures. Ultramicroscopy. 1996;63:75–79. [Google Scholar]
- 25.Bammes BE, Jakana J, Schmid MF, Chiu W. Radiation damage effects at four specimen temperatures from 4 to 100 K. J Struct Biol. 2010;169:331–341. doi: 10.1016/j.jsb.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright ER, Iancu CV, Tivol WF, Jensen GJ. Observations on the behavior of vitreous ice at approximately 82 and approximately 12 K. J Struct Biol. 2006;153:241–252. doi: 10.1016/j.jsb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 27.McMullan G, Chen S, Henderson R, Faruqi AR. Detective quantum efficiency of electron area detectors in electron microscopy. Ultramicroscopy. 2009;109:1126–1143. doi: 10.1016/j.ultramic.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milazzo AC, Leblanc P, Duttweiler F, Jin L, Bouwer JC, Peltier S, Ellisman M, Bieser F, Matis HS, Wieman H, et al. Active pixel sensor array as a detector for electron microscopy. Ultramicroscopy. 2005;104:152–159. doi: 10.1016/j.ultramic.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 29.Cambie R, Downing KH, Typke D, Glaeser RM, Jin J. Design of a microfabricated, two-electrode phase-contrast element suitable for electron microscopy. Ultramicroscopy. 2007;107:329–339. doi: 10.1016/j.ultramic.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 30.Danev R, Nagayama K. Transmission electron microscopy with Zernike phase plate. Ultramicroscopy. 2001;88:243–252. doi: 10.1016/s0304-3991(01)00088-2. [DOI] [PubMed] [Google Scholar]
- 31.Majorovits E, Barton B, Schultheiss K, Perez-Willard F, Gerthsen D, Schroder RR. Optimizing phase contrast in transmission electron microscopy with an electrostatic (Boersch) phase plate. Ultramicroscopy. 2007;107:213–226. doi: 10.1016/j.ultramic.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 32.Muller H, Jin J, Danev R, Spence J, Padmore H, Glaeser RM. Design of an electron microscope phase plate using a focused continuous-wave laser. New J Phys. 2010;12 doi: 10.1088/1367-2630/12/7/073011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 34.Stewart A, Grigorieff N. Noise bias in the refinement of structures derived from single particles. Ultramicroscopy. 2004;102:67–84. doi: 10.1016/j.ultramic.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 35.Liu H, Cheng L, Zeng S, Cai C, Zhou ZH, Yang Q. Symmetry-adapted spherical harmonics method for high-resolution 3D single-particle reconstructions. J Struct Biol. 2008;161:64–73. doi: 10.1016/j.jsb.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 36.Navaza J. On the three-dimensional reconstruction of icosahedral particles. J Struct Biol. 2003;144:13–23. doi: 10.1016/j.jsb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 37.Scheres SHW, Valle M, Carazo JM. Fast maximum likelihood refinement of electron microscopy images. Bioinformatics. 2005;21 Suppl 2:ii243–ii244. doi: 10.1093/bioinformatics/bti1140. [DOI] [PubMed] [Google Scholar]
- 38.Sigworth FJ. A maximum-likelihood approach to single-particle image refinement. J Struct Biol. 1998;122:328–339. doi: 10.1006/jsbi.1998.4014. [DOI] [PubMed] [Google Scholar]
- 39.Aldroubi A, Trus BL, Unser M, Booy FP, Steven AC. Magnification mismatches between micrographs: corrective procedures and implications for structural analysis. Ultramicroscopy. 1992;46:175–188. doi: 10.1016/0304-3991(92)90013-a. [DOI] [PubMed] [Google Scholar]
- 40.Thon F. Zur Defokussierungsabhängigkeit des Phasenkontrastes bei der elektronenmikroskopischen Abbildung. Zeitschrift für Naturforschung. 1966;21a:476–478. [Google Scholar]
- 41.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 42.Zemlin F, Weiss K, Schiske P, Kunath W, Herrmann K-H. Coma-free alignment of high resolution electron microscopes with the aid of optical diffractograms. Ultramicroscopy. 1978;3:49–60. [Google Scholar]
- 43.Smith DJ, Saxton WO, O'Keefe MA, Wood GJ, Stobbs WM. The importance of beam alignment and crystal tilt in high resolution electron microscopy. Ultramicroscopy. 1983;11 [Google Scholar]
- 44.Henderson R, Baldwin JM, Downing KH, Lepault J, Zemlin F. Structure of purple membrane from halobacterium halobium: recording, measurement and evaluation of electron micrographs at 3.5 Å resolution. Ultramicroscopy. 1986;19:147–178. [Google Scholar]
- 45.Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 46.DeRosier DJ. Correction of high-resolution data for curvature of the Ewald sphere. Ultramicroscopy. 2000;81:83–98. doi: 10.1016/s0304-3991(99)00120-5. [DOI] [PubMed] [Google Scholar]
- 47.Wolf M, DeRosier DJ, Grigorieff N. Ewald sphere correction for single-particle electron microscopy. Ultramicroscopy. 2006;106:376–382. doi: 10.1016/j.ultramic.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 48.Crowther RA, Amos LA, Finch JT, De Rosier DJ, Klug A. Three dimensional reconstructions of spherical viruses by fourier synthesis from electron micrographs. Nature. 1970;226:421–425. doi: 10.1038/226421a0. [DOI] [PubMed] [Google Scholar]
- 49.Crowther RA. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Philos Trans R Soc Lond B Biol Sci. 1971;261:221–230. doi: 10.1098/rstb.1971.0054. [DOI] [PubMed] [Google Scholar]
- 50.Estrozi LF, Navaza J. Ab initio high-resolution single-particle 3D reconstructions: the symmetry adapted functions way. J Struct Biol. 2010;172:253–260. doi: 10.1016/j.jsb.2010.06.023. [DOI] [PubMed] [Google Scholar]
- 51.Liang Y, Ke EY, Zhou ZH. IMIRS: a high-resolution 3D reconstruction package integrated with a relational image database. J Struct Biol. 2002;137:292–304. doi: 10.1016/s1047-8477(02)00014-x. [DOI] [PubMed] [Google Scholar]
- 52.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 53.Penczek PA. Fundamentals of three-dimensional reconstruction from projections. Methods Enzymol. 2010;482:1–33. doi: 10.1016/S0076-6879(10)82001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmid MF, Sherman MB, Matsudaira P, Tsuruta H, Chiu W. Scaling structure factor amplitudes in electron cryomicroscopy using X-Ray solution scattering. J Struct Biol. 1999;128:51–57. doi: 10.1006/jsbi.1999.4173. [DOI] [PubMed] [Google Scholar]
- 55.Harauz G, van Heel M. Exact filters for general geometry three dimensional reconstruction. Optik. 1986;73:146–156. [Google Scholar]
- 56.Grigorieff N. Resolution measurement in structures derived from single particles. Acta Crystallogr D Biol Crystallogr. 2000;56:1270–1277. doi: 10.1107/s0907444900009549. [DOI] [PubMed] [Google Scholar]
- 57.Huang Z, Baldwin PR, Mullapudi S, Penczek PA. Automated determination of parameters describing power spectra of micrograph images in electron microscopy. J Struct Biol. 2003;144:79–94. doi: 10.1016/j.jsb.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 58.Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 59.McClain B, Settembre E, Temple BR, Bellamy AR, Harrison SC. X-ray crystal structure of the rotavirus inner capsid particle at 3.8 A resolution. J Mol Biol. 2010;397:587–599. doi: 10.1016/j.jmb.2010.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trask SD, Dormitzer PR. Assembly of highly infectious rotavirus particles recoated with recombinant outer capsid proteins. J Virol. 2006;80:11293–11304. doi: 10.1128/JVI.01346-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reinisch KM, Nibert ML, Harrison SC. Structure of the reovirus core at 3.6 A resolution. Nature. 2000;404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- 62.Grimes JM, Burroughs JN, Gouet P, Diprose JM, Malby R, Zientara S, Mertens PP, Stuart DI. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- 63.Dryden KA, Wang G, Yeager M, Nibert ML, Coombs KM, Furlong DB, Fields BN, Baker TS. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liemann S, Chandran K, Baker TS, Nibert ML, Harrison SC. Structure of the reovirus membrane-penetration protein, Mu1, in a complex with is protector protein, Sigma3. Cell. 2002;108:283–295. doi: 10.1016/s0092-8674(02)00612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang X, Ji Y, Zhang L, Harrison SC, Marinescu DC, Nibert ML, Baker TS. Features of reovirus outer capsid protein mu1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0 Angstrom resolution. Structure. 2005;13:1545–1557. doi: 10.1016/j.str.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen XS, Garcea RL, Goldberg I, Casini G, Harrison SC. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol Cell. 2000;5:557–567. doi: 10.1016/s1097-2765(00)80449-9. [DOI] [PubMed] [Google Scholar]
- 67.Modis Y, Trus BL, Harrison SC. Atomic model of the papillomavirus capsid. EMBO J. 2002;21:4754–4762. doi: 10.1093/emboj/cdf494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harrison SC. Protein interfaces and intersubunit bonding. The case of tomato bushy stunt virus. Biophys J. 1980;32:139–153. doi: 10.1016/S0006-3495(80)84930-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harrison SC. Looking inside adenovirus. Science. 2010;329:1026–1027. doi: 10.1126/science.1194922. [DOI] [PubMed] [Google Scholar]
- 70.Chen JZ, Grigorieff N. SIGNATURE: a single-particle selection system for molecular electron microscopy. J Struct Biol. 2007;157:168–173. doi: 10.1016/j.jsb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 71.Jones TA. In: Molecular Replacement. Dodson EJ, Gover S, Wolf W, editors. Daresbury Laboratory: Science and Engineering Research Council; 1992. [Google Scholar]
- 72.Kleywegt GJ, Zou JY, Kjeldgaard M, Jones TA. In: International Tables for Crystallography. Rossmann MG, Arnold E, editors. Kluywer Academic; 2001. [Google Scholar]
- 73.Liu X, Jiang W, Jakana J, Chiu W. Averaging tens to hundreds of icosahedral particle images to resolve protein secondary structure elements using a Multi-Path Simulated Annealing optimization algorithm. J Struct Biol. 2007;160:11–27. doi: 10.1016/j.jsb.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]