Abstract
Aggressive primary tumors express transcriptional signatures that correlate with their metastatic propensity. A number of these signatures have been deployed in the clinic as risk stratification tools. However, the molecular basis of these clinically useful prognostic signatures has remained a largely unresolved area of controversy. We recently found that many prognostic signatures reflect the activity of the MYC oncogene, which in turn regulates tumor metastasis through specific effects on cancer cell invasion and migration. These findings offer a general framework for understanding the molecular basis of clinically prognostic transcriptional signatures and suggest potentially new avenues for studying metastasis.
Accumulating evidence has pointed to an intriguing model of tumor progression whereby some primary tumors are molecularly preconfigured to metastasize relatively early (rather than later) in their evolution (1). Several clinical and experimental observations support this idea: (i) many cancer patients have metastatic tumors without obvious primaries; (ii) circulating tumor cells or actual micrometastases can be found in patients with very early-stage tumors; and (iii) aggressive primary tumors express transcriptional profiles that correlate with propensity to eventual metastasis or are differentially expressed by metastatic lesions compared with more indolent tumors (1–3). Moreover, there is clear selection for oncogenic mutations that promote cancer cell proliferation and survival during tumor evolution, but the selective advantage of purely metastasis-promoting mutations remains unclear. This thinking has led to the interesting hypothesis that a primary tumor’s metastatic propensity may be dictated in large measure by molecular changes that occur early in tumorigenesis (and are reflected by the expression of prognostic signatures), rather than solely by metastasis-enabling mutations per se, which are thought to arise in rare metastatic precursors later in tumor progression (1, 4).
Over the past decade, independent groups have reported the identification of many different “poor prognosis” gene expression signatures, mainly in breast cancer, which were derived through transcriptional profiling in both human tumors and experimental systems (5). Curiously, these different signatures are independently prognostic in the same tumor data sets but overlap minimally with respect to component genes. Additionally, many signature genes relate to cell proliferation, which raises the speculation that gene expression profiles might be useful biomarkers for risk stratification in cancer patients but only indirectly relate to the molecular mechanisms that specifically regulate cancer cell metastasis. Against this backdrop, we recently found that 13 different “poor outcome” gene expression signatures are coordinately regulated by the MYC oncogene in breast cancer cells. In addition, MYC is specifically necessary for the invasion and metastasis of these cells in experimental xenografts independent of its effects on proliferation and survival (5). These results suggest a model whereby early-stage primary tumors that express poor-prognosis transcriptional signatures have high MYC activity that directly contributes to their metastatic spread.
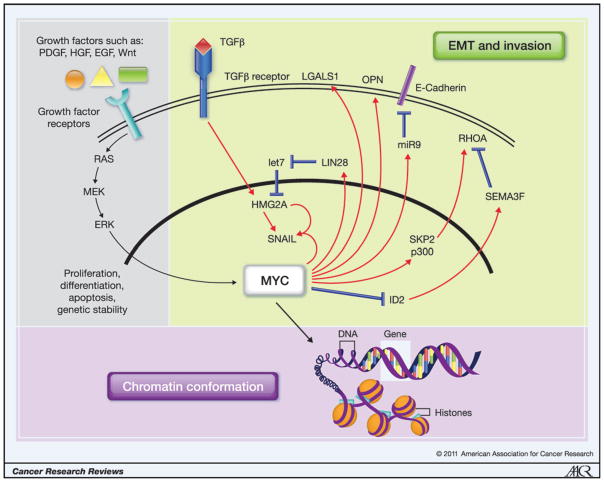
The MYC transcription factor is one of the most important somatically mutated oncogenes in human cancer. Recent studies suggest that inherited polymorphisms on 8q24 (near the MYC locus), which powerfully modify solid tumor predisposition, also influence MYC transcript expression (6). Somatic amplification and overexpression of MYC is seen in both high-grade premalignancy and invasive tumors and is associated with poor outcome in different human tumor types (7–12). Furthermore, many transforming oncogenes ultimately drive MYC expression either directly or indirectly, thus assuring that deregulation of the MYC pathway is one of the most common features of human tumorigenesis. The MYC oncoprotein can confer a selective advantage on cancer cells by promoting proliferation, cell survival, differentiation blockade, genetic instability, and angiogenesis, all of which may indirectly contribute to metastasis (Fig. 1; refs. 13–15). Cell proliferation, survival, and genetic instability promoted by MYC presumably result in more cancer cells with unstable genomes and, therefore, a higher likelihood of further mutation contributing to progression.
Figure 1.
MYC has a myriad of different functions. Its role in proliferation, differentiation, survival, and genetic stability has been studied extensively and is depicted summarily. More recent findings have outlined mechanisms by which MYC directly regulates genes involved in cell migration, invasion, and the epithelial-to-mesenchymal transition (EMT). (green-shaded box). Finally, the effect of MYC on chromatin conformation may set a “metastasis-enabling” epigenomic landscape (purple-shaded box); EGF, epidermal growth factor; ERK, extracellular signal regulated kinase (gray-shaded box). HGF, hepatocyte growth factor; MEK, MAP/ERK kinase; PDGF, platelet derived growth factor.
Aside from these indirect functions, however, it is becoming increasingly clear that MYC may also directly control cellular invasion and migration and, thus, metastasis, by regulating the expression of specific downstream programs. For example, MYC can regulate the epithelial-to-mesenchymal transition (EMT) that is necessary for cellular invasion and migration in some contexts. MYC does this by promoting TGFβ-mediated activation of the SNAIL transcription factor, both directly and indirectly though a microRNA network involving a LIN28B/let-7/HMGA2 cascade (Fig. 1; refs. 16–22). Importantly, however, we found that RNA interference (RNAi)–mediated MYC knockdown in highly metastatic MDA-MB-231 breast cancer cells disrupts cellular invasion, migration, and metastasis in vivo, independent of effects on EMT. This suggests that MYC may also regulate metastasis-promoting mechanisms beyond EMT (5). Consistent with this idea, MYC can regulate cell-cell–matrix interactions through transcriptional activation of LGALS1, which is a β-galactosidase–binding protein that promotes cell migration and invasion (23). Similarly, OPN is a MYC-regulated integrin-binding ligand that has been widely implicated in stimulating cancer cell migration and invasion (24, 25). MYC also cooperates with SKP2 to recruit MIZ1 and p300 into a transcriptional complex that activates RhoA, which is necessary for migration, invasion, and lung metastasis in vivo (26). In addition, MYC repression of ID2 inhibits expression of the secreted protein SEMA3F, which has been found to increase cell migration and invasion through RhoA activation and modulation of the actin cytoskeleton (Fig. 1; refs. 27–29). Clearly, more work is needed to comprehensively understand all MYC transcriptional targets that can directly contribute to metastasis.
Although MYC is necessary for the invasive and metastatic behavior of cancer cells, it seems to be insufficient (5, 26). Furthermore, MYC promotes but, paradoxically, can also retard migration depending on cell type (30). These and other observations suggest that MYC likely cooperates with other genes (particularly RAS pathway components) to promote both the early (e.g., invasion and migration) and late (e.g., seeding) phases of metastatic progression (15, 31–33). Systematic and unbiased approaches should increasingly reveal the spectrum of cellular and molecular contexts in which MYC promotes distant metastasis. Furthermore, MYC-driven primary tumors seem to remain dependent on this oncogene even after becoming established (34). If MYC-driven metastatic tumors also remain dependent (currently, an unanswered question), then strategies to target MYC in cancer patients with advanced disease may prove to be clinically useful.
Beyond direct transcriptional regulation of metastasis-relevant target genes, MYC may also play a more global role in regulating the metastatic phenotype. MYC is a powerful cellular reprogramming factor that determines normal and cancer cell differentiation (35). Presumably, cancer cells in an undifferentiated, more stem cell–like state might more easily migrate to and seed distant sites. Along these lines, MYC regulates the expression of different stem cell–associated transcriptional profiles (5, 33). How MYC regulates cellular differentiation is still not completely clear. However, genome-wide chromatin immunoprecipitation (ChIP) profiling studies have recently shown that MYC binds widely across the human genome (36). Other recent studies also suggest that MYC-directed transcriptional complexes containing histone-modifying enzymes may, in part, promote cellular reprogramming by uniquely inducing genome-wide changes in chromatin conformation (36–39). These findings suggest a fascinating but still speculative model. Perhaps MYC contributes to metastasis by globally altering the epigenomic landscape of cancer cells, thus providing a permissive molecular context in which additional, cooperative molecular alterations can specifically promote different aspects of the metastatic phenotype. If this speculative model proves to be correct, a deeper understanding of MYC complexes that promote metastasis in this way may provide new opportunities to target this important but currently “undruggable” cancer pathway. Further work will be required to fully explore this model.
Acknowledgments
We apologize to the many, many MYC investigators whose work we were unable to fully cite in this minireview owing to space restrictions.
Footnotes
Disclosure of Potential Conflicts of Interest
S. Ramaswamy is a recipient of a commercial research grant by Astra Zeneca.
References
- 1.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 2.Pantel K, Alix-Panabières C, Riethdorf S. Cancer micrometastases. Nat Rev Clin Oncol. 2009;6:339–51. doi: 10.1038/nrclinonc.2009.44. [DOI] [PubMed] [Google Scholar]
- 3.van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 4.Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418:823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- 5.Wolfer A, Wittner BS, Irimia D, Flavin RJ, Lupien M, Gunawardane RN, et al. MYC regulation of a “poor-prognosis” metastatic cancer cell state. Proc Natl Acad Sci U S A. 2010;107:3698–703. doi: 10.1073/pnas.0914203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–4. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aulmann S, Adler N, Rom J, Helmchen B, Schirmacher P, Sinn HP. c-myc amplifications in primary breast carcinomas and their local recurrences. J Clin Pathol. 2006;59:424–8. doi: 10.1136/jcp.2005.029264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borg A, Baldetorp B, Ferno M, Olsson H, Sigurdsson H. c-myc amplification is an independent prognostic factor in postmenopausal breast cancer. Int J Cancer. 1992;51:687–91. doi: 10.1002/ijc.2910510504. [DOI] [PubMed] [Google Scholar]
- 10.Bourhis J, Le MG, Barrois M, Gerbaulet A, Jeannel D, Duvillard P, et al. Prognostic value of c-myc proto-oncogene overexpression in early invasive carcinoma of the cervix. J Clin Oncol. 1990;8:1789–96. doi: 10.1200/JCO.1990.8.11.1789. [DOI] [PubMed] [Google Scholar]
- 11.Deming SL, Nass SJ, Dickson RB, Trock BJ. C-myc amplification in breast cancer: a meta-analysis of its occurrence and prognostic relevance. Br J Cancer. 2000;83:1688–95. doi: 10.1054/bjoc.2000.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozma L, Kiss I, Szakáll S, Ember I. Investigation of c-myc oncogene amplification in colorectal cancer. Cancer Lett. 1994;81:165–9. doi: 10.1016/0304-3835(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 13.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–99. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 14.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–56. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rapp UR, Korn C, Ceteci F, Karreman C, Luetkenhaus K, Serafin V, et al. MYC is a metastasis gene for non-small-cell lung cancer. PLoS One. 2009;4:e6029. doi: 10.1371/journal.pone.0006029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, et al. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587–91. doi: 10.1158/0008-5472.CAN-08-0264. [DOI] [PubMed] [Google Scholar]
- 17.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci U S A. 2009;106:3384–9. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dangi-Garimella S, Yun J, Eves EM, Newman M, Erkeland SJ, Hammond SM, et al. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009;28:347–58. doi: 10.1038/emboj.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith AP, Verrecchia A, Fagà G, Doni M, Perna D, Martinato F, et al. A positive role for Myc in TGFbeta-induced Snail transcription and epithelial-to-mesenchymal transition. Oncogene. 2009;28:422–30. doi: 10.1038/onc.2008.395. [DOI] [PubMed] [Google Scholar]
- 21.Thuault S, Tan EJ, Peinado H, Cano A, Heldin CH, Moustakas A. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J Biol Chem. 2008;283:33437–46. doi: 10.1074/jbc.M802016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–8. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan S, Zhou C, Lou X, Xiao Z, Zhu H, Wang Q, et al. PTTG over-expression promotes lymph node metastasis in human esophageal squamous cell carcinoma. Cancer Res. 2009;69:3283–90. doi: 10.1158/0008-5472.CAN-08-0367. [DOI] [PubMed] [Google Scholar]
- 24.Bellahcène A, Castronovo V, Ogbureke KU, Fisher LW, Fedarko NS. Small integrin-binding ligand N-linked glycoproteins (SIBLINGs): multifunctional proteins in cancer. Nat Rev Cancer. 2008;8:212–26. doi: 10.1038/nrc2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez C, Churchman M, Freeman T, Ilyas M. Osteopontin provides early proliferative drive and may be dependent upon aberrant c-myc signalling in murine intestinal tumours. Exp Mol Pathol. 2010;88:272–7. doi: 10.1016/j.yexmp.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Chan CH, Lee SW, Li CF, Wang J, Yang WL, Wu CY, et al. Deciphering the transcriptional complex critical for RhoA gene expression and cancer metastasis. Nat Cell Biol. 2010;12:457–67. doi: 10.1038/ncb2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coma S, Amin DN, Shimizu A, Lasorella A, Iavarone A, Klagsbrun M. Id2 promotes tumor cell migration and invasion through transcriptional repression of semaphorin 3F. Cancer Res. 2010;70:3823–32. doi: 10.1158/0008-5472.CAN-09-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimizu A, Mammoto A, Italiano JE, Jr, Pravda E, Dudley AC, Ingber DE, et al. ABL2/ARG tyrosine kinase mediates SEMA3F-induced RhoA inactivation and cytoskeleton collapse in human glioma cells. J Biol Chem. 2008;283:27230–8. doi: 10.1074/jbc.M804520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zippo A, De Robertis A, Serafini R, Oliviero S. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat Cell Biol. 2007;9:932–44. doi: 10.1038/ncb1618. [DOI] [PubMed] [Google Scholar]
- 30.Cappellen D, Schlange T, Bauer M, Maurer F, Hynes NE. Novel c-MYC target genes mediate differential effects on cell proliferation and migration. EMBO Rep. 2007;8:70–6. doi: 10.1038/sj.embor.7400849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adler AS, Lin M, Horlings H, Nuyten DS, van de Vijver MJ, Chang HY. Genetic regulators of large-scale transcriptional signatures in cancer. Nat Genet. 2006;38:421–30. doi: 10.1038/ng1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Podsypanina K, Politi K, Beverly LJ, Varmus HE. Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proc Natl Acad Sci U S A. 2008;105:5242–7. doi: 10.1073/pnas.0801197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–44. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–83. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 36.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–61. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–34. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–77. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc Natl Acad Sci U S A. 2007;104:13028–33. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]