Abstract
Human tuberculosis (TB) is caused by the bacillus Mycobacterium tuberculosis, a subspecies of the M. tuberculosis complex (MTC) of mycobacteria. Postgenomic dissection of the M. tuberculosis proteome is ongoing and critical to furthering our understanding of factors mediating M. tuberculosis pathobiology. Towards this end, a 32-kDa putative glyoxalase in the culture filtrate (CF) of growing M. tuberculosis (originally annotated as Rv0577 and hereafter designated CFP32) was identified, cloned, and characterized. The cfp32 gene is MTC restricted, and the gene product is expressed ex vivo as determined by the respective Southern and Western blot testing of an assortment of mycobacteria. Moreover, the cfp32 gene sequence is conserved within the MTC, as no polymorphisms were found in the tested cfp32 PCR products upon sequence analysis. Western blotting of M. tuberculosis subcellular fractions localized CFP32 predominantly to the CF and cytosolic compartments. Data to support the in vivo expression of CFP32 were provided by the serum recognition of recombinant CFP32 in 32% of TB patients by enzyme-linked immunosorbent assay (ELISA) as well as the direct detection of CFP32 by ELISA in the induced sputum samples from 56% of pulmonary TB patients. Of greatest interest was the observation that, per sample, sputum CFP32 levels (a potential indicator of increasing bacterial burden) correlated with levels of expression in sputum of interleukin-10 (an immunosuppressive cytokine and a putative contributing factor to disease progression) but not levels of gamma interferon (a key cytokine in the protective immune response in TB), as measured by ELISA. Combined, these data suggest that CFP32 serves a necessary biological function(s) in tubercle bacilli and may contribute to the M. tuberculosis pathogenic mechanism. Overall, CFP32 is an attractive target for drug and vaccine design as well as new diagnostic strategies.
The Mycobacterium tuberculosis complex (MTC) is a group of highly related pathogenic mycobacteria that include M. tuberculosis, Mycobacterium africanum (subtypes I and II), Mycobacterium bovis (along with the attenuated M. bovis bacillus Calmette-Guérin [BCG] vaccine strain), Mycobacterium bovis subsp. caprae, and Mycobacterium microti (13). The MTC taxon is extraordinary in that its members exhibit a restricted number of fixed single-nucleotide polymorphisms between subspecies but differ from one another by the presence or absence of large chromosomal deletion loci, severity of disease, and mammalian host spectra (13, 43, 66). Of the MTC members, M. tuberculosis is the predominant etiologic agent of human tuberculosis (TB).
M. tuberculosis is arguably the most successful of human pathogens in having achieved a worldwide penetrance of epidemic proportions. Estimates based on skin testing indicate that approximately one-third of the human population have been M. tuberculosis infected (21, 74). In most individuals the infection progresses to a latent phase in which there are no overt signs of disease. However, up to 10% of these persons are expected to develop life-threatening disease over the course of their lifetimes if untreated (21). In fact, TB claims up to 3 million lives each year, which is more than any other single bacterial infectious agent (74). Coupled with the emergence of drug-resistant stains and a deadly cooperation with the human immunodeficiency virus (HIV) pandemic, the incidence of TB cases worldwide continues to rise (M. Freire and G. Roscigno, Editorial, Bull. W. H. O. 80:429, 2002). Therefore, research efforts to characterize the unique biology of the tubercle bacillus, to develop new pharmacological TB interventions, and to formulate new TB vaccine strategies are of paramount importance in order to eliminate this global killer.
M. tuberculosis is remarkable in that it appears to be exquisitely adapted for human parasitization and host immune system evasion. Following inhalation of aerosolized organisms, M. tuberculosis sets up residence and propagates within the generally hostile environment of the alveolar macrophage. It avoids sterilization by the subsequent adaptive immune response that is mounted against it, and it finds sanctuary within the inflammatory response-derived granulomas meant to contain it. When immunity wanes, after years to decades of persistence, M. tuberculosis reactivates and exploits inflammation-mediated lung tissue destruction to enable its transmission to new persons (26). At present, the correlates of protection from active TB and the molecular mechanisms of infection and pathogenesis that account for the success of M. tuberculosis remain largely unknown but are likely to incorporate a complex interplay of multiple host and pathogen factors.
A key component of protective immunity to active TB is the timely and orchestrated production of proinflammatory cytokines such as tumor necrosis factor alpha, interleukin-12 (IL-12), IL-1β, and gamma interferon (IFN-γ) (16). In order to prevent overzealous proinflammatory responses and to protect against undue immune-mediated damage, counteractive immunosuppressive cytokines are also secreted as part of a balanced immune response and include transforming growth factor β and IL-10 (49). However, the premature or disproportionate secretion of inhibitory cytokines may undesirably benefit the pathogen, as elevated IL-10 levels have been associated with poor resolution of infections by HIV, human rhinovirus, Leishmania spp., and Mycobacterium leprae (36, 61, 67, 68). Recent studies have suggested that this may also be the case in M. tuberculosis infection (10, 11, 25, 46, 49).
A major advance for TB research came in 1998 with the publication of the complete genome sequence of the M. tuberculosis H37Rv laboratory strain (15). Of the approximately 4,000 open reading frames identified, close to 48% have not been assigned a function, nor have most been proven to code for expressed proteins (14). The recent advent of improved molecular tools for mycobacteria has allowed the systematic study of the M. tuberculosis genomic blueprint in order to identify genes of importance and to characterize their products (50, 60). Given the astounding success of M. tuberculosis, it is reasonable to anticipate that M. tuberculosis genes devoted to defense against host mycobacteriocidal immune mechanisms, or genes that promote disturbances in effective immune function, will be found. In fact, M. tuberculosis genes implicated in persistence, resistance to oxidative stress, and immune activation have been identified (18, 28, 30, 42, 57). Several of these putative virulence factors are secreted or released by growing M. tuberculosis into the culture filtrate (CF) compartment and are thereby strategically positioned as molecular effectors to the detriment of the host and/or for the benefit of the pathogen (28, 52, 64). A coincident characteristic of many individual CF proteins (as well as the CF as a whole) is their strong immunostimulatory capacity. This feature may be important in the M. tuberculosis life cycle strategy but may also contribute to immune control of infection. Many studies have illustrated the presence of specific antisera as well as the development of specific Th1-like responses (lymphoproliferation and/or IFN-γ secretion) and cytotoxic T-cell activity to CF proteins in TB patients and/or immunized animals (8, 9, 18, 31, 62). Indeed, the production of CF proteins is believed to account for the heightened efficacy of live, as opposed to killed, M. tuberculosis vaccines in animal models (3, 31). Containing in the range of 200 to 800 different proteins (many of which remain unidentified and whose functions are uncharacterized) (35, 53, 64), the CF presents an abundance of candidates for drug intervention, for incorporation into a TB vaccine, or to serve as TB diagnostic markers. Further systematic dissection and characterization of the constituents of CF by the TB scientific community will undoubtedly uncover useful information about the unique biology of M. tuberculosis and will provide fundamental knowledge of the immunological parameters associated with protective immunity against M. tuberculosis in humans.
In this study we detail the identification, cloning, and characterization of a 32-kDa CF protein that we have designated CFP32 (originally known as Rv0577). Comparative analyses suggest that the cfp32 gene product may be important to the biology of the MTC subspecies. Moreover, patient data suggest that CFP32 is expressed in M. tuberculosis-infected individuals and may be useful as a diagnostic, drug, and/or vaccine target. Surprisingly, levels of CFP32 in TB patient lung sputum were positively correlated with levels of IL-10 but not of IFN-γ in the same sputum sample, thereby suggesting that a link between M. tuberculosis and IL-10 may play a role in the pathogenic mechanism leading to active TB.
(This study contributed to the fulfillment of the Ph.D. requirements of R.C.H.)
MATERIALS AND METHODS
PCR and Southern blotting for CFP32.
Primer pairs suited to evaluate the cfp32 (Rv0577) locus were created using the DNASTAR program (DNASTAR, Inc., Madison, Wis.) and GenBank sequence database information (http://www.ncbi.nlm.nih.gov). These primers amplified either the upstream region of cfp32 (577proF, 5′-GTG GCT TGG CGG GCA CGG TGG AG-3′; 577proR, 5′-TTT TGG CGG CGG ACT GAT CGG TGG TCT-3′), the full coding region of cfp32 (Rv0577F, 5′-ATG CCC AAG AGA AGC GAA TAC AGG-3′ [F]; Rv0577R, 5′-CTA TTG CTG CGG TGC GGG CTT CAA-3′ [R1]), or the extended full coding region of cfp32 (577pMS3F, 5′-CCC TTA ATT AAT GTC CGC CAC CTA ACG AAA G-3′; 577pMS3R, 5′-CCC AAG CTT CTA GCA TTC TCC GAA-3′ [R2]). Each PCR mixture was prepared, each reaction was run using PCR program 1 (with an initial denaturation step of 5 min at 94°C followed by 25 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C and ending with a final elongation step for 10 min at 72°C), and results were analyzed as previously described (33). Likewise, direct sequencing of PCR fragments was performed and results were analyzed as recently described (33). PCR amplicons were sequenced using their respective amplification primers, and a minimal single overlap from two directions for each was usually achieved. Additional sequencing primers internal to cfp32 were also used (−605F, 5′-CGA ATC ATT GGC ACG TCT ACT TTG-3′; −281R, 5′-ACC ACC TTG TCC ACC ACC GCA T-3′). Southern blot analysis for cfp32 was done as previously described (37) and, as the hybridization probe, used M. tuberculosis H37Rv cfp32 PCR products generated using the Rv0577F and Rv0577R primer pair.
PAGE followed by gel staining or Western blotting for CFP32.
All polyacrylamide gel electrophoresis (PAGE) and Western blot assays were performed as follows. NuPage 12% Bis-Tris 10-well gels (Invitrogen, Carlsbad, Calif.) underwent PAGE and transfer using the Xcell II apparatus (Novex, San Diego, Calif.), per the manufacturers' instructions. In some experiments select samples were not preboiled or mixed with reducing agent (1 μl of 1 M dithiothreitol) prior to gel loading, as indicated. Full-range rainbow (Amersham, Piscataway, N.J.), midrange (Promega, Madison, Wis.), or kaleidoscope prestained (Bio-Rad, Hercules, Calif.) molecular weight protein markers were used as standards. For antibody detection of CFP32, nitrocellulose membranes were first blocked with 3% bovine serum albumin in 1× TBSt (Tris-buffered saline with 0.1% Tween 20) for 1 h following transfer. Afterwards, the membranes were probed with a CFP32 antiserum for 1 h and then washed three times with TBSt. The membranes were then probed with either anti-rabbit immunoglobulin (Ig)-horseradish peroxidase (HRP)-linked whole antibody (Amersham; used when anti-recombinant CFP32 [anti-rCFP32], anti-PepC, or anti-Pep7 was the primary antiserum) or anti-mouse Ig-HRP (Amersham) (used when IT-44 was the primary antibody), washed three times with TBSt, developed using ECL Western blot detection reagents (Amersham), and then exposed to Kodak BioMax film. Mycobacterial lysates were generated in a mini-BeadBeater (Biospec Products Inc., Bartlesville, Okla.), whereby growing cultures were spun down, the supernatant was removed, the pellet was resuspended with Tris-EDTA buffer, and six 3-mm-diameter glass beads were added to lyse the bacteria in five 30-s pulses. These lysates were subsequently heated at 80°C for 30 min and then gamma-irradiated. A total of 37 MTC strains and 29 mycobacteria other than the MTC (MOTT) isolates were evaluated for CFP32 by Western blotting including 8 strains of M. tuberculosis, 7 strains of M. bovis, 3 strains of M. bovis BCG, 8 strains of M. microti, 6 strains of M. africanum subtype I, 4 strains of M. africanum subtype II (Uganda), 1 strain of M. bovis subsp. caprae, 2 strains of Mycobacterium smegmatis, 8 strains of Mycobacterium avium subsp. avium, 2 strains of Mycobacterium avium subsp. intracellulare, 1 isolate of M. leprae, 1 strain of Mycobacterium marinum, 1 strain of Mycobacterium xenopi, 2 strains of Mycobacterium chelonae, 2 strains of Mycobacterium gordonae, 4 strains of Mycobacterium abscessus, and 6 strains of Mycobacterium fortuitum. For each strain, MTC subspecies identity was confirmed by a recently developed MTC PCR typing protocol (33) and MOTT species identity was confirmed by 16S rRNA sequencing, also as described previously (33). M. leprae lysate was kindly provided by P. Brennan as part of the Colorado State University (CSU) NIH NIAID Leprosy Research Support Contract (http://www.cvmbs.colostate.edu/mip/leprosy). Lysates of pelleted pQE31.577-transformed IPTG (isopropyl-β-d-thiogalactopyranoside)-induced Escherichia coli were prepared by a method of multiple freeze-thaws with intermittent water bath sonication in native condition lysis buffer (50 mM NaH2PO4 [pH 8.0]; 300 mM NaCl; 1 mM phenylmethylsulfonyl fluoride; 1 μg of lysozyme/ml; and 5 μg each of aprotinin, chymostatin, leupeptin, and pepstatin/ml) (Sigma, St. Louis, Mo.). The protein content of mycobacterial and E. coli lysates was quantified using the Bio-Rad protein assay and an Ultraspec 2100 Pro spectrophotometer (Amersham Pharmacia Biotech, Cambridge, United Kingdom). All M. tuberculosis subcellular components and CF fractions were generated at CSU as part of the NIH NIAID TB Research Materials and Vaccine Testing Contract (http://www.cvmbs.colostate.edu/microbiology/tb/top.htm). Stocks of the murine IT-44 monoclonal antibody (MAb) (39) are also distributed through CSU. Silver staining (Invitrogen) and 1% Coomassie blue staining of polyacrylamide gels followed standard protocols. Internal sequencing of a protein band cut from a silver-stained gel that was identified as CFP32 was done by the Rockefeller University Protein/DNA Technology Center (23). Computer analysis of CFP32 and its homologues employed the GenBank and SwissProt (http://us.expasy.org/sprot) websites. Basic summary information on CFP32 can be found in GenBank (given as Rv0577) as well as the TubercuList website (http://genolist.pasteur.fr/TubercuList/index.html) (given as TB27.3).
CFP32 cloning, expression, and purification.
A cfp32 PCR fragment representing the entire open reading frame was generated with PCR program 1 from purified M. tuberculosis H37Rv DNA by using primers that were engineered to introduce BamHI and HindIII restriction enzyme sites into the resulting PCR product (SMC-1, 5′-GAA AGG ATG AGG ATC CCC AAG AGA AGC G-3′, and SMC-2, 5′-CGG GAT GCT CAA GCT TGC TGC GGT GC-3′). By standard procedures, the amplified product was restriction digested, ligated into the pQE31 vector (Qiagen, Valencia, Calif.) to create the pQE31.577 plasmid, and introduced into M15 E. coli, and the sequence was confirmed. The production of N-terminal hexahistidine (His)-tagged rCFP32 followed the methodology described in the QiaExpressionist handbook (Qiagen) but was further optimized by growing the bacteria in Terrific Broth (Sigma) and inducing the pQE31.577 transformants with 0.5 mM IPTG (Sigma) for 4.5 h and shaking at 30°C. The predicted amino acid sequence of rCFP32 is RGS-6×H-TD-(CFP32)-A. His-tagged rCFP32 was purified by nickel affinity chromatography, using nickel-nitrilotriacetic acid spin columns (Qiagen), under native conditions, per the manufacturer's protocol. A single difference was that nickel-nitrilotriacetic acid-bound rCFP32 was washed three times using buffer containing 1 mM imidazole prior to elution. The rCFP32 was then washed free of the imidazole and concentrated using Centriplus centrifugal filter devices (30-kDa cutoff) (Millipore, Bedford, Mass.). PAGE, followed by 1% Coomassie blue staining and/or silver staining, was done to qualify the purity of the preparation. A standard Bio-Rad protein assay was done to quantify yield. The identity of the recombinant protein was verified by electrospray tandem mass spectrometry of rCFP32 digested with trypsin and interrogation of the mass spectrometry data against the M. tuberculosis genome by using Sequest software (22). Rabbit antisera were generated by a commercial provider (Covance Research Products, Denver, Pa.). Candidate rabbits for immunization with rCFP32, or CFP32-derived synthetic peptides, were prescreened for serum reactivity to M. tuberculosis whole-cell lysate by Western blotting, and only rabbits with low to absent reactivity were chosen. The Pep7 immunogen was generated by a commercial provider (Sigma Genosys, Houston, Tex.) while PepC was kindly provided by Shibo Jiang, New York Blood Center. These synthetic peptides were covalently linked to keyhole limpet hemocyanin prior to injection.
Enzyme-linked immunosorbent assay (ELISA) detection of CFP32 and human antibody to CFP32.
For the detection of human anti-CFP32 serum specificity, two different ELISAs were fashioned. In the first (Cornell laboratory), the IT-44 murine MAb (1:105 in phosphate-buffered saline [PBS], 50 μl per well) was used to coat a 96-well ELISA plate (Corning International, Corning, N.Y.) and was incubated overnight at 4°C. PBSt (PBS with 0.1% Tween 20) was used to wash the plate four times followed by 2.5 h of incubation with 200 μl of blocking buffer (PBS with 10% fetal calf serum) and with shaking at room temperature. Next, rCFP32 (2.5 ng/ml in blocking buffer, 100 μl per well) was incubated for 2.5 h, with gentle shaking at room temperature, and subsequently washed four times with PBSt. Duplicate samples of each test human serum from a Brazilian cohort (1:5 × 104 in blocking buffer, 100 μl per well) were then incubated for 2 h, with gentle shaking at room temperature, and subsequently washed four times with PBSt. Biotinylated anti-human Ig (1:104 in blocking buffer, 100 μl per well) (Amersham) was then input and incubated for 1 h, with gentle shaking at room temperature, and washed four times with PBSt. Extravidin peroxidase conjugate (1:2 × 103 in PBS, 100 μl per well) (Sigma) was then applied to the plate, shaken gently for 2 h at room temperature, and subsequently washed four times with PBSt. 3,3′,5,5′-Tetramethylbenzidine (TMB) (Sigma) acted as the enzymatic substrate (100 μl per well). Once the blue color had sufficiently developed, the reaction was stopped using 0.5 M H2SO4 (100 μl) and read at 450 nm with an EL 340 Biokinetics Reader (BioTek Instruments Inc., Winooski, Vt.). The absorbance values for each donor sample were then averaged. The detection of human anti-CFP32 antisera from a cohort of patients in India, by the New York University laboratory, was performed as previously described (59) using rCFP32 (2 μg/ml in PBS, 50 μl per well) to coat a 96-well ELISA plate and capture the specific antibodies. The international standard of ≥10-mm induration following the injection of 5 TU of M. tuberculosis purified protein derivative (PPD) was used to define a positive skin test. To get a measure of CFP32 in the lungs of TB patients, a variation on the ELISA to detect anti-CFP32 antisera (Cornell laboratory) was used. Patients living in Brazil who presented at the Pulmonary Service with “lung disease suggestive of TB” and who failed to provide a spontaneous sputum sample or for whom a sample was negative for acid-fast bacilli (AFB) underwent sputum induction using 3% saline in an ultrasonic nebulizer. The induced sputum remaining from diagnostic workup was treated with dithiothreitol (Sigma) and centrifuged, and the supernatant was stored at −80°C prior to use. For the CFP32 ELISA, 50 μl of sputum per well, one sample per donor, was input in place of a single set amount of rCFP32. Duplicate twofold dilutions of rCFP32 (5 × 103 to 5 × 101 pg/ml) were also used to establish a standard curve for CFP32 at this stage. As a final difference, anti-rCFP32 (1:104, with gentle shaking at room temperature overnight) was used as the second antiserum (as opposed to the human antisera), thereby necessitating the use of anti-rabbit Ig-HRP and TMB substrate for detection. For these experiments, TB case patients were defined as having a positive solid medium culture or treatment response with resolution of clinical and radiological features of TB. Suspected TB cases were defined as patients with clinical and radiological features compatible with TB for whom cultures were negative, contaminated, or not available and who had insufficient follow-up or had prior TB without sufficient follow-up. Non-TB cases with other lung diseases (OLD) were defined as those patients who were AFB smear and TB culture negative, for whom another diagnosis was established, and/or who showed clinical improvement after a short course of non-TB antibiotics. The detection of CFP32 by the testing laboratory was independent of knowledge of the clinical classification of each patient. For the quantification of lung cytokine levels, ELISAs were performed upon the same lung sputum samples as those evaluated for CFP32. For these experiments, an anti-IL-10 antibody pair (Pierce Endogen, Rockford, Ill.) was used in an otherwise identical protocol as given for the evaluation of sputum CFP32 levels. IFN-γ was measured using a commercial kit (Immunotech, Marseille, France) and converted to picograms per milliliter using the relationship 1 U of IFN-γ = 33.33 pg of IFN-γ. The quantification of CFP32 in M. tuberculosis subcellular compartments followed the ELISA protocol for sputum CFP32 measurement. All serum and sputum donors signed informed consent papers, and the study was approved by the Internal Review Boards of Cornell University, Hospital Universitário Clementino Fraga Filho, and New York University.
RESULTS AND DISCUSSION
Identification of a novel M. tuberculosis CF protein.
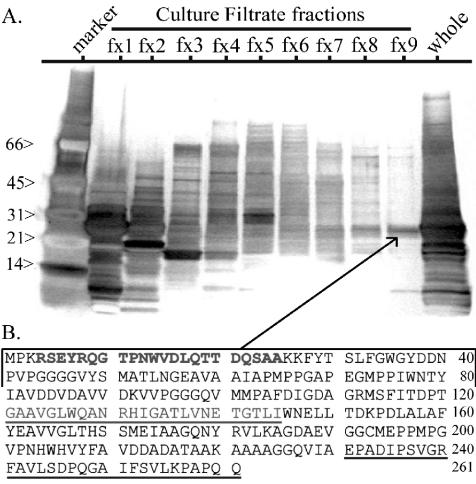
To identify a novel extracellular M. tuberculosis antigen of prospectivepathological and/or immunological importance, CF proteins were first separated by anion-exchange chromatography into analyzed pools of fractions. Of these pooled fractions, PAGE under nondenaturing conditions followed by silver staining revealed that CF-fraction pool 9 (fx9) contained a predominant band at approximately 24 kDa (Fig. 1A). To determine the identity of the major CF-fx9 protein, PAGE and silver staining were repeated for CF-fx9 alone, and the same predominant band was excised (data not shown). N-terminal sequence analysis of the gel fragment failed; however, internal protein sequencing identified a peptide that was identical to amino acids 4 to 25 of the predicted product of the M. tuberculosis gene annotated as Rv0577 (Fig. 1B). Rv0577 was a hypothetical gene of unknown function that was identified upon completion of the M. tuberculosis H37Rv genome sequence (15). Rv0577 also corresponds to the MT0606 locus of M. tuberculosis strain CDC1551 (GenBank accession no. AE000516). In being a novel CF protein the Rv0577 gene product was of sufficient interest that we decided to pursue its characterization. In keeping with previous convention (54, 73), and based upon the PAGE mobility of its gene product under denaturing conditions (see below), the Rv0577 gene is hereafter designated cfp32 and the protein that it encodes is designated CFP32. While this study was ongoing, a separate group independently identified CFP32 by microsequencing CF spots in silver-stained two-dimensional polyacrylamide gels (given as TB27.3), thereby confirming the synthesis and export of CFP32 to the CF (55).
FIG. 1.
Identification of CFP32 (Rv0577) from fractionated M. tuberculosis CF. (A) Silver-stained gel of CF fractions. The CF of growing M. tuberculosis H37Rv was fractionated by anion-exchange chromatography (using QAE Sepharose resin and an increasing NaCl concentration), and 15 μl of each fraction pool (fx), as well as 1 μg of unfractionated CF (whole), was subjected to nondenaturing PAGE (without preboiling) followed by silver staining. Molecular mass standards (Bio-Rad; values in kilodaltons) are provided in the lane labeled marker. (B) Amino acid sequence of CFP32. The predominant band in CF-fx9 was excised and internally sequenced to obtain a peptide (boldface) matching the hypothetical M. tuberculosis H37Rv gene Rv0577. Synthetic peptides, based upon the underlined amino acid sequences, were used to derive the anti-Pep7 (amino acids 121 to 145) and anti-PepC (amino acids 231 to 161) rabbit antisera.
Information predicted by the sequences of cfp32 and CFP32.
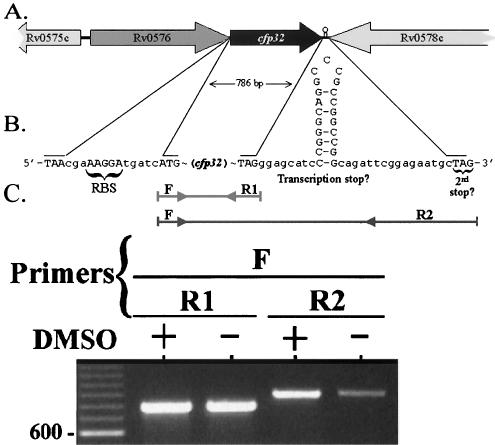
The gene for CFP32 is transcribed in the forward direction (Fig. 2A), and the start of cfp32 is preceded by an AAGGA putative Shine-Dalgarno ribosomal binding site (RBS) (Fig. 2B). The region upstream of cfp32 also contains several putative regulatory elements that are homologous to previously described mycobacterial −10 and −35 RNA polymerase contact sites (n = 3 and 5, respectively), including three −35 sites identified for the CF virulence factor katG (44; data not shown). The predicted TAG stop codon of cfp32 is followed by 15 intervening codons and a second in-frame TAG. The biological significance of such an arrangement of two stop codons is not known but has been noted previously for the glutamine synthetase gene of M. tuberculosis, E. coli, and Salmonella enterica serovar Typhimurium (27). Rv0576 is the hypothetical open reading frame found upstream of cfp32 (Fig. 2A). This element could cotranscribe with cfp32, as it is the only other local gene also predicted to be transcribed in the forward direction. However, the presence of an RBS for cfp32 suggests that one level of its regulation is monocistronic. Downstream of cfp32 is the inversely transcribed PE-PGRS gene Rv0578c as well as a putative stem-loop structure(s) that may act as the transcriptional stop signal for both cfp32 and Rv0578c (Fig. 2B). Evidence for the presence of secondary DNA structure in the intergenic region of cfp32 and Rv0578c was provided in PCR amplification experiments (Fig. 2C). Herein, a single forward primer (F) was used in combination with either of two reverse primers that are complementary to sequences flanking each side of the putative stem-loop. For further comparison, the amplification was done in the presence or absence of dimethyl sulfoxide (DMSO), a PCR recipe additive that helps to open secondary structure and improve PCR efficiency. As a result, the PCR product band intensity was significantly reduced using the reverse primer that was 3′ to the putative stem-loop (R2; Fig. 2C) compared to that with the reverse primer that was 5′ to the stem-loop (R1). In addition, the amplification efficiency was further reduced in the absence of DMSO in the case of the F-R2 but not F-R1 primer pair, thereby indicating that amplification using R2 is inhibited by secondary DNA structure.
FIG. 2.
Characterization of the cfp32 locus. The predicted coding region of cfp32 is 786 bp long and located at nucleotide coordinates 671166 to 671951 (relative to the M. tuberculosis H37Rv genome sequence, accession no. AL123456). (A) Illustration of genes in the vicinity of cfp32. (B) Depiction of the DNA sequences upstream and downstream of cfp32. Shown are the putative RBS, ATG start codon, TAG stop codon, and a second in-frame TAG stop codon, as well as a potential stem-loop structure-transcription stop signal for cfp32. (C) PCR evaluation of the region 3′ of cfp32 indicates the presence of secondary DNA structure by differences in PCR amplicon intensity. PCR products and a 100-bp ladder (in the first lane) were visualized by agarose gel electrophoresis and ethidium bromide staining. The F sense primer (Rv0577F) was used in combination with either the R1 (Rv0577R; product size, 786 bp) or R2 (577pMS3R; product size, 838 bp) antisense primer. Amplification was also done in the presence (+) or absence (−) of DMSO. An additional 2.5 μl of water was included in the reaction mixtures that purposely excluded DMSO. One representative example of four experiments is shown.
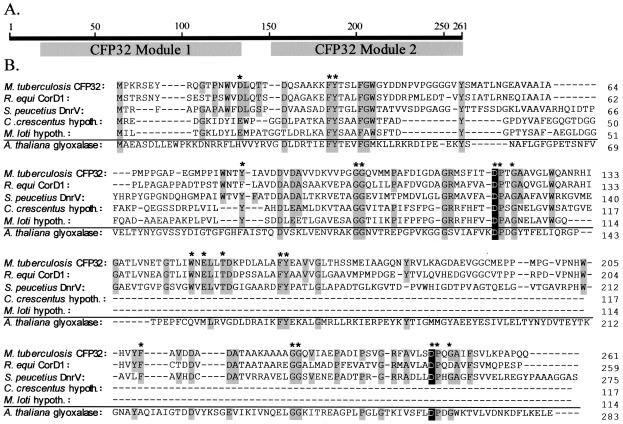
In silico modeling revealed several intriguing insights into the nature of the cfp32 gene product. Analysis for functional regions indicated that CFP32 is a bimodular protein with two homologous domains (N-terminal residues ∼2 to 129 and C-terminal residues ∼130 to 261; 26% identity) each with structural similarity to members of the glyoxalase-dioxygenase superfamily of enzymes (GenBank; Fig. 3A). CFP32 may therefore be a bifunctional enzyme and catalyze more than one reaction. GenBank BLAST searches found that CFP32 shows significant pairwise homology (15 to 58% identity) to many other unimodular and bimodular polypeptides from a variety of microorganisms (described for Fig. 3B). As with CFP32, many of these polypeptides are of unknown function and annotated as “probable hydrolases” or “hypothetical proteins.” Notably, CFP32 homologues were most plentiful in Streptomyces spp., while CorD1 of Rhodococcus equi (a close phylogenetic relative of M. tuberculosis and an intracellular pathogen that causes a TB-like pulmonary disease in foals and immunocompromised patients [47]) had the highest percent identity (58%). Of additional note were the Streptomyces spp. dnrV and sgaA gene product homologues of CFP32. The dnrV-encoded protein plays a role in the synthesis of the polyketide antibiotic doxorubicin (40), and the sgaA gene encodes a regulatory factor of growth and osmotic stress responses, as well as streptomycin production and resistance (4). By analogy, CFP32 may therefore have similar physiological activities. Remarkably, the alignment of CFP32 homologues revealed several highly conserved amino acids, several of which were also present in a surprising number of ostensibly paralogous glycosyl hydrolases (one example is given in Fig. 3B). Of these residues, the tyrosines and aspartic acids may be important to the catalytic mechanism with the most significant being CFP32 (module 1) Asp118 and CFP32 (module 2) Asp252. These aspartic acids were each in the context of a DPXG motif analogous to that for the determined enzymatic nucleophile (the residue that forms the enzyme-substrate intermediate during cleavage) of the well-characterized class II (family 38) α-mannosidases (32). Compare human Golgi α-mannosidase II (PRSGWQIDPFGHSA), jack bean α-mannosidase (PRAGWAIDPFGHSP), CFP32 module 1 (GRMSFITDPTGAAV), and CFP32 module 2 (GRFAVLSDPQGAIF) whereby the conserved amino acids are in boldface and the nucleophiles (known and putative) are underlined. Perhaps residues Asp118 and Asp252 serve similar mechanistic roles for CFP32 and its homologues.
FIG. 3.
Alignment of the bimodular CFP32 and its homologues with a divergent glyoxalase reveals conserved amino acids that may be related to the catalytic mechanism. The CFP32 polypeptide is predicted to contain 261 amino acids, to have a molecular mass of 27.3 kDa, to have an isoelectric point (pI) of 4.24, and to be a compact globular protein (SwissProt). (A) Cartoon to illustrate the two predicted glyoxylase domains of each CFP32 module. (B) M. tuberculosis CFP32 was aligned with its homologues (encoded by sequences with GenBank accession numbers in parentheses; 15 to 58% overall range of homology). The alignment results for CFP32 with two bimodular [R. equi (CorD1, CAC44898) and Streptomyces peucetius (DnrV, AAD04716)] and two unimodular [Mesorhizobium loti (BAB53970) and Caulobacter crescentus (AAK25386)] representative homologues are illustrated. Not shown are the additional CFP32 homologues from M. tuberculosis (Rv0911, CAB08509), Streptomyces spp. (SgaA, BAA14012; BAA08202; CAA15810; CAB42934; CAB45588; CAB55527; CAB92885; CAB95980; CAC08431), Corynebacterium glutamicum (CAC26380), M. loti (BAB48973), Vibrio cholerae (AAF96246; AAF96546), C. crescentus (AAK23809), Bacillus halodurans (BAB04023), Pseudomonas aeruginosa (AAG05061), Myxococcus xanthus (AAL56603), Agrobacterium tumefaciens (AAK86662; AAK87322; AAL41869), Brucella melitensis (AAL54026), and Sinorhizobium meliloti (CAC47416). A contrasting glyoxalase from Arabidopsis thaliana (BAB17665) that had low sequence identity (13%) to CFP32 is also provided for comparison. Amino acids that were highly conserved among the set of CFP32-like proteins are shaded. Glutamic acids that substitute for conserved aspartic acids are also shaded. Asterisks above residues indicate those that are present almost without exception. Homologous residues in the A. thaliana glyoxylase are also specified by shading. Putative aspartic acid nucleophiles are shaded black. DnrV, dnrV gene product; CorD1, corD1 gene product; hypoth., hypothetical protein.
Development of antisera to CFP32 and confirmation of CFP32 as the IT-44-reactive antigen.
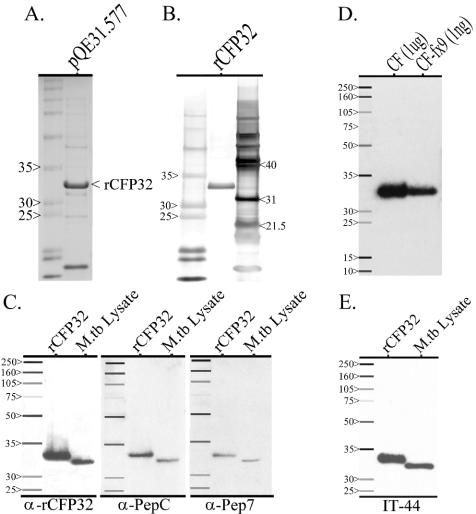
The cfp32 gene was cloned from M. tuberculosis H37Rv and expressed in E. coli, and rCFP32 was purified by nickel column affinity chromatography. The IPTG-induced pQE31.577 transformant expressed rCFP32 at a band size of ∼33 kDa (Fig. 4A). This band was absent from the uninduced transformant and was absent from both the induced and uninduced E. coli transformed with the naked pQE31 plasmid (data not shown). Soluble rCFP32 of high purity, as determined by PAGE and silver staining (Fig. 4B), was readily obtained, supporting the predicted soluble nature of CFP32. Mass spectrometry of trypsin-digested rCFP32 derived four peptide sequences (18 to 36 amino acids long), each of which perfectly matched separate stretches of amino acids in the expected sequence of CFP32 (data not shown). Rabbits were then immunized either with the purified rCFP32 or with CFP32-based synthetic peptides. Peptide 7 (Pep7) is identical to an internal length of amino acids while peptide C (PepC) parallels the C terminus of CFP32 (Fig. 1B). In Western blotting, each of the three rabbit-raised antisera (anti-rCFP32, anti-PepC, and anti-Pep7) recognized rCFP32 at a band size of 33 kDa (Fig. 4C). Importantly, the preimmunization sera of these rabbits did not show any reactivity in parallel Western blots (data not shown). The trio of anti-CFP32 antisera also recognized a band at ∼32 kDa from the whole-cell lysate of M. tuberculosis H37Rv that is presumably CFP32 (Fig. 4C). Subsequent Western blotting of the whole CF fraction, as well as CF-fx9 (from whence CFP32 was first identified), also showed a 32-kDa band (Fig. 4D). The enrichment of CFP32 in CF-fx9 is given by its relative band strength in Western blotting (at 103-fold-less input CF-fx9 sample compared to whole CF). The difference between expected (27.3 kDa) and observed (32 kDa) molecular masses of CFP32 in PAGE under denaturing conditions has been noted previously for other CF factors (73) and may be due to anomalous migration and/or unidentified posttranslational modifications such as myristoylation, glycosylation, or phosphorylation. Overall, the combined data confirm the correct cloning and exogenous expression of CFP32 from fractionated CF and argue for the specificity of the developed antisera.
FIG. 4.
Cloning of CFP32 and the derivation of anti-CFP32 antisera. For each of the following, all samples were boiled prior to being loaded in the gel, and molecular mass protein markers (Amersham; values in kilodaltons) are shown in the first lane. (A) Coomassie blue-stained polyacrylamide gel of lysate from IPTG-induced pQE31.577-transformed M15 E. coli expressing rCFP32. The rCFP32 band appears at ∼33 kDa. A similar band was absent from parallel IPTG-induced M15 E. coli lysates that were either wild type or transformed with the pQE31 vector (data not shown). (B) Silver-stained gel following PAGE of His-tagged purified rCFP32. Additional protein molecular mass markers (Promega; values in kilodaltons) are shown in the third lane. (C) Each of the three rabbit-derived antisera recognized both CFP32 and rCFP32. Separate parallel sets of purified rCFP32 (10 ng) and M. tuberculosis (M.tb) lysate (1 μg) were probed with either anti-rCFP32 (1:103), anti-PepC (1:103), or anti-Pep7 (1:250) antiserum in a Western blot. (D) Antiserum raised against purified rCFP32 recognized, and was specific for, M. tuberculosis CFP32. Samples of CF (1 μg) and CF-fx9 (1 ng) were probed with the rabbit-derived anti-rCFP32 antiserum (1:103) in a Western blot. The M. tuberculosis CFP32 band appears at ∼32 kDa. (E) CFP32 is the IT-44 MAb-reactive antigen. IT-44 is a mouse-derived IgG2a MAb raised upon mouse challenge with M. tuberculosis CF (39). Western blotting, similar to the preceding, was done using IT-44 (1:2.5 × 104) to probe for CFP32.
IT-44 (also known as HBT7) is an IgG2a murine MAb that was derived from the immunization of inbred mouse strains with the CF of M. tuberculosis H37Rv (39). A GenBank submission of unpublished data from T. Oettinger (accession no. AJ007737) identified the IT-44-reactive antigen as being the gene product of cfp32 (given as the cfp30B gene). IT-44 was also shown previously to react with three spots in two-dimensional PAGE of M. tuberculosis CF (64). However, the proteins were clustered at ∼32 kDa and migrated within a narrow pI range of 4.75 to 4.93, thereby suggesting that the antibody was reacting with multiple isoforms of the same antigen. As a result of this information, IT-44 was obtained and was evaluated for CFP32 reactivity by Western blotting. Bands for rCFP32 and CFP32 were seen in the same position as in the Western blots probed with rabbit anti-rCFP32 antisera (Fig. 4E) while IT-44 Western blot reactivity could be blocked by blot preincubation with anti-rCFP32 (data not shown), thereby verifying CFP32 as the IT-44-reactive antigen. This finding has been independently confirmed in CF mapping studies (53, 55). It should also be noted that Western blot assays probing for CFP32, similar to previous silver stain gel results (Fig. 1A), suggested that CFP32 and rCFP32 exist in two states: the respective linearized 32- or 33-kDa form that was seen when samples were prepared under denaturing conditions (by being heated to 100°C for 5 min in the presence of dithiothreitol) and a predominant ∼24-kDa form that was visible in parallel nondenatured samples (data not shown). It was therefore thought that native CFP32 maintains a compacted hydrophodynamic volume that is unfolded upon boiling, the likes of which were also noted previously for CFP25 (73). However, the CFP32 sequence contains but a single cysteine residue, and so forces other than intramolecular disulfide bonds must maintain the globular three-dimensional structure of monomeric CFP32.
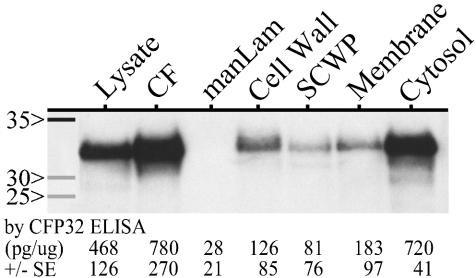
Distribution of CFP32 among M. tuberculosis subcellular compartments.
To localize CFP32, M. tuberculosis H37Rv subcellular compartments were evaluated for the presence of CFP32 by Western blotting with the developed antisera (Fig. 5). On a per-microgram basis, the greatest quantity of CFP32 was found in the CF followed by a very strong CFP32 band in the cytosolic and whole-cell lysate fractions. Small amounts were also detected in the cell wall, soluble cell wall proteins, and membrane fractions but not in the purified mannosylated lipoarabinomannan. At least one additional lot of each component was tested by Western blotting and gave a similar result (data not shown). ELISA measurement of CFP32 levels in the illustrated components supported the Western blot data, indicating relative amounts of CFP32 in each by band intensity (Fig. 5). These data suggest a directed movement of CFP32 from the cytosol to the CF despite the lack of a clear generalized signal peptide for bacterial secretory proteins in the CFP32 N terminus (51; data not shown). It is further noteworthy that the original sequencing of the CF-fx9 CFP32 band did not indicate the occurrence of N-terminal cleavage associated with export signal peptides. Even so, there are several other known CF protein genes that do not code for the classical signal peptides, including superoxide dismutase (28), glutamine synthetase (27), and CFP29 (54) as well as ESAT-6 and CFP10 (65). Whether CFP32 or other such proteins are actively exported, excreted, or released during cell division or autolysis is unresolved but may involve an uncharacterized mycobacterial secretory mechanism (27, 28, 70). Moreover, by analogy to other CF proteins (27, 28), CFP32 may serve intracellular, in addition to extracellular, functions, thus explaining the necessity of its partial cytoplasmic retention.
FIG. 5.
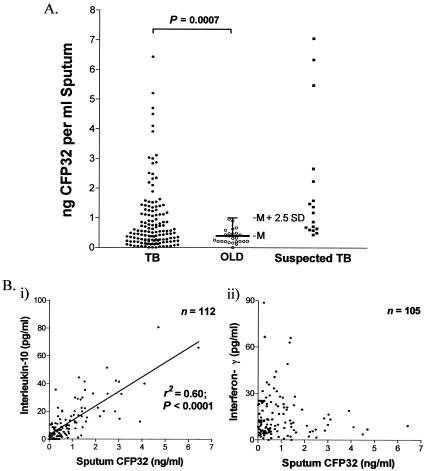
CFP32 localizes predominantly to the CF and cytosol fractions of M. tuberculosis. Western blotting was done to probe the lysate, CF, mannosylated lipoarabinomanan (manLam) glycolipid, cell wall, soluble cell wall proteins (SCWP), membrane, and cytosol components of M. tuberculosis (at 1 μg each) for the presence of CFP32 by using the anti-rCFP32 antiserum (1:103). Molecular mass protein markers (Amersham; values in kilodaltons) are shown in the first lane. The amount of CFP32, as measured by ELISA (average for duplicate samples in three experiments), in each sample is given below each respective lane (in picograms per microgram of sample ± standard error [SE]). M. tuberculosis PPD was negative for CFP32 by Western blotting (data not shown) and by ELISA was measured as having 51 ± 28 pg of CFP32 per μg of sample. CF-fx9 was also tested by ELISA and had 610 ± 53 ng of CFP32 per μg of sample.
CFP32 is MTC restricted.
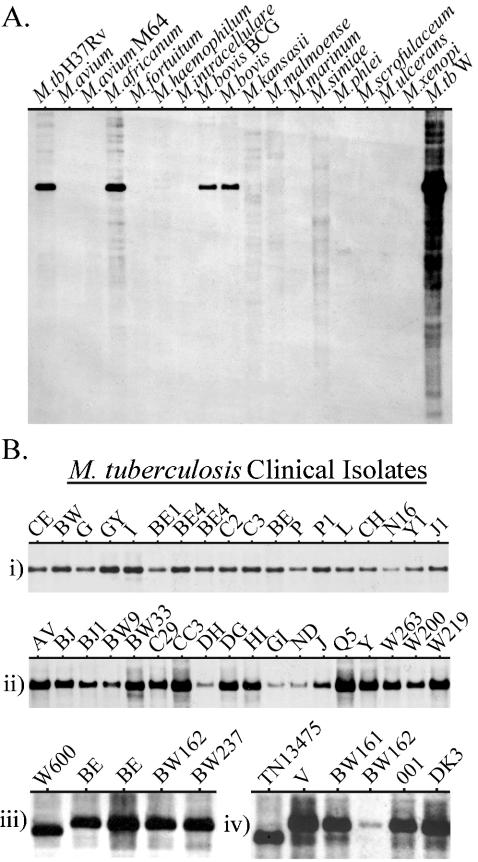
For CFP32 to be characterized as a unique biofactor of tuberculous mycobacteria, it should be present in all members of the MTC and absent from MOTT. In a previous study, 8 MTC subspecies (representing 72 strains) and 12 MOTT species (comprising 46 strains) were evaluated for the presence of cfp32 by PCR (given as Rv0577 and using primers Rv0577F and Rv0577R [33]). A cfp32 PCR fragment was observed only in the MTC groupings, indicating that cfp32 is an MTC-restricted gene. To determine the degree of polymorphism in cfp32 among the MTC subspecies, sequence analysis of PCR products representing the full and/or extended full cfp32 coding sequence from M. tuberculosis (n = 8), M. africanum (subtypes I and II, n = 4 and 4, respectively), M. bovis (n = 4), M. bovis BCG (n = 3), M. bovis subsp. caprae (n = 1), and M. microti (n = 4) was done. Likewise, the 321 bp of the putative cfp32 upstream promoter region was also PCR amplified and sequenced from M. tuberculosis (n = 5), M. africanum (subtypes I and II, n = 2 and 4, respectively), M. bovis (n = 1), M. bovis BCG (n = 2), M. bovis subsp. caprae (n = 1), and M. microti (n = 3). In all cases, the entire cfp32 locus (1,160 bp, representing nucleotides 670843 to 672002 of the M. tuberculosis H37Rv genome sequence, accession no. AL123456) was completely nonpolymorphic relative to the M. tuberculosis H37Rv genome sequence (data not shown). This observation is in keeping with previous studies that have noted a remarkable paucity of single-nucleotide polymorphisms in the structural genes of the MTC subspecies (66).
Southern blotting was employed next to verify that cfp32 is restricted to the MTC organisms. Of the evaluated species, only M. tuberculosis strains H37Rv and W, as well as the additional MTC subspecies M. africanum subtype I, M. bovis, and M. bovis BCG, were positive for a single copy of cfp32, while all 13 MOTT species and strains evaluated were negative (Fig. 6A). M. smegmatis was also repeatedly evaluated and found to be negative for cfp32 by Southern blotting (data not shown). Moreover, a cfp32 homologue could not be found in the M. smegmatis or the M. leprae genome sequences (http://www.tigr.org and http://www.sanger.ac.uk). Further cfp32 Southern blotting probed a comprehensive range of M. tuberculosis clinical isolates (n = 70) previously coded by IS6110-restriction fragment length polymorphism pattern classification (Fig. 6B subpanels i to iv) (37). Included in this evaluation were 36 strains prototypic for their particular IS6110 fingerprint (Fig. 6B subpanels i and ii). Remarkably, every M. tuberculosis strain was positive for a single band that ran at approximately the same location for all but two strains, for which it ran slightly lower than the others (Fig. 6B subpanels iii and iv). This difference most likely relates to the emergence of a new a PvuII cutting site outside cfp32 since sequencing of the strain TN13475 cfp32 did not uncover any polymorphisms. As such, it is impressive that cfp32 was completely conserved within the MTC given that subspecies- and strain-defining large chromosomal deletions are increasingly found in the MTC genomes (13, 33, 43). These deletions are emerging as potentially significant determinants of MTC pathobiological diversity but do not appear to include cfp32. Therefore, the complete conservation of cfp32 and its sequence for the tested isolates and its absence from MOTT species suggest that this gene may play an important role that is unique to M. tuberculosis and the other MTC groupings.
FIG. 6.
Southern blot analysis for cfp32. (A) The cfp32 gene is MTC restricted by Zoo blotting. DNA from an assortment of MTC subspecies (n = 5; namely, M. tuberculosis [M.tb] strains H37Rv and W, M. africanum subtype I, M. bovis, and M. bovis BCG) and mycobacteria other than MTC (MOTT; n = 13) was evaluated using cfp32 PCR fragments as the probe in Southern blotting. (B) Each clinical isolate of M. tuberculosis tested possesses the cfp32 gene. Subpanels i and ii illustrate the cfp32 Southern blot results for 36 M. tuberculosis isolates with unique IS6110-restriction fragment length polymorphism patterns prototypical of their lineages. An additional 35 M. tuberculosis clinical isolates were also evaluated (72 M. tuberculosis strains tested in total), examples of which are shown in subpanels iii and iv.
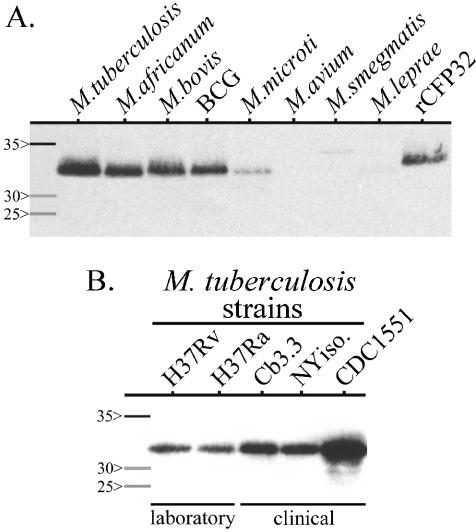
To probe for the mycobacterial expression of CFP32, Western blotting was done against a panel of MTC subspecies and strains (n = 37), as well as a range of MOTT species and strains (n = 29), which are listed in Materials and Methods. Upon completion, a CFP32 band was detected only from M. tuberculosis and the other MTC subspecies and not from any of the 10 MOTT species that were evaluated (Fig. 7; select strains are illustrated). The combined data support the idea that cfp32 is an expressed MTC-restricted gene and are in keeping with an integral role for CFP32 in the lifestyle of tubercle bacilli.
FIG. 7.
Western blot analysis for CFP32. (A) CFP32 is MTC restricted. Mycobacterial lysates (7 μg of each sample) and purified rCFP32 (10 ng) were probed by Western blotting with the anti-rCFP32 antiserum (1:103). A total of 37 MTC isolates (M. tuberculosis H37Rv and one strain each of M. africanum subtype I, M. bovis, M. bovis BCG, and M. microti are shown) and 29 MOTT isolates (one isolate each of M. avium subsp. avium, M. smegmatis, and M. leprae is illustrated) were tested. A breakdown by MOTT species and MTC subspecies is given in Materials and Methods. (B) Both laboratory and clinical isolates of M. tuberculosis express CFP32. The lysates of M. tuberculosis strains (3 μg of each sample) were probed by Western blotting with anti-rCFP32 antiserum (1:103). For both panels, parallel silver-stained gels (with 10-fold-more sample per isolate) confirmed that approximately the same amount of protein was loaded for each Mycobacterium isolate illustrated (data not shown). Molecular mass protein markers (Amersham; values in kilodaltons) are shown in the first lane of each blot.
Detection of specific antisera to CFP32 in clinical samples by ELISA.
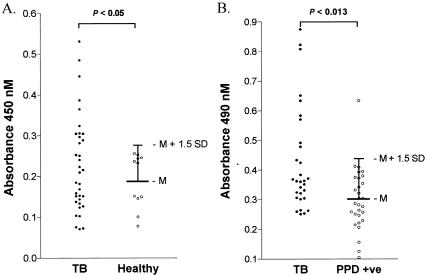
CFP32 is immunogenic for mice as given by the development of the murine IT-44 MAb following immunization with M. tuberculosis CF (39). As an indirect indicator that CFP32 is expressed in TB patients, the human serologic response to CFP32 was evaluated. Sera from a cohort of patients with active TB from Brazil (n = 35) and their healthy household contacts (n = 11; four PPD skin test positive, seven PPD skin test negative) were tested for antibodies that recognize rCFP32. Altogether, 34% (12 of 35) of TB case patients had detectable antibodies to CFP32 while none of the healthy controls were positive (P < 0.05, Fisher's exact test) (Fig. 8A); PPD status did not segregate the healthy household contacts. Notably, only half of these patients had a documented chest X-ray in their medical record, and of these, only 25% of the CFP32 antiserum-positive patients had cavitary TB, thereby indicating that cavitary TB status did not increase the likelihood of having a positive anti-CFP32 serologic response. The anti-CFP32 positivity rate was similar in those with and those without a documented chest X-ray. To extend these observations to a population from India, a modified ELISA (without the primary coating MAb) was used to test sera from AFB smear-positive cavitary TB patients (n = 30) and PPD skin test-positive healthy controls (n = 29). In this set, 30% (9 of 30) of the cavitary TB patients and 3% (1 of 29) of the PPD skin test-positive controls were reactive to rCFP32 (P < 0.013, Fisher's exact test) (Fig. 8B). Hence, there was comparatively limited variation between the two TB patient populations. Combined, 32% of TB case patients (n = 65) and only 2.5% of healthy controls (n = 40) exhibited a significant serological response to E. coli-expressed rCFP32 (P < 0.003, Fisher's exact test). These data are in the range of those of other studies that examined humoral immunity to recombinant M. tuberculosis proteins, whereby, depending upon the antigen evaluated and its method of production, the sera of 12 to 58% of TB patients were found to contain specific antibodies (38, 41). Overall, the available data indicate that the spectrum of M. tuberculosis antigens recognized humorally varies dramatically between patients (41, 58). One contributing factor is the expansion of the repertoire of humoral specificities to M. tuberculosis antigens as TB progresses (2, 58). For example, serum antibodies to the IT-44-reactive antigen (i.e., CFP32) have been identified in cavitary TB patients but not in noncavitary TB patients or PPD-positive individuals by Western blotting (58). (The apparent inconsistency with regard to our detection of anti-CFP32 specificities in noncavitary TB patients may well be a factor of the different means of evaluation.) Moreover, the CFP32 spot was one of only 26 serum-reactive CF spots that were seen in the study by Samanich et al. (58). As such, CFP32 appears to be a promising humoral antigen for inclusion in a multiantigen serodiagnostic test and may provide a useful indicator of worsening disease. It is also important that significantly fewer patients recognize certain M. tuberculosis proteins when they are expressed in E. coli compared to the counterpart native proteins (59). Since E. coli-expressed rCFP32 was used in these evaluations as the antibody capture antigen, this may have been a factor in reducing the sensitivity of our assays for CFP32 humoral specificity.
FIG. 8.
Antiserum specificity to CFP32 is detectable in a significant proportion of human TB patients. (A) Cohort of patients living in Brazil. Sera from 35 active TB case patients, along with the sera of 11 healthy household contacts (seven PPD skin test negative and four PPD skin test positive), were used in an ELISA to identify human humoral specificity for CFP32. The serologic reactivity of the healthy controls was used to set the cutoff value above which samples were deemed positive (mean [M] A450 + 1.5 standard deviation). The results of statistical analysis of the data are shown (P < 0.05, Fisher's exact test). (B) Cohort of patients living in India. Sera from 30 active TB case patients, as well as from 29 PPD-positive controls, were used in a variant ELISA to confirm the existence of human humoral response to CFP32. The serologic reactivity of the healthy PPD-positive controls was used to set the cutoff value (mean A490 + 1.5 standard deviation). The results of statistical analysis of the data are shown (P < 0.013, Fisher's exact test).
Detection of CFP32 in lung sputum samples of TB patients by ELISA and positive correlation of CFP32 levels with IL-10.
If immunogenic, a candidate in vivo-expressed M. tuberculosis antigen should be present at the site of disease. To detect lung CFP32, the induced sputa from patients in Brazil, who presented for diagnostic workups for “lung disease or suspected TB,” were tested for the presence of CFP32 by ELISA. This cohort included patients with TB (n = 41), suspected TB (n = 16), or OLD (n = 18), as defined in Materials and Methods. Sequential samples were taken at days 0, 15, 30, 60, and 180 following first presentation with one to five visits per patient. A significant number of TB patients had detectable amounts of CFP32 in their sputum. In contrast to the TB cases, none of the patients with OLD had detectable CFP32 in their sputum, resulting in a specificity of 100%. In total, 56% (23 of 41) of TB case patients were positive for CFP32 in at least one sample (P < 0.0001, Fisher's exact test), of which 32% (13 of 41) were positive at study entry for CFP32 (P = 0.0057, Fisher's exact test). Overall, 59% (10 of 17) of cavitary TB patients were CFP32 sputum positive in at least one sample and 30% (42 of 140) of all samples from TB patients were positive for CFP32 (P = 0.0007, Fisher's exact test) (Fig. 9A). Among TB cases, AFB smear was positive in 54% (22 of 41) while culture was positive in 80% (33 of 41). Sputum CFP32 was detected in at least one sample in 61% (20 of 33) of culture-positive TB cases and 50% (3 of 6) culture-negative (n = 5) and culture-unavailable (n = 1) TB cases. When cross-correlated with AFB smear, 64% (14 of 22) of AFB smear-positive and 47% (9 of 19) of AFB smear-negative case patients had detectable sputum CFP32. There were only five TB case patients who were AFB and culture negative and whose diagnosis was based on treatment response with resolution of clinical and radiological features of TB. Of these case patients, 40% (two of five) were positive for CFP32. In the suspected TB category, for whom follow-up data were not available to establish a diagnosis, CFP32 was positive in 56% (9 of 16) of cases. For these experiments the standard curve of the CFP32 ELISA was consistently linear and sensitive to the level of ∼5 to 10 pg/ml (data not shown). The data strongly support the idea that CFP32 is present in the diseased lung. Although the function of CFP32 here remains unknown, we speculate that it may contribute to the pathobiology of M. tuberculosis. However, as proposed for other M. tuberculosis CF proteins (5, 12, 52), the actions of CFP32 could include both direct enzymatic activity upon host cells or structures and/or bacterial components and antigenicity-based local tissue damage via immunohyperstimulation. By virtue of its expression in vivo and given that CFP32 could be detected in the suspected TB subset of patients as well as a proportion of AFB smear-negative and/or culture-negative patients, these data further present CFP32 as a strong candidate antigen for inclusion in a next-generation diagnostic strategy as a marker of increasing bacterial burden or as an indication of the effectiveness of TB pharmacologic therapy.
FIG. 9.
CFP32 is detectable in the lungs of a significant number of TB patients and is positively correlated with IL-10 but not IFN-γ levels. (A) Excess induced lung sputum not used for diagnostic purposes was tested for the presence of CFP32 by ELISA. Donors were either TB case patients (TB; defined as having a positive solid medium culture or treatment response with resolution of clinical and radiological features of TB) (n = 41; 140 samples), suspected TB case patients (Suspected TB; defined as patients with clinical and/or radiological features compatible with TB for whom cultures were negative, contaminated, or not available and who had insufficient follow-up or had prior TB without sufficient follow-up) (n = 16; 17 samples), or non-TB case patients with other lung diseases (OLD; defined as those patients who were AFB smear and TB culture negative, for whom another diagnosis was established, and/or who showed clinical improvement after a short course of non-TB antibiotics) (n = 18; 25 samples). The mean (M) value for the non-TB group was used to set the cutoff value (M + 2.5 standard deviations) above which samples were deemed positive. Statistical analyses found a significant difference between TB and non-TB cases (P = 0.0007, Fisher's exact test). (B) CFP32 levels were then correlated with IL-10 and IFN-γ in matched sputum samples by ELISA; n, number of samples. Linear regression analyses revealed that 60% of the variance in the measured amounts of sputum CFP32 corresponds to variation in IL-10 levels (r2 = 0.60, P < 0.0001).
Previously, the coexpression of mRNA for IFN-γ with IL-10, IL-2, and inducible nitric oxide synthase, by lung cells from patients with active pulmonary TB, was described [48; M. D. Bonecini-Almeida, J. R. Lapa e Silva, S. Nicholson, J. Geng, N. Boechat, C. Linhares, L. Rego, and A. L. Kritski, abstract from the American Thoracic Society Annual Meeting 1997, Am. J. Respir. Crit. Care Med. 155(Suppl.):A441, 1997]. To further dissect the in vivo human immunologic parameters associated with TB, ELISA was done to quantify the IL-10 and IFN-γ in the same induced sputum samples from TB patients (n = 34) previously assayed for CFP32. A significant correlation between CFP32 and IL-10 in the sputum of patients was found by linear regression analysis (n = 112 samples; r2 = 0.60, P < 0.0001) (Fig. 9B subpanel i). No convincing association was identified between CFP32 and IFN-γ (n = 125 samples) (Fig. 9B subpanel ii) nor between IL-10 and IFN-γ (n = 110 samples; data not shown). IFN-γ is regarded as a pivotal cytokine in the protective immune response against M. tuberculosis infection, acting as the major mediator of macrophage activation and as a crucial component in the development of specific counter-M. tuberculosis adaptive immunity (16). IL-10, on the other hand, is a pleiotropic immunosuppressive cytokine that opposes many IFN-γ-mediated effects including macrophage-mediated mycobacteriocidal activity (10, 19, 24). Indeed, evidence is accumulating linking IL-10 to the failure in immunity that results in the progression to active TB. For example, infection studies with either IL-10 gene-knockout mice or IL-10 transgenic mice have shown that IL-10 is an inhibitor of anti-tubercle bacillus responses (34, 45, 46, 71). In humans, healthy persons reactive to PPD produce high concentrations of IFN-γ from M. tuberculosis antigen-stimulated peripheral blood mononuclear cells (PBMCs) while TB patients with severe disease, and without reactivity to PPD (i.e., anergized), exhibit impaired IFN-γ production in association with increased IL-10 (11, 17). Moreover, increased levels of IL-10, in the presence of IFN-γ, have been detected in the serum of TB patients, as well as from ex vivo M. tuberculosis antigen-stimulated PBMCs of TB patients (20, 49, 63, 72). In this report, we found a novel association of bacterial physiological activity (as reflected by CFP32 antigen levels) and IL-10 production in the lungs of patients with the failure of counter-M. tuberculosis immunity (which was also notably coincident with continued IFN-γ production). These data contrast with those of Barnes et al. (7), who found elevated IFN-γ in association with IL-10 in the pleural fluid of patients with tuberculous pleuritis (a form of TB that resolves without chemotherapy). Together these data suggest that the immunosuppressive actions of IL-10 may come to predominate and eliminate the protective immune system-activating properties of IFN-γ in the lungs of persons with advanced TB. The mechanism underlying the elevated IL-10 levels is likely multifactorial and involves contributions from both the host and the pathogen. In this regard, naïve human PBMCs, monocytes, and dendritic cells are known to produce IL-10 when stimulated with M. tuberculosis or with its cell wall constituents (6, 10, 29, 69). Hence, as an immune evasion strategy, M. tuberculosis may deliberately induce the production of IL-10 and thereby depress cellular responses to IFN-γ and promote M. tuberculosis intramacrophage survival (10). In relation to this idea, increased local IL-10 is also thought to promote the development of the IL-10-producing CD4+ T regulatory 1 (Tr1) cells (1). In fact, the majority of bronchoalveolar lavage-derived CD4+ T-cell clones from TB patients are reminiscent of Tr1 cells (25; although in that study they also produced IFN-γ), and Tr1-like cells have been implicated in the PPD anergy of TB patients (11, 17). Therefore, Tr1 cells may act in their turn to further stifle local anti-M. tuberculosis innate and T-cell-mediated adaptive immune responses and potentiate a positive feedback loop of IL-10 secretion that supports M. tuberculosis persistence and/or reactivation. Since IL-10 is also known to promote B-cell production of antibody (56), the increased lung IL-10 levels may be responsible for the expansion of anti-M. tuberculosis serum specificities and enhanced antibody titers as TB progresses (2, 56). Therefore, lung IL-10 level bears further investigation as an immunological correlate for the development of pulmonary TB.
In summary, this work adds cfp32 to the growing list of M. tuberculosis genes proven to code for an expressed protein. This CFP32 protein appears to be a bimodular glyoxalase localized to both the cytosolic and CF compartments of M. tuberculosis. Although the biological role(s) of CFP32 remains to be elucidated, the accumulated data suggest that CFP32 is an important biofactor since it is MTC restricted, the nonpolymorphic cfp32 gene is present in all M. tuberculosis strains that have been evaluated, and it is expressed in the lungs of a significant proportion of TB patients in addition to being a recognized humoral antigen. That CFP32 levels in the lungs of active TB patients correlated with measured IL-10, but not IFN-γ, levels supports the hypothesis that M. tuberculosis-stimulated local IL-10 secretion precipitates the immunodysregulation that contributes to the success of M. tuberculosis as a human pathogen. Determining the role of CFP32, if any, in this particular pathogenic strategy of M. tuberculosis is a priority interest of our laboratory.
Acknowledgments
We thank Howard Doo, Kelley Sookraj, Krishna Menon, Patricia Lago, Vera Batista, and Brianna Holland for technical assistance and Albert Ko for aiding the statistical analyses. We are also grateful to Lee W. Riley, Sabine Ehrt, and Warren D. Johnson for suggestions, support, and encouragement.
Funding support was provided by NIH grants R0-1 AI39606 and R0-1 HL61960 (J.L.H.), NIH NIAID contract N01 AI-75320 (J.T.B.), NIH Fogarty International Center Training grant (FICTG) D43 TW00018, a grant from the Coordenação de Aperfeiçoamento de Pessoal de Nivel Superior (CAPES; Ministry of Education-Brazil), and a grant from the Laura Cook Hull Trust Fund (LCHTF) (Warren D. Johnson, Principal Investigator). R.C.H. was supported by the LCHTF, H.Z. was a FICTG trainee, and L.C.O.L. was an FICTG and CAPES trainee. S.L. was supported by a VA merit award. M.B.C., A.L.K., and J.R.L.S. were funded in Brazil by the following grants: Brazilian TB Research Network 62.0055/01-4-PADCT III/MILLENIUM (CNPq/Brazilian Research Council and World Bank; M.B.C., A.L.K., J.R.L.S.), “Excellence Research Nuclei for TB Control” 66.1028/1998-4 (PRONEX/Brazilian Research Council; A.L.K., J.R.L.S.), “Scientists of Our State” 2000 and 2003 (Rio de Janeiro Research Council/FAPERJ; A.L.K., J.R.L.S.), “Small Grants Program” (Rio de Janeiro Research Council/FAPERJ; M.B.C.), and “Small Grants Program” (Fundação Universitária José Bonifácio/FUJB; J.R.L.S.).
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Akbari, O., R. H. DeKruyff, and D. T. Umetsu. 2001. Pulmonary dendritic cells producing IL-10 mediate tolerance induced by respiratory exposure to antigen. Nat. Immunol. 2:725-731. [DOI] [PubMed] [Google Scholar]
- 2.Amara, R. R., and V. Satchidanandam. 1996. Analysis of a genomic DNA expression library of Mycobacterium tuberculosis using tuberculosis patient sera: evidence for modulation of host immune response. Infect. Immun. 64:3765-3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen, P. 1997. Host responses and antigens involved in protective immunity to Mycobacterium tuberculosis. Scand. J. Immunol. 45:115-131. [DOI] [PubMed] [Google Scholar]
- 4.Ando, N., K. Ueda, and S. Horinouchi. 1997. A Streptomyces griseus gene (sgaA) suppresses the growth disturbance caused by high osmolality and a high concentration of A-factor during early growth. Microbiology 143:2715-2723. [DOI] [PubMed] [Google Scholar]
- 5.Armitige, L. Y., C. Jagannath, A. R. Wanger, and S. J. Norris. 2000. Disruption of the genes encoding antigen 85A and antigen 85B of Mycobacterium tuberculosis H37Rv: effect on growth in culture and in macrophages. Infect. Immun. 68:767-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnes, P. F., D. Chatterjee, J. S. Abrams, S. Lu, E. Wang, M. Yamamura, P. J. Brennan, and R. L. Modlin. 1992. Cytokine production induced by Mycobacterium tuberculosis lipoarabinomannan. Relationship to chemical structure. J. Immunol. 149:541-547. [PubMed] [Google Scholar]
- 7.Barnes, P. F., S. Lu, J. S. Abrams, E. Wang, M. Yamamura, and R. L. Modlin. 1993. Cytokine production at the site of disease in human tuberculosis. Infect. Immun. 61:3482-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhaskar, S., S. P. Khanna, and R. Mukherjee. 2000. Isolation, purification and immunological characterization of novel low molecular weight protein antigen CFP 6 from culture filtrate of M. tuberculosis. Vaccine 18:2856-2866. [DOI] [PubMed] [Google Scholar]
- 9.Boesen, H., B. N. Jensen, T. Wilcke, and P. Andersen. 1995. Human T-cell responses to secreted antigen fractions of Mycobacterium tuberculosis. Infect. Immun. 63:1491-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonecini-Almeida, M. G., S. Chitale, I. Boutsikakis, J. Geng, H. Doo, S. He, and J. L. Ho. 1998. Induction of in vitro human macrophage anti-Mycobacterium tuberculosis activity: requirement for IFN-gamma and primed lymphocytes. J. Immunol. 160:4490-4499. [PubMed] [Google Scholar]
- 11.Boussiotis, V. A., E. Y. Tsai, E. J. Yunis, S. Thim, J. C. Delgado, C. C. Dascher, A. Berezovskaya, D. Rousset, J. M. Reynes, and A. E. Goldfeld. 2000. IL-10-producing T cells suppress immune responses in anergic tuberculosis patients. J. Clin. Investig. 105:1317-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brodin, P., K. Eiglmeier, M. Marmiesse, A. Billault, T. Garnier, S. Niemann, S. T. Cole, and R. Brosch. 2002. Bacterial artificial chromosome-based comparative genomic analysis identifies Mycobacterium microti as a natural ESAT-6 deletion mutant. Infect. Immun. 70:5568-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camus, J. C., M. J. Pryor, C. Medigue, and S. T. Cole. 2002. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology 148:2967-2973. [DOI] [PubMed] [Google Scholar]
- 15.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentiles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jegels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 16.Collins, H. L., and S. H. Kaufmann. 2001. The many faces of host responses to tuberculosis. Immunology 103:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delgado, J. C., E. Y. Tsai, S. Thim, A. Baena, V. A. Boussiotis, J. M. Reynes, S. Sath, P. Grosjean, E. J. Yunis, and A. E. Goldfeld. 2002. Antigen-specific and persistent tuberculin anergy in a cohort of pulmonary tuberculosis patients from rural Cambodia. Proc. Natl. Acad. Sci. USA 99:7576-7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demissie, A., P. Ravn, J. Olobo, T. M. Doherty, T. Eguale, M. Geletu, W. Hailu, P. Andersen, and S. Britton. 1999. T-cell recognition of Mycobacterium tuberculosis culture filtrate fractions in tuberculosis patients and their household contacts. Infect. Immun. 67:5967-5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dickensheets, H. L., S. L. Freeman, M. F. Smith, and R. P. Donnelly. 1997. Interleukin-10 upregulates tumor necrosis factor receptor type-II (p75) gene expression in endotoxin-stimulated human monocytes. Blood 90:4162-4171. [PubMed] [Google Scholar]
- 20.Dlugovitzky, D., A. Torres-Morales, L. Rateni, M. A. Farroni, C. Largacha, O. Molteni, and O. Bottasso. 1997. Circulating profile of Th1 and Th2 cytokines in tuberculosis patients with different degrees of pulmonary involvement. FEMS Immunol. Med. Microbiol. 18:203-207. [DOI] [PubMed] [Google Scholar]
- 21.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 22.Eng, J. K., A. L. McCormack, and J. R. Yates. 1994. A approach to correlate tandem mass-spectral data of peptides with amino-acid sequences in a protein database. J. Am. Soc. Mass. Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez, J., L. Andrews, and S. M. Mische. 1994. An improved procedure for enzymatic digestion of polyvinylidene difluoride-bound proteins for internal sequence analysis. Anal. Biochem. 218:112-117. [DOI] [PubMed] [Google Scholar]
- 24.Flesch, I. E., J. H. Hess, I. P. Oswald, and S. H. Kaufmann. 1994. Growth inhibition of Mycobacterium bovis by IFN-gamma stimulated macrophages: regulation by endogenous tumor necrosis factor-alpha and by IL-10. Int. Immunol. 6:693-700. [DOI] [PubMed] [Google Scholar]
- 25.Gerosa, F., C. Nisii, S. Righetti, R. Micciolo, M. Marchesini, A. Cazzadori, and G. Trinchieri. 1999. CD4+ T cell clones producing both interferon-gamma and interleukin-10 predominate in bronchoalveolar lavages of active pulmonary tuberculosis patients. Clin. Immunol. 92:224-234. [DOI] [PubMed] [Google Scholar]
- 26.Glickman, M. S., and W. R. Jacobs, Jr. 2001. Microbial pathogenesis of Mycobacterium tuberculosis: dawn of a discipline. Cell 104:477-485. [DOI] [PubMed] [Google Scholar]
- 27.Harth, G., and M. A. Horwitz. 1997. Expression and efficient export of enzymatically active Mycobacterium tuberculosis glutamine synthetase in Mycobacterium smegmatis and evidence that the information for export is contained within the protein. J. Biol. Chem. 272:22728-22735. [DOI] [PubMed] [Google Scholar]
- 28.Harth, G., and M. A. Horwitz. 1999. Export of recombinant Mycobacterium tuberculosis superoxide dismutase is dependent upon both information in the protein and mycobacterial export machinery. A model for studying export of leaderless proteins by pathogenic mycobacteria. J. Biol. Chem. 274:4281-4292. [DOI] [PubMed] [Google Scholar]
- 29.Henderson, R. A., S. C. Watkins, and J. L. Flynn. 1997. Activation of human dendritic cells following infection with Mycobacterium tuberculosis. J. Immunol. 159:635-643. [PubMed] [Google Scholar]
- 30.Heym, B., Y. Zhang, S. Poulet, D. Young, and S. T. Cole. 1993. Characterization of the katG gene encoding a catalase-peroxidase required for the isoniazid susceptibility of Mycobacterium tuberculosis. J. Bacteriol. 175:4255-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horwitz, M. A., B. W. Lee, B. J. Dillon, and G. Harth. 1995. Protective immunity against tuberculosis induced by vaccination with major extracellular proteins of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 92:1530-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howard, S., S. He, and S. G. Withers. 1998. Identification of the active site nucleophile in jack bean α-mannosidase using 5-fluoro-β-l-gulosyl fluoride. J. Biol. Chem. 273:2067-2072. [DOI] [PubMed] [Google Scholar]
- 33.Huard, R. C., L. C. de Oliveira Lazzarini, W. R. Butler, D. van Soolingen, and J. L. Ho. 2003. A PCR-based method to differentiate the subspecies of the Mycobacterium tuberculosis complex on the basis of genomic deletions. J. Clin. Microbiol. 41:1637-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobs, M., N. Brown, N. Allie, R. Gulert, and B. Ryffel. 2000. Increased resistance to mycobacterial infection in the absence of interleukin-10. Immunology 100:494-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jungblut, P. R., U. E. Schaible, H. J. Mollenkopf, U. Zimny-Arndt, B. Raupach, J. Mattow, P. Halada, S. Lamer, K. Hagens, and S. H. Kaufmann. 1999. Comparative proteome analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG strains: towards functional genomics of microbial pathogens. Mol. Microbiol. 33:1103-1117. [DOI] [PubMed] [Google Scholar]
- 36.Kane, M. M., and D. M. Mosser. 2001. The role of IL-10 in promoting disease progression in leishmaniasis. J. Immunol. 166:1141-1147. [DOI] [PubMed] [Google Scholar]
- 37.Kurepina, N. E., S. Sreevatsan, B. B. Plikaytis, P. J. Bifani, N. D. Connell, R. J. Donnelly, D. van Soolingen, J. M. Musser, and B. N. Kreiswirth. 1998. Characterization of the phylogenetic distribution and chromosomal insertion sites of five IS6110 elements in Mycobacterium tuberculosis: non-random integration in the dnaA-dnaN region. Tuber. Lung Dis. 79:31-42. [DOI] [PubMed] [Google Scholar]
- 38.Lim, R. L., L. K. Tan, W. F. Lau, M. C. Ming, R. Dunn, H. P. Too, and L. Chan. 2000. Cloning and expression of immunoreactive antigens from Mycobacterium tuberculosis. Clin. Diagn. Lab. Immunol. 7:600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ljungqvist, L., A. Worsaae, and I. Heron. 1988. Antibody responses against Mycobacterium tuberculosis in 11 strains of inbred mice: novel monoclonal antibody specificities generated by fusions, using spleens from BALB.B10 and CBA/J mice. Infect. Immun. 56:1994-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lomovskaya, N., S. L. Otten, Y. Doi-Katayama, L. Fonstein, X. C. Liu, T. Takatsu, A. Inventi-Solari, S. Filippini, F. Torti, A. L. Colombo, and C. R. Hutchinson. 1999. Doxorubicin overproduction in Streptomyces peucetius: cloning and characterization of the dnrU ketoreductase and dnrV genes and the doxA cytochrome P-450 hydroxylase gene. J. Bacteriol. 181:305-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lyashchenko, K., R. Colangeli, M. Houde, H. Al Jahdali, D. Menzies, and M. L. Gennaro. 1998. Heterogeneous antibody responses in tuberculosis. Infect. Immun. 66:3936-3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKinney, J. D., K. Honer zu Bentrup, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 43.Mostowy, S., D. Cousins, J. Brinkman, A. Aranaz, and M. A. Behr. 2002. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J. Infect. Dis. 186:74-80. [DOI] [PubMed] [Google Scholar]
- 44.Mulder, M. A., H. Zappe, and L. M. Steyn. 1997. Mycobacterial promoters. Tuber. Lung Dis. 78:211-223. [DOI] [PubMed] [Google Scholar]
- 45.Murray, P. J., L. Wang, C. Onufryk, R. I. Tepper, and R. A. Young. 1997. T cell-derived IL-10 antagonizes macrophage function in mycobacterial infection. J. Immunol. 158:315-321. [PubMed] [Google Scholar]
- 46.Murray, P. J., and R. A. Young. 1999. Increased antimycobacterial immunity in interleukin-10-deficient mice. Infect. Immun. 67:3087-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Navas, J., B. Gonzalez-Zorn, N. Ladron, P. Garrido, and J. A. Vazquez-Boland. 2001. Identification and mutagenesis by allelic exchange of choE, encoding a cholesterol oxidase from the intracellular pathogen Rhodococcus equi. J. Bacteriol. 183:4796-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicholson, S., M. D. Bonecini-Almeida, J. R. Lapa e Silva, C. Nathan, Q. W. Xie, R. Mumford, J. R. Weidner, J. Calaycay, J. Geng, N. Boechat, C. Linhares, W. Rom, and J. L. Ho. 1996. Inducible nitric oxide synthase in pulmonary alveolar macrophages from patients with tuberculosis. J. Exp. Med. 183:2293-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Othieno, C., C. S. Hirsch, B. D. Hamilton, K. Wilkinson, J. J. Ellner, and Z. Toossi. 1999. Interaction of Mycobacterium tuberculosis-induced transforming growth factor beta and interleukin-10. Infect. Immun. 67:5730-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parish, T., B. G. Gordhan, R. A. McAdam, K. Duncan, V. Mizrahi, and N. G. Stoker. 1999. Production of mutants in amino acid biosynthesis genes of Mycobacterium tuberculosis by homologous recombination. Microbiology 145:3497-3503. [DOI] [PubMed] [Google Scholar]
- 51.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raynaud, C., G. Etienne, P. Peyron, M. A. Laneelle, and M. Daffe. 1998. Extracellular enzyme activities potentially involved in the pathogenicity of Mycobacterium tuberculosis. Microbiology 144:577-587. [DOI] [PubMed] [Google Scholar]
- 53.Rosenkrands, I., A. King, K. Weldingh, M. Moniatte, E. Moertz, and P. Andersen. 2000. Towards the proteome of Mycobacterium tuberculosis. Electrophoresis 21:3740-3756. [DOI] [PubMed] [Google Scholar]
- 54.Rosenkrands, I., P. B. Rasmussen, M. Carnio, S. Jacobsen, M. Theisen, and P. Andersen. 1998. Identification and characterization of a 29-kilodalton protein from Mycobacterium tuberculosis culture filtrate recognized by mouse memory effector cells. Infect. Immun. 66:2728-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosenkrands, I., K. Weldingh, S. Jacobsen, C. V. Hansen, W. Florio, I. Gianetri, and P. Andersen. 2000. Mapping and identification of Mycobacterium tuberculosis proteins by two-dimensional gel electrophoresis, microsequencing and immunodetection. Electrophoresis 21:935-948. [DOI] [PubMed] [Google Scholar]
- 56.Rousset, F., E. Garcia, T. Defrance, C. Peronne, N. Vezzio, D. H. Hsu, R. Kastelein, K. W. Moore, and J. Banchereau. 1992. Interleukin 10 is a potent growth and differentiation factor for activated human B lymphocytes. Proc. Natl. Acad. Sci. USA 89:1890-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruan, J., G. St. John, S. Ehrt, L. Riley, and C. Nathan. 1999. noxR3, a novel gene from Mycobacterium tuberculosis, protects Salmonella typhimurium from nitrosative and oxidative stress. Infect. Immun. 67:3276-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Samanich, K. M., J. T. Belisle, M. G. Sonnenberg, M. A. Keen, S. Zolla-Pazner, and S. Laal. 1998. Delineation of human antibody responses to culture filtrate antigens of Mycobacterium tuberculosis. J. Infect. Dis. 178:1534-1538. [DOI] [PubMed] [Google Scholar]
- 59.Samanich, K. M., M. A. Keen, V. D. Vissa, J. D. Harder, J. S. Spencer, J. T. Belisle, S. Zolla-Pazner, and S. Laal. 2000. Serodiagnostic potential of culture filtrate antigens of Mycobacterium tuberculosis. Clin. Diagn. Lab. Immunol. 7:662-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sambandamurthy, V. K., X. Wang, B. Chen, R. G. Russell, S. Derrick, F. M. Collins, S. L. Morris, and W. R. Jacobs. 2002. A pantothenate auxotroph of Mycobacterium tuberculosis is highly attenuated and protects mice against tuberculosis. Nat. Med. 8:1171-1174. [DOI] [PubMed] [Google Scholar]
- 61.Sieling, P. A., J. S. Abrams, M. Yamamura, P. Salgame, B. R. Bloom, T. H. Rea, and R. L. Modlin. 1993. Immunosuppressive roles for IL-10 and IL-4 in human infection. In vitro modulation of T cell responses in leprosy. J. Immunol. 150:5501-5510. [PubMed] [Google Scholar]
- 62.Smith, S. M., M. R. Klein, A. S. Malin, J. Sillah, K. Huygen, P. Andersen, K. P. McAdam, and H. M. Dockrell. 2000. Human CD8+ T cells specific for Mycobacterium tuberculosis secreted antigens in tuberculosis patients and healthy BCG-vaccinated controls in The Gambia. Infect. Immun. 68:7144-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song, C. H., H. J. Kim, J. K. Park, J. H. Lim, U. O. Kim, J. S. Kim, T. H. Paik, K. J. Kim, J. W. Suhr, and E. K. Jo. 2000. Depressed interleukin-12 (IL-12), but not IL-18, production in response to a 30- or 32-kilodalton mycobacterial antigen in patients with active pulmonary tuberculosis. Infect. Immun. 68:4477-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sonnenberg, M. G., and J. T. Belisle. 1997. Definition of Mycobacterium tuberculosis culture filtrate proteins by two-dimensional polyacrylamide gel electrophoresis, N-terminal amino acid sequencing, and electrospray mass spectrometry. Infect. Immun. 65:4515-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sorensen, A. L., S. Nagai, G. Houen, P. Andersen, and A. B. Andersen. 1995. Purification and characterization of a low-molecular-mass T-cell antigen secreted by Mycobacterium tuberculosis. Infect. Immun. 63:1710-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stockl, J., H. Vetr, O. Majdic, G. Zlabinger, E. Kuechler, and W. Knapp. 1999. Human major group rhinoviruses downmodulate the accessory function of monocytes by inducing IL-10. J. Clin. Investig. 104:957-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stylianou, E., P. Aukrust, D. Kvale, F. Muller, and S. S. Froland. 1999. IL-10 in HIV infection: increasing serum IL-10 levels with disease progression—down-regulatory effect of potent anti-retroviral therapy. Clin. Exp. Immunol. 116:115-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thoma-Uszynski, S., S. M. Kiertscher, M. T. Ochoa, D. A. Bouis, M. V. Norgard, K. Miyake, P. J. Godowski, M. D. Roth, and R. L. Modlin. 2000. Activation of toll-like receptor 2 on human dendritic cells triggers induction of IL-12, but not IL-10. J. Immunol. 165:3804-3810. [DOI] [PubMed] [Google Scholar]
- 70.Tullius, M. V., G. Harth, and M. A. Horwitz. 2001. High extracellular levels of Mycobacterium tuberculosis glutamine synthetase and superoxide dismutase in actively growing cultures are due to high expression and extracellular stability rather than to a protein-specific export mechanism. Infect. Immun. 69:6348-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Turner, J., M. Gonzalez-Juarrero, D. L. Ellis, R. J. Basaraba, A. Kipnis, I. M. Orme, and A. M. Cooper. 2002. In vivo IL-10 production reactivates chronic pulmonary tuberculosis in C57BL/6 mice. J. Immunol. 169:6343-6351. [DOI] [PubMed] [Google Scholar]
- 72.Verbon, A., N. Juffermans, S. J. Van Deventer, P. Speelman, H. Van Deutekom, and T. Van Der Poll. 1999. Serum concentrations of cytokines in patients with active tuberculosis (TB) and after treatment. Clin. Exp. Immunol. 115:110-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weldingh, K., I. Rosenkrands, S. Jacobsen, P. B. Rasmussen, M. J. Elhay, and P. Andersen. 1998. Two-dimensional electrophoresis for analysis of Mycobacterium tuberculosis culture filtrate and purification and characterization of six novel proteins. Infect. Immun. 66:3492-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.World Health Organization. 1996. Report of the tuberculosis epidemic. World Health Organization, Geneva, Switzerland.