Abstract
Actinobacillus pleuropneumoniae, the causative agent of porcine pleuropneumonia, is capable of persisting in oxygen-deprived surroundings, namely, tonsils and sequestered necrotic lung tissue. Utilization of alternative terminal electron acceptors in the absence of oxygen is a common strategy in bacteria under anaerobic growth conditions. In an experiment aimed at identification of genes expressed in vivo, the putative catalytic subunit DmsA of anaerobic dimethyl sulfoxide reductase was identified in an A. pleuropneumoniae serotype 7 strain. The 90-kDa protein exhibits 85% identity to the putative DmsA protein of Haemophilus influenzae, and its expression was found to be upregulated under anaerobic conditions. Analysis of the unfinished A. pleuropneumoniae genome sequence revealed putative open reading frames (ORFs) encoding DmsB and DmsC proteins situated downstream of the dmsA ORF. In order to investigate the role of the A. pleuropneumoniae DmsA protein in virulence, an isogenic deletion mutant, A. pleuropneumoniae ΔdmsA, was constructed and examined in an aerosol infection model. A. pleuropneumoniae ΔdmsA was attenuated in acute disease, which suggests that genes involved in oxidative metabolism under anaerobic conditions might contribute significantly to A. pleuropneumoniae virulence.
Actinobacillus pleuropneumoniae, the causative agent of porcine pleuropneumonia, is able to persist in host tissues for weeks or months after infection, surviving in tonsils as well as in sequestered necrotic lung tissue (11, 14, 20). In necrotic tissue, the oxygen supply is scarce, and therefore, strategies for respiration under reduced-oxygen or anaerobic conditions, such as utilization of alternative electron acceptors, are required. To date, the metabolism of A. pleuropneumoniae under these conditions has not been the subject of extensive studies, although a putative anaerobic regulator protein, HlyX, has been identified (19, 31).
Escherichia coli possesses an enzyme complex that allows the use of dimethyl sulfoxide (DMSO) and various other substrates as terminal electron acceptors in cell respiration under anaerobic conditions. The DMSO reductase complex, which has been extensively studied in E. coli (44), consists of three subunits, DmsA, DmsB, and DmsC. The DmsA protein (87 kDa) is the catalytic subunit containing a molybdopterin cofactor; DmsB (23 kDa) is an electron carrier containing four iron-sulfur clusters, and DmsC (30 kDa) serves as a membrane anchor for the other two subunits. The corresponding genes, dmsABC, are organized in an operon in E. coli (8). DMSO reductases have been identified in several other organisms, including Rhodobacter capsulatus (33), Rhodobacter sphaeroides (26), and Haemophilus influenzae (30). H. influenzae possesses a putative dmsABC operon containing an open reading frame (ORF) coding for a 90-kDa putative DmsA protein.
In E. coli, the enzyme complex is situated at the cytoplasmic face of the inner membrane, with the DmsA and DmsB subunits facing the cytoplasm and the DmsC protein embedded in the membrane (10). In R. capsulatus, DMSO reductase consists of a single periplasmic polypeptide containing a molybdenum cofactor with a molecular mass of 82 kDa (33).
Substrates of the DMSO reductase include a wide variety of S and N oxides, including DMSO, trimethylamine N-oxide, and adenosine N-oxide, as well as sodium chlorate and hydroxylamine (43). E. coli DMSO reductase is expressed under anaerobic conditions (9) and upon addition of ferrous sulfate, which serves as an iron source for the DmsB protein, to culture medium (43). Expression of DMSO reductase is regulated by the FNR(fumarate and nitrate reduction) protein (12). The A. pleuropneumoniae HlyX protein is an FNR homologue that is able to complement E. coli fnr mutants, and it also induces a latent E. coli hemolysin (19, 31). DMSO reductase has not been associated with virulence to date.
In the study described here, A. pleuropneumoniae DMSO reductase was identified in a subtracted A. pleuropneumoniae AP76 cDNA library obtained after induction of A. pleuropneumoniae cultures with bronchoalveolar lavage fluid (BALF) from pigs infected with A. pleuropneumoniae (23). Following characterization of the DmsA subunit, an isogenic dmsA deletion mutant was constructed, and the influence of the DmsA protein was investigated in an aerosol infection experiment.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
The strains, plasmids, and primers used in this work are listed in Table 1.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Characteristics | Reference or sourcea |
|---|---|---|
| Strains | ||
| E. coli DH5αF′ | F′ endA1 hsdR17 [rK− mK−supE44 thi-1 recA1 gyrA (NalrrelA1 Δ(lacZYA-argF′)]U169 deoR [φ80dlacΔ(lacZ)M15] | 38 |
| E. coli β2155 | thr B1004 pro thi strA hsdS lacZΔM15 (F′ lacZΔM15 laqIqtraD36 proA+proB+) ΔdapA::erm (Ermr) recA::RP4-2-tet (Tcr)::Mu-km (Kmr) λ pir | 13 |
| E. coli TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara leu)7697 galU galK rpsL (Strr) endA1 nupG | TOPO TA |
| A. pleuropneumoniae AP76 | A. pleuropneumoniae serotype 7b | 2 |
| ATCC 27088 | Serotype 1 reference strain | ATCC |
| ATCC 27089 | Serotype 2 reference strain | ATCC |
| ATCC 27090 | Serotype 3 reference strain | ATCC |
| ATCC 33378 | Serotype 4 reference strain | ATCC |
| ATCC 33377 | Serotype 5A reference strain | ATCC |
| ATCC 33590 | Serotype 6 reference strain | ATCC |
| WF83 | Serotype 7 reference strain | Rosendal |
| 405 | Serotype 8 reference strain | Nielsen |
| CVJ13261 | Serotype 9 reference strain | Nielsen |
| D13039 | Serotype 10 reference strain | Nielsen |
| 56153 | Serotype 11 reference strain | Nielsen |
| 8329 | Serotype 12 reference strain | Nielsen |
| A. pleuropneumoniae ΔdmsA | Unmarked dmsA-negative knockout mutant of A. pleuropneumoniae AP76 | This study |
| Plasmids | ||
| pBMK1 | Transconjugation vector based on pBluescript SK with mobRP4, polycloning site, Tn903-derived kanamycin resistance determinant, and transcriptional fusion of the omlA promoter with the sacB gene | 37 |
| pCR 2.1-TOPO | Topoisomerase I enhanced E. coli cloning vector carrying ampicillin and kanamycin resistance determinants, as well as a lacZ gene for blue-white selection | Amersham Biosciences |
| pGH432/433 lacI | E. coli vector carrying an ampicillin resistance determinant, the lacI gene, and a tac promoter | 17 |
| pGEX1λT | E. coli expression vector carrying an ampicillin resistance determinant, devised to construct GST fusion proteins | Amersham Biosciences |
| pIC20R | E. coli cloning plasmid carrying an ampicillin resistance determinant | 32 |
| pUC19 | E. coli cloning plasmid carrying an ampicillin resistance determinant | Amersham Biosciences |
| pRN5 | pCR 2.1-TOPO plasmid containing 597-bp partial dmsA ORF | This study |
| pDM5-116 | pGH433 carrying the dmsA gene on an 8,700-bp insert in the BglII restriction site | This study |
| pDM5-117 | BglII deletion of pRN5-116 | This study |
| pDM5-118 | Ligation of an EcoRI-NsiI fragment from pDM5-117 into pBluescript cut with EcoRI and PstI | This study |
| pDM5-119 | SwaI-NdeI deletion of pDM5-118 after filling of cohesive ends with the Klenow fragment | This study |
| pDM800 | Ligation of a SalI-XbaI fragment from pDM5-119 into pBMK1 | This study |
| pDMP1 | Ligation of a BamHI-EcoRI fragment from pDM5-118 into pGEX1λT | This study |
| Primers | ||
| M13 forward | 5′ CAG GAA ACA GCT ATG AC 3′ | Amersham Biosciences |
| M13 reverse | 5′ GTA AAA CGA CGG CCA G 3′ | Amersham Biosciences |
| RBgl12 | 5′ GAT CTG CGG TGA 3′ | 29 |
| RBgl24 | 5′ AGC ACT CTC CAG CCT CTC ACC GCA 3′ | 29 |
| JBgl12 | 5′ GAT CTG TTC ATG 3′ | 29 |
| JBgl24 | 5′ ACC GAC GTC GAC TAT CCA TGA ACA 3′ | 29 |
| oGH1 | 5′ TGT GTG GAA TTG TGA GCG 3′ (downstream primer for the multiple cloning site of pGH plasmids) | 17 |
| oGH2 | 5′ GTC CCA CTC CCT GCC TCT 3′ (upstream primer for the multiple cloning site of pGH plasmids) | 17 |
| oRN5-1 | 5′ TAT TTC ACA AGG CTG GGG AC 3′ (forward) (internal primer for RDA fragment RN5) | This study |
| oRN5-2 | 5′ CCG TCC ATA AGA ACA TTG GG 3′ (reverse) (internal primer for fragment RN5) | This study |
| oDMSAdel1 | 5′ TTG AAA TAT CCG ATG AAA CGT 3′ (downstream primer comprising positions 327 to 348 of the dmsA homologue) | This study |
| oDMSAdel2 | 5′ TCA TAT TGG CGA CAT AAG CAT C 3′ (upstream primer comprising positions 1593 to 1614 of the dmsA homologue) | This study |
| oDMSAseq11 fw | 5′ GAG CAT TAC GGT GAC CCA TT 3′ (downstream primer comprising positions 2019 to 2038 of the dmsA homologue) | This study |
| oDMSA2 | 5′ CCA TCT GCT TGT TGA TTC CA 3′ (upstream primer comprising positions 2631 to 2650 of the H. influenzae dmsABC sequence) | This study |
TOPO TA, TOPO TA cloning kit (Invitrogen, Groningen, The Netherlands). ATCC, American Type Culture Collection, Manassas, Va.; Rosendal, S. Rosendal, University of Guelph, Guelph, Ontario, Canada; Nielsen, R. Nielsen, State Veterinary Serum Laboratory, Copenhagen, Denmark.
Strain AP76 was kindly provided by the Western College of Veterinary Medicine, Saskatoon, Canada.
Preparation of antisera.
Serum directed against the DmsA protein was raised in rabbits by using an initial intracutaneous injection and two subcutaneous booster injections of 100 μg of dissolved recombinant glutathione S-transferase (GST)-DmsA fusion protein in Emulsigen-Plus (MVP Inc., Ralston, Nebr.). Convalescent-phase sera directed against A. pleuropneumoniae serotypes 2, 3, 5, 6, and 9 were obtained from naturally infected herds and were kindly provided by Innovative Veterinaerdiagnostik GmbH, Hannover, Germany; serotype specificity was determined by complement fixation with total cell antigen (34).
Media and growth conditions.
E. coli strains were cultured in Luria-Bertani medium supplemented with the appropriate antibiotics (ampicillin, 100 μg/ml; kanamycin, 50 μg/ml); for cultivation of E. coli β2155 (Δdap), diaminopimelic acid (1 mM; Sigma Chemical Company, Deisenhofen, Germany) was added. A. pleuropneumoniae strains were cultured in PPLO medium (Difco GmbH, Augsburg, Germany) supplemented with NAD (10 μg/ml; Merck, Darmstadt, Germany), l-glutamine (100 μg/ml; Serva, Heidelberg, Germany), l-cysteine hydrochloride (260 μg/ml; Sigma), l-cystine dihydrochloride (10 μg/ml; Sigma), dextrose (1 mg/ml), and Tween 80 (0.1%). For anaerobic culture, media were preincubated overnight in anaerobic jars by using the AnaeroGen system (Oxoid GmbH, Wesel, Germany) before inoculation with liquid A. pleuropneumoniae cultures (10% of the total culture volume). Cultures were then incubated with stirring at 37°C for 5 h. Iron restriction was induced by addition of 2,2-dipyridyl (Sigma) to a final concentration of 100 μM, and excess-iron conditions were achieved by addition of ferric citrate to a final concentration of 20 μM or addition of ferrous sulfate to a final concentration of 50 μM; BALF-induced modulation was achieved by addition of 5 ml of BALF to an equal volume of a liquid A. pleuropneumoniae culture. For comparison of different growth conditions under aerobic conditions, one culture was divided into an appropriate number of samples, and then the samples were incubated with shaking for 45 min. For selection of A. pleuropneumoniae transconjugants, kanamycin (25 μg/ml) was added. The medium used for counterselection was prepared as described previously (41).
Manipulation of DNA.
DNA-modifying enzymes were purchased from New England Biolabs (Bad Schwalbach, Germany) and were used according to the manufacturer's instructions. Taq polymerase was purchased from GIBCO BRL Life Technologies (Karlsruhe, Germany). DNA for PCR and Southern blotting, as well as plasmid DNA, were prepared by standard protocols (39). Transformation, gel electrophoresis, PCR, and Southern blotting were performed by using standard procedures (39), and pulsed-field gel electrophoresis (PFGE) of A. pleuropneumoniae DNA was performed as described previously (36).
Construction of recombinant plasmids.
To construct plasmid pDMP1 expressing a GST-DmsA fusion protein, an EcoRI-NsiI fragment from plasmid pDM5-116 containing 1,980 bp of the dmsA ORF was cloned into pBluescript SK, resulting in plasmid pDM5-118, and this was followed by ligation of a BamHI-EcoRI fragment from pDM5-118 into pGEX1λT.
Transconjugation plasmid pDM800 was constructed by deleting a 954-bp SwaI-NdeI fragment from the dmsA ORF in plasmid pDM5-118 by endonuclease restriction, filling in the cohesive ends with the Klenow fragment, and relegation, which resulted in plasmid pDM5-119 containing a dmsA in-frame deletion; this was followed by ligation of a SalI-XbaI fragment from pDM5-119 into pBMK1. The deletion was verified by nucleotide sequencing.
Construction of A. pleuropneumoniae cDNA and genomic libraries.
Construction of a subtracted cDNA library of A. pleuropneumoniae and construction of a genomic library for identification of larger fragments containing cDNA sequences have been described previously (5). Briefly, BALF from A. pleuropneumoniae-infected pigs was used to induce genes expressed in vivo in liquid cultures of A. pleuropneumoniae. Then cDNA obtained from these bacteria (tester cDNA) and cDNA obtained from bacteria grown under standard conditions (driver cDNA) were restricted with DpnII, ligated to an oligonucleotide adapter consisting of a 12-mer (RBgl12 fragment) and a 24-mer (RBgl24), and amplified by PCR (representation product) (24). The representation products were again restricted with DpnII, a new adapter (JBgl12-JBgl24) was ligated to only tester fragments, and one round of subtractive hybridization followed by PCR with oligonucleotide JBgl24 as the primer was performed with an excess of driver cDNA as described previously for cDNA representational difference analysis (24). Difference products were cloned by using a TOPO TA cloning kit (Invitrogen), and clones that hybridized with tester cDNA and genomic A. pleuropneumoniae DNA but not with driver cDNA in a Southern blot analysis were chosen for nucleotide sequencing and sequence analyses with the HUSAR program package (Heidelberg UNIX Sequence Analysis Resources; Deutsches Krebsforschungszentrum, Heidelberg, Germany). Radiolabeled PCR products of these clones were then used to identify larger fragments of corresponding ORFs in an A. pleuropneumoniae genomic library by colony blotting.
Transconjugation and analysis of transconjugants and deletion mutants.
Transconjugation was performed as described previously (37). Kanamycin-resistant colonies were analyzed by colony blotting by using a [32P]dCTP-labeled kanamycin resistance determinant (Kmr). Counterselection to obtain unmarked deletion mutants was performed as previously described (41), and colonies were tested by PCR analysis by using primers oDMSAdel1 and oDMSAdel2 (Table 1). Colonies with the correct PCR profile were confirmed by Southern blot analysis by using the radiolabeled oDMSAdel1-oDMSAdel2 PCR product as a probe, by PFGE, by nucleotide sequence analysis, and by Western blotting.
Fractionation of bacteria, preparation of protein aggregates, electrophoresis, and Western blotting.
Protein aggregates were prepared as previously described (16). Total membrane protein fractions were prepared as described by Hancock and Nikaido (21), and periplasmic protein fractions were prepared as described by Ames et al. (1). A. pleuropneumoniae whole-cell lysates and protein aggregates were analyzed by discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10.8% acrylamide and 0.29% bisacrylamide) and Western blotting by using a Protean II Minigel system (Bio-Rad) as described previously (16).
Virulence studies.
Virulence of the A. pleuropneumoniae dmsA mutant strain was assessed in an aerosol infection model that has been described previously (3, 4, 25) by using 16 A. pleuropneumoniae seronegative outbred pigs that were 8 to 9 weeks old. The pigs were randomly assigned to two groups and cared for in accordance with the principles outlined in the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes (ETS123). The pigs were examined clinically at least once a day or as needed. Body temperatures were recorded in charts for each pig along with clinical symptoms like depression, dyspnea, and coughing. To assess the clinical pulmonary condition as noted during BALF sampling, a clinical score based on the findings of Kipper was employed (27). The scores are described in Table 2. Postmortem analyses, as well as bacteriological and serological examinations, were performed as previously described (4, 18, 22, 28). Lung tissues were immersion fixed in formalin and embedded in paraffin, and 5-μm-thick sections were stained with hematoxylin and eosin. Statistical calculations (Wilcoxon signed-rank test) were performed by using the WinStat Add-In for Microsoft Excel (R. Fitch Software, Staufen, Germany).
TABLE 2.
Clinical scoring scheme for endoscopic evaluation
| Score | Amt of secretion | Quality of secretion | Color of mucosa | Vascular injection | Mucosal swelling |
|---|---|---|---|---|---|
| 0 | None | None | Light pink | Low-medium | None, carina not swollen, symmetrical |
| 1 | Low | Serous | Hyperemic | High | Deformed bronchial lumen or swollen carina or deformed bronchial subdivisions |
| 2 | Medium | Mucous | Deformed bronchial lumen and swollen carina or deformed bronchial subdivisions | ||
| 3 | High | Purulent or bloody |
Nucleotide sequence accession number.
The dmsA nucleotide sequence identified in this study has been deposited in the GenBank database under accession number AY138463.
RESULTS
Identification and characterization of A. pleuropneumoniae DMSO reductase.
By subtracting cDNA prepared from bacteria grown under standard culture conditions from cDNA of bacteria grown with addition of BALF and subsequent cloning of cDNA fragments into the TOPO 2.1 vector, a subtracted A. pleuropneumoniae cDNA library was constructed. One 597-bp fragment, designated RN5, contained a continuous ORF whose product exhibited similarity to the catalytic subunit of DMSO reductase, DmsA, of H. influenzae and E. coli (8, 30).
Using the radiolabeled PCR product of primers oRN5-1 and 5-2 as a probe, we screened a genomic A. pleuropneumoniae library in order to identify the entire ORF encoding DmsA. One clone, designated pDM5-116, was found to contain an incomplete ORF lacking the 3′ end of the dmsA gene. Using sequence data for the H. influenzae dmsA gene (GenBank accession no. P45004), we constructed oligonucleotide primer oDMSA2 and used it to amplify the 3′ end of the putative dmsA ORF from A. pleuropneumoniae AP76 genomic DNA. Sequence analysis of the complete nucleotide sequence revealed a 2,418-bp ORF encoding a protein that was 805 amino acids long and had a calculated molecular mass of 90 kDa. The predicted A. pleuropneumoniae DmsA protein is 85% identical to the putative DmsA protein of H. influenzae and 74% identical to the E. coli DmsA protein.
Five base pairs upstream of the start codon of dmsA, a Shine-Dalgarno consensus sequence (GGAG) was identified, and 106 bases upstream of the dmsA start codon, a single putative binding site for the FNR homologue of A. pleuropneumoniae HlyX, with the sequence T-T-G-A-T-X-X-X-X-A-T-C-A-G, was located. This sequence is a close match with the proposed consensus sequence for FNR, T-T-G-A-T-X-X-X-X-A-T-C-A-A(19).
On the A. pleuropneumoniae genomic map (36), the A. pleuropneumoniae dmsA gene is located on fragments APA5, ASC1, and NOT2. Thus, dmsA is colocated on a 200-kb fragment with the outer membrane lipoprotein gene omlA of A. pleuropneumoniae. In a BLAST search of the incomplete genomic sequence which has recently become available for A. pleuropneumoniae serotype 7 (GenBank accession no. NC_004427) with the A. pleuropneumoniae AP76 dmsA ORF, putative dmsB and dmsC reading frames were identified downstream of the dmsA ORF, and these reading frames encoded a putative 23-kDa DmsB protein and a putative 30-kDa DmsC protein.
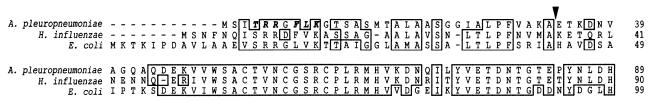
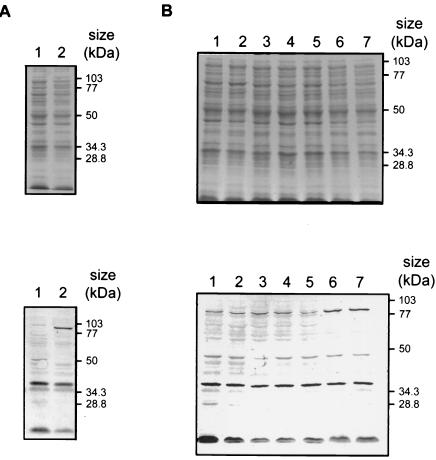
At the amino terminus of the A. pleuropneumoniae DmsA protein sequence, a twin arginine leader sequence was identified (Fig. 1). This leader sequence conforms to the conserved consensus sequence (S/T)-R-R-X-F-L-K (6) that is typical of proteins that are exported or targeted to the membrane via the twin arginine translocation pathway (40). Using the SignalP V1.1 server (35), we predicted that a signal peptidase cleavage site is situated between amino acid 33 (alanine) and amino acid 34 (glutamic acid) (Fig. 1). DmsA protein expression in vitro was examined by using a polyclonal rabbit serum raised against a DmsA-GST fusion protein expressed from plasmid pDMP1. Low-level DmsA expression was detected in A. pleuropneumoniae AP76 under standard culture conditions. Expression was clearly enhanced under anaerobic conditions (Fig. 2A), and a slight enhancement of expression was also seen upon addition of ferrous sulfate to the culture medium under aerobic conditions (Fig. 2B), while iron depletion and addition of ferric citrate had no influence on aerobic DmsA expression. Enhancement of DmsA expression upon exposure to BALF was not observed (Fig. 2B). The presence of the dmsA gene in A. pleuropneumoniae serotype reference strains 1 to 12 was examined by PCR by using primers oDMSAdel1 and oDMSAdel2. A product of the same size was amplified from A. pleuropneumoniae AP76 and from all 12 reference strains (data not shown). Furthermore, using sera from convalescent pigs infected with A. pleuropneumoniae serotypes 2, 3, 5, 6, 7, and 9, we detected a DmsA-GST fusion protein in Western blots with increased signal strength compared to the signal strength of a protein in preimmune sera (data not shown). This finding implies that the A. pleuropneumoniae AP76 DmsA protein is expressed in vivo.
FIG. 1.
PrettyPlot alignment of amino-terminal ends of DmsA proteins from A. pleuropneumoniae, H. influenzae, and E. coli. The TAT pathway consensus sequence T-R-R-X-F-L-K is indicated by boldface italic type, and the putative signal peptidase cleavage site for A. pleuropneumoniae DmsA is indicated by an arrowhead. The PrettyPlot presentation of the amino-terminal end of the mature DmsA protein shows the high degree of homology typical for the entire protein.
FIG. 2.
Expression of the A. pleuropneumoniae DmsA protein as assessed by Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (top panels) and corresponding Western blotting (bottom panels). (A) Expression upon growth under standard conditions (lane 1) and anaerobic conditions (lane 2). (B) Effects of different iron sources and BALF upon growth under aerobic conditions. Lane 1, standard conditions; lane 2, iron restriction; lane 3, addition of BALF; lanes 4 and 5, addition of ferric citrate at final concentrations of 20 and 50 μM, respectively; lanes 6 and 7, addition of ferrous citrate at final concentrations of 20 and 50 μM, respectively.
In order to determine the localization of DmsA in the bacterium (i.e., periplasmic [as in R. capsulatus] or membrane bound [as in E. coli]), whole-cell lysate, total membranes, periplasm, and cytoplasm were prepared and assessed by Western blot analyses. DmsA appeared to be associated with the membrane of A. pleuropneumoniae, as the total-membrane fraction contained most of the DmsA protein, whereas only traces were detected in the periplasm (data not shown).
Construction of isogenic mutant.
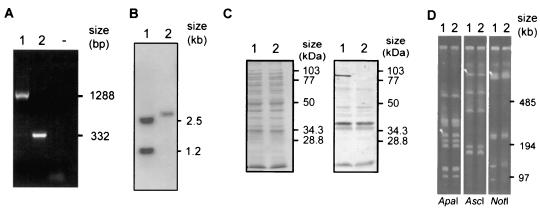
A deletion in the dmsA gene was introduced into A. pleuropneumoniae AP76 by transconjugation of plasmid pDM800 and subsequent sucrose counterselection as described previously (41). An isogenic deletion mutant designated A. pleuropneumoniae ΔdmsA was obtained and verified by PCR (Fig. 3A), Southern blot analysis (Fig. 3B), and Western blotting (Fig. 3C). The absence of gross genomic rearrangements was shown by PFGE (Fig. 3D). In vitro, no consistent differences between the growth of the deletion mutant and the growth of the A. pleuropneumoniae AP76 parent strain were observed under anaerobic conditions.
FIG. 3.
Characterization of the A. pleuropneumoniae dmsA deletion mutant with A. pleuropneumoniae AP76 (lanes 1) and A. pleuropneumoniae ΔdmsA (lanes 2). (A) PCR performed with primers oDMSAdel1 and oDMSAdel2. Lane − contained no DNA template. (B) Southern blot analysis of genomic DNA digested with EcoRI and BspMI, performed with the oDMSAdel1-oDMSAdel2 PCR product as the radiolabeled probe. A BspMI restriction site is located in the deleted fragment. (C) Coomassie blue-stained gel (left) and Western blot (right) developed with polyclonal rabbit antiserum. (D) PFGE analysis, showing that no gross genomic rearrangements occurred. The arrowheads in lanes 1 (parent strain) indicate the fragments that hybridized with a dmsA-derived probe.
Virulence studies.
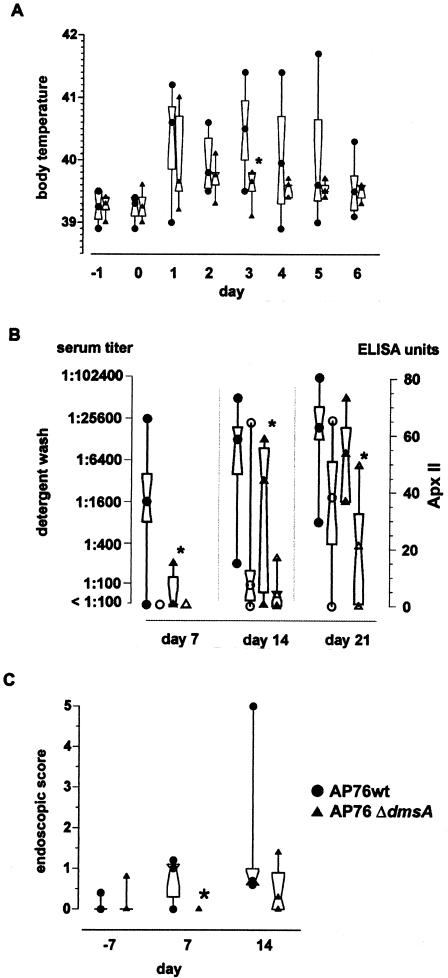
A. pleuropneumoniae ΔdmsA was used in an aerosol infection model and compared to A. pleuropneumoniae wild-type strain AP76. The challenge doses were 1.28 × 105 bacteria (aerosolized for four pigs) for the A. pleuropneumoniae wild-type strain AP76-treated group and 1.7 × 105 bacteria for the A. pleuropneumoniae ΔdmsA-treated group. Body temperatures and clinical symptoms like depression, inappetence, and coughing were monitored daily. The body temperatures exceeded 40°C in six of eight animals in the A. pleuropneumoniae wild-type strain AP76-treated group and in two of eight animals in the A. pleuropneumoniae ΔdmsA-treated group. Overall, the body temperatures in the A. pleuropneumoniae ΔdmsA-treated group were lower than those in the A. pleuropneumoniae wild-type strain AP76-treated group on days 2 to 4 postinfection, and the difference was significant on day 3 postinfection (Fig. 4A). BALF and serum samples were obtained 1 week before and 1 and 3 weeks after challenge. The enzyme-linked immunosorbent assay (ELISA) titers were significantly lower (P < 0.05) in the group infected with A. pleuropneumoniae ΔdmsA (Fig. 4B). The clinical findings of an endoscopy analysis were documented by using the guidelines shown in Table 2. The pigs infected with the deletion mutant A. pleuropneumoniae ΔdmsA had fewer symptoms of inflammation than the pigs infected with the parent strain A. pleuropneumoniae wild-type strain AP76. The difference in endoscopic scores between the A. pleuropneumoniae wild-type strain AP76-treated and A. pleuropneumoniae ΔdmsA-treated groups was statistically significant (P < 0.05) on day 7 postinfection (Fig. 4C). The challenge strains were reisolated on days 7 and 21 postinfection from BALF from several pigs in both challenge groups (Table 3). Upon necropsy, lung lesions were diagnosed as multifocal coagulative and liquefaction necroses up to 4 cm in diameter. Lung tissue samples were examined histologically; samples obtained from the two groups were affected by lesions that were indistinguishable in terms of quality or severity. Lesions were sequestered by a fibrous demarcation estimated to be between 300 and 500 μm thick. The sequestered tissue consisted of remnants of parenchymal and inflammatory cells, mostly neutrophils and macrophages. The fibrous tissue walls contained mostly active fibroblasts, collagen fibers, and lymphocytes, as well as plasma cells. No significant difference was detected between the two (challenge) groups.
FIG. 4.
Virulence of A. pleuropneumoniae ΔdmsA in an aerosol infection model. •, A. pleuropneumoniae wild-type strain AP76 (AP76wt); ▴, A. pleuropneumoniae ΔdmsA (AP76 ΔdmsA). The central symbol in each hourglass shape indicates the geometric mean, the hinges indicate the values in the middle half of the data, and the top and bottom symbols indicate the maximum and minimum values. The asterisks indicate statistical significance (P < 0.05) as determined by the Wilcoxon signed-rank test. (A) Body temperatures of pigs over the course of 8 days. Day 0 was the day of infection. (B) Humoral immune responses of pigs challenged with the A. pleuropneumoniae parent strain and the isogenic mutant 7 days before and 7 and 21 days after challenge. The antibody response was assessed with two ELISAs by using the recombinant ApxIIA protein (Apx ELISA) (open symbols) or a detergent extract (extract ELISA) (solid symbols) as the solid-phase antigen. The immune response was expressed in ELISA units (based on an external standard) for the standardized Apx ELISA, and serum activities of more than 30 ELISA units were considered positive (28); for the extract ELISA, the immune response was expressed as the serum titer compared to the data for an internal control (18). (C) Endoscopic scores 7 days before and 7 and 21 days after challenge.
TABLE 3.
Virulence of A. pleuropneumoniae parent and isogenic mutant strains following aerosol challenge
| A. pleuropneu-moniae challenge strain | Challenge dose (CFU, aero-solized) | Serological
response
|
No. of animals with lung lesions/Total no. | Arithmetic mean of lung lesion score | No. of animals with reisolation of
A. pleuropneumoniae
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Detergent wash (titer) | ApxIIA (u) | From
BALF
|
At postmortem analysis
|
|||||||
| Day 7 after challenge | Day 21 after challenge | Tonsils | Tracheobronchial lymph nodes | Lungs | ||||||
| AP76 parent | 1.28 × 105 | 30,500 | 36.2 ± 20.4 | 7/8 | 7.09 ± 5.32a | 3 | 4 | 2 | 1 | 7 |
| AP76 | 1.7 × 105 | 13,800 | 19.6 ± 17.5 | 7/8 | 6.63 ± 6.9b | 3 | 3 | 1 | 1 | 6 |
One animal which was euthanized on day 9 postinfection due to severe respiratory symptoms was excluded from the analysis.
One animal showing severe E. coli superinfection was excluded from the analysis.
A. pleuropneumoniae was consistently reisolated from pneumonic lesions in pure culture by using surface smears showing sparse to confluent growth; reisolation from tonsils and tracheobronchial lymph nodes resulted in sparse growth only in 3 and 2 of the 16 pigs, respectively. No consistent difference was observed between the data for the two groups or, when BALF was used, between the data for days 7 and 21 within each group (Table 3).
DISCUSSION
Based on the hypothesis that some virulence-associated genes of A. pleuropneumoniae might be expressed only in vivo, it was the goal of this study to identify such genes and to investigate their role in infection. Starting with BALF-induced (23) RNA, a subtracted cDNA library was constructed (5). From this library, the A. pleuropneumoniae DMSO reductase catalytic subunit gene, dmsA, was identified, and analyses of the unfinished A. pleuropneumoniae genome sequence revealed putative dmsB and dmsC ORFs downstream of the dmsA gene. Expression of the DmsA protein was characterized, and its role in A. pleuropneumoniae virulence was assessed in an aerosol infection experiment, which showed that deletion of the dmsA gene results in attenuation of A. pleuropneumoniae in the acute phase of infection.
The genomic organization of the DMSO reductase operon, dmsABC, in A. pleuropneumoniae is consistent with findings obtained for E. coli (10) and H. influenzae (30). Upstream of the A. pleuropneumoniae dmsA gene, a consensus sequence for a single binding site for HlyX, the A. pleuropneumoniae homologue of the anaerobic transcriptional regulator FNR of E. coli, was identified, and enhanced expression under anaerobic conditions (Fig. 2A, lane 2) and enhanced expression under aerobic conditions upon addition of ferrous sulfate were observed for A. pleuropneumoniae DMSO reductase (Fig. 2B, lanes 6 and 7), which corresponds to findings obtained for E. coli (10). The failure to demonstrate induction of DmsA expression by addition of BALF under aerobic conditions (Fig. 2B, lane 3) may have been due to the inability of standard Western blot examination to detect slight differences. The finding that convalescent-phase sera from pigs infected with A. pleuropneumoniae serotypes 2, 3, 5, 6, 7, and 9 showed an increased level of detection compared to the level of detection obtained with preimmune sera implied that the DmsA protein might be expressed in vivo, causing a booster effect for the humoral immune response already present against homologous DmsA proteins from other bacteria, like E. coli.
Additionally, the consensus sequence for the twin arginine translocation (TAT) pathway (6) was identified in the signal peptide sequence of A. pleuropneumoniae DmsA (Fig. 1). This pathway, also termed the membrane targeting and translocation pathway (40), is sec independent and typically functions to transport folded, cofactor-containing proteins through the cytoplasmic membrane into the periplasm or, as is the case for E. coli DmsA, directs these proteins to the inner face of the cytoplasmic membrane (6). Proteins transported via this pathway contain a typical twin arginine motif in the signal sequence, the consensus sequence is (S/T)-R-R-X-F-L-K, and the twin arginine residues are invariable (6). TAT leader sequences have been identified in genes of E. coli, H. influenzae, and other species (7). To our knowledge, this is the first report of a twin arginine motif in A. pleuropneumoniae; a membrane targeting and translocation mechanism has not been characterized in this organism.
DMSO reductase allows bacteria to utilize DMSO and other compounds as alternative terminal electron acceptors under oxygen-deprived conditions. Since tissue destruction in A. pleuropneumoniae disease can be quite extensive due to the action of the Apx toxins and gas exchange over the alveoles is the nearly exclusive source of oxygen for lung tissue, the hypothesis that A. pleuropneumoniae encounters oxygen-deficient conditions in these parts of the lung appears to be plausible. DMSO reductase function has been well studied in E. coli (44); however, it has not previously been associated with virulence.
In Mycobacterium bovis BCG, a similar mechanism in which nitrate is used as a terminal electron acceptor has been shown to be involved in virulence. Anaerobic nitrate reductase is an analogue of DMSO reductase in that nitrate serves as the terminal electron acceptor under anaerobic conditions. An M. bovis BCG mutant lacking nitrate reductase was attenuated in immunodeficient mice (15, 42). Due to the similarity of the oxygen-restricted conditions inside granulomas observed in tuberculosis and inside the sequestered pulmonary necroses observed in chronic porcine pleuropneumonia, it was hypothesized that in A. pleuropneumoniae infection, DMSO reductase might be advantageous for survival of A. pleuropneumoniae in sequestered necrotic lung tissue, as it should facilitate respiration rather than fermentation under anaerobic conditions. Therefore, impairment of DMSO reductase function might result in reduced virulence and/or reduced persistence of A. pleuropneumoniae in lung tissue.
In vivo characterization of DmsA involved the construction of an isogenic deletion mutant, A. pleuropneumoniae ΔdmsA, and use of this mutant in an animal infection experiment. The mutant was shown to be attenuated in acute disease, as assessed by body temperatures and endoscopy scores (Fig. 4A and C). In addition, animals infected with the deletion mutant had lower antibody titers than animals infected with the wild-type strain (Fig. 4B). However, A. pleuropneumoniae ΔdmsA and the parent strain did not differ in the ability to persist in host tissues, and no difference was found between lung lesions in the two groups macroscopically or histologically.
A characteristic of A. pleuropneumoniae disease is high fever following rapid multiplication of bacteria in affected tissues. The milder course of disease observed in this study, which was supported by body temperature curves and endoscopic findings, may have been due to slower growth of A. pleuropneumoniae caused by absence of DMSO reductase in early stages of infection. The finding that there were no consistent differences between the reisolation rates and histological examination data for altered lung tissue in the A. pleuropneumoniae AP76 wild-type strain AP76-treated and A. pleuropneumoniae ΔdmsA-treated groups implies that the absence of the DmsA function is compensated for in part by other terminal electron acceptors, such as nitrate or fumarate, or by other mechanisms during the course of infection. These findings imply that introduction of a dmsA deletion into A. pleuropneumoniae may be useful for construction of a live vaccine strain, as such a deletion attenuates the clinical symptoms of acute infection, provided that further attenuation and reduction of persistence are achieved by other deletions.
Acknowledgments
This work was supported by grant GE522/3-2 and Sonderforschungsbereich 587 (project A4) from the Deutsche Forschungsgemeinschaft (DFG), Bonn, Germany. I.J. is a fellow of graduate college 745 (project A1) of the DFG.
Editor: B. B. Finlay
REFERENCES
- 1.Ames, G. F., C. Prody, and S. Kustu. 1984. Simple, rapid, and quantitative release of periplasmic proteins by chloroform. J. Bacteriol. 160:1181-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, C., A. A. Potter, and G. F. Gerlach. 1991. Isolation and molecular characterization of spontaneously occurring cytolysin-negative mutants of Actinobacillus pleuropneumoniae serotype 7. Infect. Immun. 59:4110-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baltes, N., I. Hennig-Pauka, and G. F. Gerlach. 2002. Both transferrin binding proteins are virulence factors in Actinobacillus pleuropneumoniae serotype 7 infection. FEMS Microbiol. Lett. 209:283-287. [DOI] [PubMed] [Google Scholar]
- 4.Baltes, N., W. Tonpitak, G. F. Gerlach, I. Hennig-Pauka, A. Hoffmann-Moujahid, M. Ganter, and H. J. Rothkotter. 2001. Actinobacillus pleuropneumoniae iron transport and urease activity: effects on bacterial virulence and host immune response. Infect. Immun. 69:472-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baltes, N., W. Tonpitak, I. Hennig-Pauka, A. D. Gruber, and G. F. Gerlach. 2003. Actinobacillus pleuropneumoniae serotype 7 siderophore receptor FhuA is not required for virulence. FEMS Microbiol. Lett. 220:41-48. [DOI] [PubMed] [Google Scholar]
- 6.Berks, B. C. 1996. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 22:393-404. [DOI] [PubMed] [Google Scholar]
- 7.Berks, B. C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 35:260-274. [DOI] [PubMed] [Google Scholar]
- 8.Bilous, P. T., S. T. Cole, W. F. Anderson, and J. H. Weiner. 1988. Nucleotide sequence of the dmsABC operon encoding the anaerobic dimethylsulphoxide reductase of Escherichia coli. Mol. Microbiol. 2:785-795. [DOI] [PubMed] [Google Scholar]
- 9.Bilous, P. T., and J. H. Weiner. 1985. Dimethyl sulfoxide reductase activity by anaerobically grown Escherichia coli HB101. J. Bacteriol. 162:1151-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bilous, P. T., and J. H. Weiner. 1988. Molecular cloning and expression of the Escherichia coli dimethyl sulfoxide reductase operon. J. Bacteriol. 170:1511-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiers, K., I. van Overbeke, P. De Laender, R. Ducatelle, S. Carel, and F. Haesebrouck. 1998. Effects of endobronchial challenge with Actinobacillus pleuropneumoniae serotype 9 of pigs vaccinated with inactivated vaccines containing the Apx toxins. Vet. Q. 20:65-69. [DOI] [PubMed] [Google Scholar]
- 12.Cotter, P. A., and R. P. Gunsalus. 1989. Oxygen, nitrate, and molybdenum regulation of dmsABC gene expression in Escherichia coli. J. Bacteriol. 171:3817-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dehio, C., and M. Meyer. 1997. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J. Bacteriol. 179:538-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenwick, B., and S. Henry. 1994. Porcine pleuropneumonia. J. Am. Vet. Med. Assoc. 204:1334-1340. [PubMed] [Google Scholar]
- 15.Fritz, C., S. Maass, A. Kreft, and F. C. Bange. 2002. Dependence of Mycobacterium bovis BCG on anaerobic nitrate reductase for persistence is tissue specific. Infect. Immun. 70:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerlach, G. F., C. Anderson, A. A. Potter, S. Klashinsky, and P. J. Willson. 1992. Cloning and expression of a transferrin-binding protein from Actinobacillus pleuropneumoniae. Infect. Immun. 60:892-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerlach, G. F., S. Klashinsky, C. Anderson, A. A. Potter, and P. J. Willson. 1992. Characterization of two genes encoding distinct transferrin-binding proteins in different Actinobacillus pleuropneumoniae isolates. Infect. Immun. 60:3253-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goethe, R., O. F. Gonzales, T. Lindner, and G. F. Gerlach. 2000. A novel strategy for protective Actinobacillus pleuropneumoniae subunit vaccines: detergent extraction of cultures induced by iron restriction. Vaccine 19:966-975. [DOI] [PubMed] [Google Scholar]
- 19.Green, J., and M. L. Baldwin. 1997. HlyX, the FNR homologue of Actinobacillus pleuropneumoniae, is a [4Fe-4S]-containing oxygen-responsive transcription regulator that anaerobically activates FNR-dependent class I promoters via an enhanced AR1 contact. Mol. Microbiol. 24:593-605. [DOI] [PubMed] [Google Scholar]
- 20.Haesebrouck, F., K. Chiers, I. van Overbeke, and R. Ducatelle. 1997. Actinobacillus pleuropneumoniae infections in pigs: the role of virulence factors in pathogenesis and protection. Vet. Microbiol. 58:239-249. [DOI] [PubMed] [Google Scholar]
- 21.Hancock, R. E., and H. Nikaido. 1978. Outer membranes of gram-negative bacteria. XIX. Isolation from Pseudomonas aeruginosa PAO1 and use in reconstitution and definition of the permeability barrier. J. Bacteriol. 136:381-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hannan, P. C., B. S. Bhogal, and J. P. Fish. 1982. Tylosin tartrate and tiamutilin effects on experimental piglet pneumonia induced with pneumonic pig lung homogenate containing mycoplasmas, bacteria and viruses. Res. Vet. Sci. 33:76-88. [PubMed] [Google Scholar]
- 23.Hennig, I., B. Teutenberg-Riedel, and G. F. Gerlach. 1999. Downregulation of a protective Actinobacillus pleuropneumoniae antigen during the course of infection. Microb. Pathog. 26:53-63. [DOI] [PubMed] [Google Scholar]
- 24.Hubank, M., and D. G. Schatz. 1994. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res. 22:5640-5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobsen, M. J., J. P. Nielsen, and R. Nielsen. 1996. Comparison of virulence of different Actinobacillus pleuropneumoniae serotypes and biotypes using an aerosol infection model. Vet. Microbiol. 49:159-168. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, J. L., N. R. Bastian, and K. V. Rajagopalan. 1990. Molybdopterin guanine dinucleotide: a modified form of molybdopterin identified in the molybdenum cofactor of dimethyl sulfoxide reductase from Rhodobacter sphaeroides forma specialis denitrificans. Proc. Natl. Acad. Sci. USA 87:3190-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kipper, S. 1990. Bronchoscopy in pigs, and microscopical and cytological examination of bronchoalveolar fluids. Ph.D dissertation. Veterinary School of Hannover, Hannover, Germany. (In German).
- 28.Leiner, G., B. Franz, K. Strutzberg, and G. F. Gerlach. 1999. A novel enzyme-linked immunosorbent assay using the recombinant Actinobacillus pleuropneumoniae ApxII antigen for diagnosis of pleuropneumonia in pig herds. Clin. Diagn. Lab. Immunol. 6:630-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lisitsyn, N., N. Lisitsyn, and M. Wigler. 1993. Cloning the differences between two complex genomes. Science 259:946-951. [DOI] [PubMed] [Google Scholar]
- 30.Loosmore, S. M., J. M. Shortreed, D. C. Coleman, D. M. England, and M. H. Klein. 1996. Sequences of the genes encoding the A, B and C subunits of the Haemophilus influenzae dimethylsulfoxide reductase complex. Gene 169:137-138. [DOI] [PubMed] [Google Scholar]
- 31.MacInnes, J. I., J. E. Kim, C. J. Lian, and G. A. Soltes. 1990. Actinobacillus pleuropneumoniae hlyX gene homology with the fnr gene of Escherichia coli. J. Bacteriol. 172:4587-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marsh, J. L., M. Erfle, and E. J. Wykes. 1984. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene 32:481-485. [DOI] [PubMed] [Google Scholar]
- 33.McEwan, A. G., S. J. Ferguson, and J. B. Jackson. 1991. Purification and properties of dimethyl sulphoxide reductase from Rhodobacter capsulatus. A periplasmic molybdoenzyme. Biochem. J. 274:305-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicolet, J., P. A. de Meuron, and P. Bachmann. 1974. Sur l'hémophilose du porc IV. L' épreuve de déviation du complément, un teste d' épistage des infections à Haemophilus parahaemolyticus. Schweiz. Arch. Tierheilkd. 113:119-200. [PubMed] [Google Scholar]
- 35.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 36.Oswald, W., D. V. Konine, J. Rohde, and G. F. Gerlach. 1999. First chromosomal restriction map of Actinobacillus pleuropneumoniae and localization of putative virulence-associated genes. J. Bacteriol. 181:4161-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oswald, W., W. Tonpitak, G. Ohrt, and G. Gerlach. 1999. A single-step transconjugation system for the introduction of unmarked deletions into Actinobacillus pleuropneumoniae serotype 7 using a sucrose sensitivity marker. FEMS Microbiol. Lett. 179:153-160. [DOI] [PubMed] [Google Scholar]
- 38.Raleigh, F. A., K. Lech, and R. Brent. 1989. Current protocols in molecular biology, p. 1.4.1-1.4.14. In F. M. Ausubel et al. (ed.), Current protocols in molecular biology. Publishing Associates and Wiley Interscience, New York, N.Y.
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 40.Sargent, F., E. G. Bogsch, N. R. Stanley, M. Wexler, C. Robinson, B. C. Berks, and T. Palmer. 1998. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 17:3640-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tonpitak, W., N. Baltes, I. Hennig-Pauka, and G. F. Gerlach. 2002. Construction of an Actinobacillus pleuropneumoniae serotype 2 prototype live negative-marker vaccine. Infect. Immun. 70:7120-7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber, I., C. Fritz, S. Ruttkowski, A. Kreft, and F. C. Bange. 2000. Anaerobic nitrate reductase (narGHJI) activity of Mycobacterium bovis BCG in vitro and its contribution to virulence in immunodeficient mice. Mol. Microbiol. 35:1017-1025. [DOI] [PubMed] [Google Scholar]
- 43.Weiner, J. H., D. P. MacIsaac, R. E. Bishop, and P. T. Bilous. 1988. Purification and properties of Escherichia coli dimethyl sulfoxide reductase, an iron-sulfur molybdoenzyme with broad substrate specificity. J. Bacteriol. 170:1505-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiner, J. H., R. A. Rothery, D. Sambasivarao, and C. A. Trieber. 1992. Molecular analysis of dimethylsulfoxide reductase: a complex iron-sulfur molybdoenzyme of Escherichia coli. Biochim. Biophys. Acta 1102:1-18. [DOI] [PubMed] [Google Scholar]