This article presents an overview of protein-engineering methods designed to enhance crystallizability and discusses a number of examples of their successful application.
Keywords: protein engineering
Abstract
Until recently, protein crystallization has mostly been regarded as a stochastic event over which the investigator has little or no control. With the dramatic technological advances in synchrotron-radiation sources and detectors and the equally impressive progress in crystallographic software, including automated model building and validation, crystallization has increasingly become the rate-limiting step in X-ray diffraction studies of macromolecules. However, with the advent of recombinant methods it has also become possible to engineer target proteins and their complexes for higher propensity to form crystals with desirable X-ray diffraction qualities. As most proteins that are under investigation today are obtained by heterologous overexpression, these techniques hold the promise of becoming routine tools with the potential to transform classical crystallization screening into a more rational high-success-rate approach. This article presents an overview of protein-engineering methods designed to enhance crystallizability and discusses a number of examples of their successful application.
1. Introduction
Following the dawn of recombinant technology brought about by the groundbreaking overexpression of synthetic genes coding for insulin and somatostatin in Escherichia coli (Goeddel et al., 1979 ▶; Itakura et al., 1977 ▶) and the subsequent discovery of the polymerase chain reaction (PCR; Mullis et al., 1986 ▶; Saiki et al., 1985 ▶, 1988 ▶), macromolecular crystallography was freed of its longstanding dependence on purified native protein samples for crystallization. Heterologous expression made it possible to generate samples of proteins and complexes that are found in only small or trace amounts in living cells and to engineer large and unstable proteins so that isolated domains or modified forms can be made available for crystallization. At the same time, the effort required for protein purification was dramatically reduced by the use of fusion proteins and affinity tags (Brewer et al., 1991 ▶; Sassenfeld, 1990 ▶; Malhotra, 2009 ▶). As a consequence, the overwhelming majority of samples used today for crystallization are recombinantly derived proteins. However, even though material for crystallization is more easily available, the preparation of single well diffracting crystals of the target macromolecule is still a time-consuming challenge.
Historically, two complementary approaches to protein crystallization were developed in parallel. Firstly, natural variations in the amino-acid sequences of homologues from different species were exploited to identify a target with suitable crystallization properties during the purification procedure (Kendrew et al., 1954 ▶; Campbell et al., 1972 ▶). The second approach was to extensively screen a specific target protein against a range of diverse precipitating agents, buffers and additives until the right conditions for crystallization were identified (Carter & Carter, 1979 ▶; Jancarik & Kim, 1991 ▶). These strategies remain the pillars of contemporary macromolecular crystallization. However, as the palette of molecular biology techniques expanded to include site-directed mutagenesis, ligation-independent cloning and other tools, it became possible to modify proteins with relative ease with the specific purpose of enhancing their propensity to crystallize or improving the diffraction quality of the resulting crystals. The early proof-of-principle of these capabilities was the crystallization of an engineered variant of human H-ferritin in which a single-site mutation, K86Q, was introduced to duplicate a crystal contact mediated by Cd2+ ions in the crystal structure of the homologous rat L-ferritin (Lawson et al., 1991 ▶).
In this review, current progress in the methodologies of protein engineering used to enhance the crystallizability of targets that are recalcitrant to crystallization in their wild-type form is discussed. This burgeoning field is very broad and includes both general strategies that apply to a range of targets and many diverse approaches that only apply to specific proteins or protein families. Thus, owing to space limitations, the focus is on those techniques that have either been demonstrated to be of general utility or are at a point in their development to clearly have the potential to become widely used in the future. Understandably, only representative examples are provided.
2. Microscopic aspects of protein crystallization
Proteins are inherently dynamic entities, a property that greatly hinders their crystallizability. Not surprisingly, it has been estimated that even for the stable and relatively small single-domain prokaryotic proteins fewer than one in four will yield X-ray-quality crystals when using a routine screening process (Canaves et al., 2004 ▶; Price et al., 2009 ▶). In order to rationally modify proteins to enhance their crystallizability, it is first necessary to understand the physical properties that make most proteins resistant to crystallization.
Protein crystals are nucleated ab initio at supersaturation levels in the 200–1000% range (McPherson, 1982 ▶). Nucleation is believed to proceed via a two-step process: clusters of solute molecules form first and upon reaching critical size reorganize into three-dimensionally ordered nuclei (Georgalis et al., 1997 ▶; Vekilov, 2004 ▶; Erdemir et al., 2009 ▶). Subsequent transfer of protein molecules from solution onto the growing crystal surface is driven by relatively small negative changes in Gibbs free energy (ΔG°), from approximately −10 to −100 kJ mol−1, at ambient temperature (Vekilov, 2003 ▶). Interestingly, enthalpy changes are generally negligible during crystallization (Yau et al., 2000 ▶; Petsev et al., 2001 ▶; Gliko et al., 2005 ▶), so that entropic phenomena dominate (Vekilov et al., 2002 ▶; Vekilov, 2003 ▶; Derewenda & Vekilov, 2006 ▶). The microscopic effects underlying the entropy changes, both favorable and unfavorable, involve the protein itself as well as the solvent. Protein packing, which results in an ordered three-dimensional lattice and loss of translational and rotational degrees of freedom, is unfavorable and produces an energy barrier in the 30–100 kJ mol−1 range at room temperature (Finkelstein & Janin, 1989 ▶; Tidor & Karplus, 1994 ▶). Similarly, incorporation into the growing crystal and ordering of any intrinsically unstructured elements, such as flexible termini or loops and side chains, at the point of crystal contacts further increases the entropic cost. However, the release of ordered solvent molecules from the surfaces involved in crystal contacts, which is estimated to be in the 25–150 kJ mol−1 range, may sufficiently compensate for these entropy losses and ultimately provide the driving force for crystal growth (Vekilov et al., 2002 ▶; Vekilov, 2003 ▶).
Based on these considerations, it is evident that a protein must satisfy certain criteria in order to crystallize. Firstly, it must have a molecular surface that confers adequate solubility under initial conditions to reach the necessary supersaturation level for nucleation. Furthermore, it should have few, if any, intrinsically unstructured fragments such as extended N- or C-termini or long and solvent-exposed loops which may impede crystallization. Finally, the protein should have distinct ‘sticky’ patches on the surface with a structured layer of solvent molecules, allowing the ordering of nascent nuclei by mediating thermodynamically viable specific crystal contacts.
The notion that protein crystallization involves specific and anisotropic intermolecular interactions, as opposed to random contacts, is relatively new. Early analyses of intermolecular contacts in protein crystals concluded that crystallization is a stochastic process generated by mostly random contacts (Janin & Rodier, 1995 ▶; Janin, 1997 ▶; Carugo & Argos, 1997 ▶). However, more recent stringent statistical analyses using a larger database strongly suggested that crystal contacts are generated by anisotropic interactions that favor small hydrophobic residues and disfavor large polar side chains with high conformational entropy (Cieślik & Derewenda, 2009 ▶). This view is also supported by a large-scale comparison of the amino-acid sequences of crystallizable and noncrystallizable proteins, which established that crystallization propensity is negatively correlated with the prevalence of residues with high side-chain entropy (Price et al., 2009 ▶). Finally, molecular-dynamics simulations of the intermolecular interactions of lysozyme in solution show that they are anisotropic and that their magnitude and nature depend on the physical chemistry of the participating interfaces, suggesting that the nucleation phenomenon is initiated in a nonstochastic fashion (Pellicane et al., 2008 ▶).
Understanding the physical principles that govern crystallization at the microscopic level provides the singular underpinning to rationally engineer target proteins to enhance their crystallizability either by improving their solution properties or by increasing their propensity to engage in weak but specific interactions that organize the transformation of nascent clusters into nuclei and drive subsequent crystal growth.
3. Engineering proteins with enhanced solubility
The solubility of a protein is the primary essential prerequisite for its crystallization. It should be noted that the expression ‘low solubility’ is often used indiscriminately to describe quite different phenomena, including a propensity to aggregate and precipitate upon overexpression owing to misfolding, amyloid formation and finally genuine low in vitro solubility, i.e. low protein concentration in equilibrium with the solid phase, of otherwise fully folded and stable proteins (Trevino et al., 2008 ▶). Here, the strategies and methods that specifically address the latter case are discussed, i.e. precipitation at low concentrations of properly folded proteins.
It has been well established that even single-site mutations of surface residues can dramatically affect the solubility of a protein and its crystallizability (McElroy et al., 1992 ▶). Consequently, the intuitively obvious approach is to mutate solvent-exposed hydrophobic amino acids to hydrophilic residues. In this way, the low solubility of the catalytic domain of HIV-1 integrase was addressed by screening 29 mutants in which hydrophobic residues were systematically mutated to hydrophilic amino acids; of the variants tested, the single-site mutant F185K showed a dramatically improved solubility and ultimately yielded X-ray-quality crystals (Dyda et al., 1994 ▶; Jenkins et al., 1995 ▶). In the case of leptin, the product of the obese gene, the solubility-enhancing W100E mutation proved to be critical for crystallization of the protein (Zhang et al., 1997 ▶). Recently, a screen of several variants of human apolipoprotein D identified a triple mutant (W99H, I118S, L120S) which was much more soluble than the wild-type protein and which was ultimately used to obtain well diffracting crystals (Nasreen et al., 2006 ▶; Eichinger et al., 2007 ▶).
While engineering enhanced solubility using site-directed mutagenesis is potentially a powerful approach, in the absence of structural information it is a challenge to predict which hydrophobic residues are solvent-exposed and might therefore constitute useful targets for mutagenesis. Moreover, even if structural information is available for a homolog or the target itself, it may not be clear what type of mutation actually works best, forcing the investigator to rely on extensive screening. This uncertainty arises from the fact that hydrophobicity scales for individual amino acids cannot be used directly to evaluate the increase or decrease of protein solubility as a consequence of a specific mutation. Furthermore, there have been few rigorous studies of the effects of specific mutations on protein solubility. A notable example is a study on ribonuclease SA in which the solvent-exposed Thr76 was replaced by 19 other amino acids and the solubility of all of the variants was carefully evaluated (Trevino et al., 2007 ▶). Those variants that contained Asp, Arg, Glu and Ser were the most soluble. Unexpectedly, even though a lysine might be expected to confer higher solubility than a serine or alanine, the T76S mutation actually led to a significantly higher solubility than T76K, while the T76A variant was only marginally less soluble than T76K (Trevino et al., 2007 ▶). The authors of the study concluded that mutating Asn and Gln to their respective acids may constitute the most robust strategy of enhancing solubility. Interestingly, one of the first examples of rational enhancement of solubility, i.e. the study of trimethoprim-resistant type S1 hydrofolate reductase (Dale et al., 1994 ▶), used this very strategy: the amide-containing side chains were systematically substituted with carboxylic amino acids and one specific variant, a double mutant N48E, N130D, was found to exhibit markedly increased solubility and ultimately yielded crystals that were suitable for crystallographic analysis.
Somewhat ironically, large charged residues such as glutamate that confer higher solubility on the target protein may at the same time impede crystallization because they increase the total surface side-chain entropy, making the surface recalcitrant to engaging in crystal contact-mediating interactions. Thus, variants engineered for increased solubility may simultaneously show a decreased propensity to crystallize.
Some of the above uncertainties can be overcome with an alternative approach of directed evolution and phenotypic selection methods, in which soluble mutants are directly selected from vast protein libraries (Farinas et al., 2001 ▶; Farinas, 2006 ▶; Pédelacq et al., 2002 ▶; Waldo, 2003 ▶; Cabantous et al., 2005 ▶). Several different variations of this method have been reported (Waldo, 2003 ▶). For example, the target protein may be fused to the N-terminus of a reporter protein such as the green fluorescent protein (GFP; Waldo et al., 1999 ▶) or direct detection methods can be employed to identify soluble variants (Peabody & Al-Bitar, 2001 ▶). While elegant and potentially very effective, directed evolution has not yet been widely adopted for the generation of crystallizable proteins.
Solubility problems are not always caused by excessively exposed hydrophobic surfaces. In some cases, the root of the problem is aggregation caused by exposed free cysteines. Reduced cysteines can be identified by alkylation with N-ethylmaleimide or iodoacetamide under anaerobic conditions and subsequent electrospray mass spectrometry (Niessing et al., 2004 ▶). Several examples illustrate how this approach is helpful in generating samples that are suitable for crystallization. In mitogen-activated protein (MAP) kinase p38α, a single-site mutation (C162S) prevented aggregation and yielded a crystallizable variant (Patel et al., 2004 ▶). Similarly, a double mutant (C95K, C142S) of foot-and-mouth disease virus 3C protease showed none of the aggregation problems that plagued the wild-type protein and was subsequently crystallized (Birtley & Curry, 2005 ▶). It is noteworthy that in this case an alternative strategy involving mutations of the exposed hydrophobic residues Met81, Leu82 and Val140 did not eliminate aggregation (Birtley & Curry, 2005 ▶). In a number of cases aggregation problems were traced to multiple free cysteines. In She2p, an RNA-binding protein, four cysteines (Cys14, Cys68, Cys106 and Cys180) were mutated to serines in order to overcome oxidation and aggregation (Niessing et al., 2004 ▶). In an extreme case, that of human maspin, which is a serpin with antitumor activities, all unpaired cysteines were mutated (C20S, C34A, C183S, C205S, C214S, C297S, C373S) in an effort to obtain a soluble crystallizable variant (Al-Ayyoubi et al., 2004 ▶).
4. Optimization of target constructs
The N- and C-termini of proteins are often flexible and unstructured (Thornton & Sibanda, 1983 ▶), creating a potential entropic impediment to crystallization. Initially, the preferred way to circumvent this problem was to use limited proteolysis to trim off the ends, leaving the stable core of the target protein. This strategy remains useful, particularly in its in situ version, in which trace amounts of proteases are added directly to crystallization screens (Dong et al., 2007 ▶; Wernimont & Edwards, 2009 ▶). However, on the downside it introduces the possibility of heterogeneity in the sample owing to incomplete proteolysis. An alternative route is to first identify the smallest functional fragment of the target protein and to then design and overexpress an appropriately modified gene. A number of options are possible. The simplest is the direct prediction of intrinsically disordered regions from the amino-acid sequence alone (Obradovic et al., 2003 ▶; He et al., 2009 ▶). The functional core units can also be identified experimentally by mass spectrometry following limited proteolysis (Cohen et al., 1995 ▶). Alternatively, deuterium–hydrogen exchange coupled to mass spectrometry (DXMS) may be used to identify fast-exchanging amides that map to unstructured fragments (Hamuro et al., 2003 ▶; Pantazatos et al., 2004 ▶; Sharma et al., 2009 ▶).
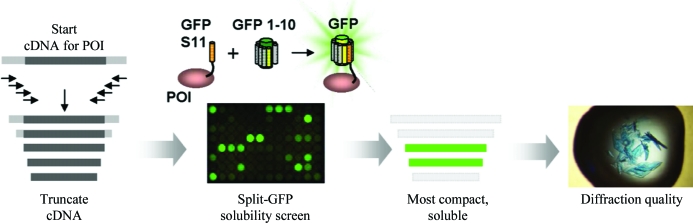
Importantly, the choice of optimal N- and C-termini may also critically influence the solubility of the target protein. For example, in the case of MAPKAP kinase 2, 16 truncation variants were assayed, all of which contained the catalytic domain, and were shown to have dramatically differing solubilities and propensities for crystallization (Malawski et al., 2006 ▶). Similarly, a series of truncations were screened in order to identify a soluble and crystallizable variant of a three-domain fragment of the Vav1 guanine nucleotide-exchange factor (Brooun et al., 2007 ▶). In both these cases only a limited number of rationally designed constructs were screened. However, to increase the prospects of success it is also possible to utilize much larger libraries of variants and screen them in vivo using the high-throughput split-GFP complementation assay (Fig. 1 ▶; Cabantous & Waldo, 2006 ▶).
Figure 1.
A domain-trapping strategy to engineer soluble variants of a protein of interest (POI) for crystallization using the split-GFP complementation methodology. (Figure courtesy of Dr Geoff Waldo, LANL.)
Another troublesome problem associated with flexible termini is their occasional propensity to form multiple intermolecular contacts, leading to crystal forms that contain multiple copies of the target protein in the asymmetric unit. This has been observed, for example, for Plasmodium falciparum peptide deformylase, in which removal of three residues from the N-terminus reduced the number of subunits in the asymmetric unit from ten to two (Robien et al., 2004 ▶).
In addition to the disordered N- and C-termini, target proteins may contain internal unstructured regions such as subdomains or loops which can be removed or shortened to reduce conformational heterogeneity. For example, the construct used in the successful crystallization of the HIV gp120 envelope glycoprotein had two flexible loops which were replaced with Gly-Ala-Gly linkages to obtain a crystallizable variant (Kwong et al., 1998 ▶, 1999 ▶). In the case of 8R-lipoxygenase the replacement of a flexible Ca2+-dependent membrane-insertion loop consisting of five amino acids by a Gly-Ser dipeptide resulted in crystals that diffracted to a resolution 1 Å higher than the wild-type protein (Neau et al., 2007 ▶). An interesting variation of this approach was introduced for the preparation of crystals of the β-subunit of the signal recognition particle receptor. A 26-residue flexible loop was removed, but instead of replacing it with a shorter sequence the authors connected the native N- and C-termini of the protein using a heptapeptide GGGSGGG, thus creating a circular permutation of the polypeptide chain (Schwartz et al., 2004 ▶).
Given that the majority of eukaryotic proteins contain at least one stretch of 40 or more disordered residues (Vucetic et al., 2003 ▶), optimization of crystallization targets by removal of these sequences is likely to become a routine strategy.
5. The use of fusion proteins for crystallization
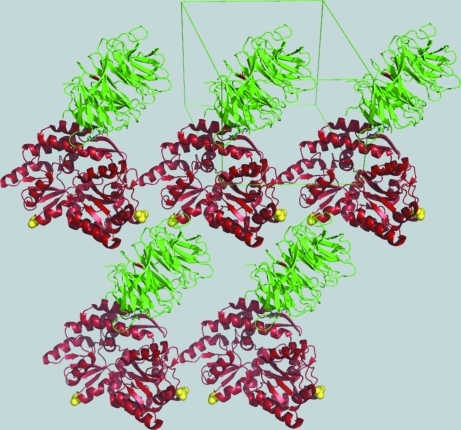
Tags are routinely used in heterologous protein expression in order to enhance folding and solubility and to facilitate purification (Uhlen et al., 1992 ▶; Malhotra, 2009 ▶). They are either short oligopeptides, such as a hexahistidine, with unique affinity properties or well expressed and highly soluble proteins, such as GST (glutathione S-transferase), MBP (maltose-binding protein) or thioredoxin. The tags are inserted into the expression vectors downstream or upstream of the target protein and are often separated from it by a protease-sensitive linker sequence. They are cleaved proteolytically following expression and partial purification of the fusion protein and removed, leaving the isolated target ready for crystallization. However, in some cases the target protein may not be adequately soluble after cleavage or may resist crystallization. One of the possible solutions is to use the intact fusion protein in the crystallization screens in the hope that the carrier protein will both confer solubility on the construct and mediate crystal contacts. Not surprisingly, the canonical carrier proteins, all of which crystallize fairly easily on their own, constitute the obvious first choice. Using this strategy, the DNA-binding domain of DNA replication-related element-binding factor (DREF) was crystallized in fusion with Escherichia coli GST (Kuge et al., 1997 ▶) and the U2AF homology motif (UHM) domain of splicing factor Puf60 was crystallized as a fusion with thioredoxin (Corsini et al., 2008 ▶). A key problem limiting the utility of this technique is the inherent flexibility of a two-domain fusion protein, which is detrimental to its crystallizability. A possible solution to this problem is shortening the linker between the two proteins until a relatively rigid construct is identified (Smyth et al., 2003 ▶). This approach was successfully pioneered for maltose-binding protein (MBP), which was used as a fusion chaperone to crystallize the human T-cell leukemia virus type 1 gp21 ectodomain fragment (Center et al., 1998 ▶). The same strategy was employed in the crystallization of the ZP-N domain of ZP3 (Monne et al., 2008 ▶), the islet amyloid polypeptide (IAPP; Wiltzius et al., 2009 ▶) and the MATα1 homeodomain (Ke & Wolberger, 2003 ▶). Recently, a genetically modified version of MBP (see below) was used as an N-terminal fusion chaperone to crystallize the signal transduction regulator RACK1 from Arabidopsis thaliana (Ullah et al., 2008 ▶; Fig. 2 ▶). Thus, MBP remains the most successful fusion chaperone for protein crystallization, even though the absolute number of proteins crystallized in this way is still limited.
Figure 2.
An example of the use of a fused carrier protein in crystallization: the crystal structure of the RACK1 protein (green) crystallized in fusion with an engineered variant of the maltose-binding protein (MBP; red); the major crystal contacts are mediated by MBP (PDB code 3dm0; Ullah et al., 2008 ▶). The yellow spheres show alanines introduced by site-directed mutagenesis (see text for further details). Figs. 2 and 4 were generated using PyMOL (http://www.pymol.org).
In addition to the canonical fusion chaperones, which were originally designed as affinity tags, other carrier proteins can be used to assist crystallization. For example, a module made up of two sterile α motif (SAM) domains has been engineered to polymerize in response to a pH drop and was shown to drive the crystallization of 11 target proteins in a pilot study (Nauli et al., 2007 ▶). In another example, barnase, a secreted ribonuclease from Bacillus amyloliquefaciens, was recently used as a carrier protein for crystallization of the disulfide-rich protein McoEeTI (Niemann et al., 2006 ▶).
An alternative to N- or C-terminal fusions is an insertion fusion, in which a carrier protein is inserted into a loop in the sequence of a poorly soluble target. To date, this approach has exclusively been used in membrane-protein crystallization and was initially pioneered for the E. coli lactose permease, in which cytochrome b 562, flavodoxin and T4 lysozyme were tested as carrier proteins inserted into one of the loops (Privé et al., 1994 ▶; Engel et al., 2002 ▶). In this specific case none of these variants actually yielded useful crystals and the structure of lactose permease was eventually solved using crystals obtained using a variant containing the C154G mutation which stabilized a single conformation in complex with a lactose analogue (Abramson et al., 2003 ▶). In contrast, a similar insertion fusion with T4 lysozyme replacing the third intracellular loop of the β2-adrenergic receptor was highly successful and yielded good-quality crystals that allowed structure determination at 2.4 Å resolution (Cherezov et al., 2007 ▶; Rosenbaum et al., 2007 ▶). This spectacular result attests to the potential of insertion-fusion proteins, but the method is not trivial as the constructs must be carefully evaluated for both structural and functional consequences of the insertion and a number of variants may have to be screened before a suitable one is identified.
6. Noncovalent crystallization chaperones
Noncovalent crystallization chaperones, i.e. engineered binding proteins that produce noncovalent complexes with target macromolecules, constitute an exciting alternative to fusion carrier proteins. Complexes with such chaperones often exhibit enhanced solubility and/or crystallizability in comparison to the isolated targets. The Fab and Fv fragments of antibodies are most commonly used for this purpose (Kovari et al., 1995 ▶; Hunte & Michel, 2002 ▶; Prongay et al., 1990 ▶; Ostermeier et al., 1995 ▶; Jiang et al., 2003 ▶; Dutzler et al., 2003 ▶; Lee et al., 2005 ▶). In its canonical version, the technique requires animal immunization with subsequent purification of hybridoma-derived antibodies and their proteolytic digestion to obtain pure homogeneous Fab fragments (Karpusas et al., 2001 ▶; Kovari et al., 1995 ▶). Alternatively, the Fab fragment can be directly sequenced and a synthetic gene can be used for E. coli expression, although this is not trivial owing to the presence of disulfides and two separate polypeptide chains in a Fab molecule. To overcome this bottleneck, a more efficient method of recombinant production of antibody fragments using mammalian HEK 293T has recently been proposed (Nettleship et al., 2008 ▶). Another possibility is the use of so-called nanobodies, i.e. single-chain fragments derived from camelid antibodies (Koide, Tereshko et al., 2007 ▶; Lam et al., 2009 ▶; Korotkov et al., 2009 ▶). However, this strategy requires immunization of camels or llamas, which is not technically easy.
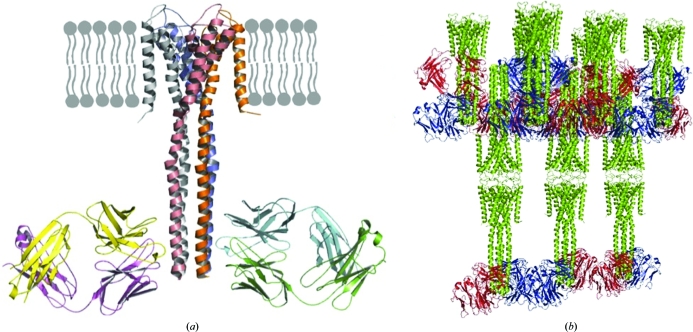
Regardless of the specific strategy, the use of hybridoma technology and animal immunization is always time-consuming and expensive. In principle, a more efficient approach is to carry out in vitro selection of Fab fragments using phage display (Lee et al., 2004 ▶) or ribosome display (Lipovsek & Pluckthun, 2004 ▶). However, since a typical antibody–antigen interface involves ∼30 amino acids, the total number of possible sequences of a given template Fab significantly exceeds the available combinatorial libraries. Consequently, traditional phage-display libraries greatly diminish diversity at the mutated sites, which explains why synthetic antibodies were initially weaker binders than natural ones (Hawkins et al., 1992 ▶; Koide, 2009 ▶). This problem was successfully overcome using a different type of phage-display library based on a ‘reduced genetic code’ and comprised of only a few amino acids, e.g. four, which produces high-affinity binders based on a single Fab scaffold (Fellouse et al., 2004 ▶; Lee et al., 2004 ▶). In contrast to natural antibodies, such synthetic Fab fragments can be generated against unique conformations, complexes or weak antigens such as RNA. Among recent examples are the crystallization and structure determination of the closed form of the full-length KcsA potassium channel with its cognate synthetic Fab (Uysal et al., 2009 ▶; Fig. 3 ▶) and the crystallographic study of the ΔC209 P4-P6 domain of the Tetrahymena group I intron, a structured RNA molecule (Ye et al., 2008 ▶).
Figure 3.
Phage-display-generated Fab fragments as crystallization chaperones: the structure of the KcsA channel in the closed conformation in complex with a synthetic Fab (PDB code 3eff; Uysal et al., 2009 ▶). (a) A diagram showing how the Fab binds to the cytosolic portion in reference to the transmembrane domain. (b) The crystal structure of the complex showing how the synthetic Fab molecules mediate the major crystal contacts. (Figure courtesy of Dr Anthony Kossiakoff, University of Chicago.)
The in vitro display methods also allow the engineering of non-antibody scaffolds as alternative protein binders and crystallization chaperones (Koide, 2009 ▶). For example, a fibronectin type III domain (FN3) scaffold was successfully used to generate binders with a reduced genetic code phage-display library (Koide, Gilbreth et al., 2007 ▶; Gilbreth et al., 2008 ▶). A similar approach was used for DARPins, i.e. designed ankyrin-repeat proteins (Sennhauser & Grütter, 2008 ▶), based on ribosome-display selection (Lipovsek & Pluckthun, 2004 ▶; Sennhauser & Grütter, 2008 ▶). Several new protein structures have been solved as complexes with DARPin chaperones, including polo-like kinase 1 (Bandeiras et al., 2008 ▶), the trimeric integral membrane multidrug transporter AcrB (Sennhauser et al., 2007 ▶) and the receptor-binding protein (RBP, the BppL trimer) of the baseplate complex of the lactococcal phage TP901-1 (Veesler et al., 2009 ▶).
7. Removal of post-translational modifications
A number of proteins undergo post-translational modifications which can adversely affect crystallization. By far the most ubiquitous is N- and O-glycosylation, primarily of membrane-associated, secreted and lysosomal proteins. In a number of cases successful crystallization of glycoproteins purified from natural sources has been reported and carbohydrate groups have often been found to be ordered and occasionally sequestered between the protein molecules, thus even contributing in a positive way to crystallization (Mark et al., 2003 ▶; Aleshin et al., 1994 ▶). In general terms, however, the flexible and heterogeneous carbohydrate moieties, particularly the oligosaccharides linked by N-glycosylation, can account for a significant fraction of the surface area of the protein and can therefore be detrimental to crystallization. The preparation of recombinant proteins in E. coli eliminates these post-translational modifications and may sometimes solve the problem (Mohanty et al., 2009 ▶), but N-glycosylation is often required for appropriate folding and solubility, so this approach is not always possible. However, if a eukaryotic expression system is a necessity, the problem can often be resolved by mutating the asparagines within the relevant glycosylation motifs (Asn-X-Thr/Ser), e.g. to aspartates, as was performed in the case of the extracellular domain of the metabotropic glutamate receptor expressed in insect cells (Muto et al., 2009 ▶), or to glutamines, as was performed for the human testis angiotensin-converting enzyme (Gordon et al., 2003 ▶). Alternatively, glycosylation at these sites can be eliminated by mutation of the Thr/Ser residues in the glycosylation motif to alanine or other amino acids, as described for rat cathepsin B (Lee et al., 1990 ▶), or valine, as was the case with the Ebola virus glycoprotein (Lee et al., 2008 ▶, 2009 ▶). Similarly, potentially glycosylated threonines or serines in O-glycosylated glycoproteins can be mutated to other amino acids (Horan et al., 1998 ▶) to avoid or reduce glycosylation.
Other post-translational modifications occur less frequently. Prenylation and N-myristoylation can occur at the C- and N-termini, respectively. Expression in E. coli, often using truncated versions of target proteins, is a common remedy (Pai et al., 1990 ▶).
8. Stabilization of protein targets
There is currently no clear consensus regarding a possible correlation between the thermostability of a protein and its propensity to form crystals. It is often assumed on the basis of somewhat anecdotal evidence that thermostable proteins are more readily crystallizable and therefore if a specific target protein is recalcitrant to crystallization then a homologue from a thermophilic organism should instead be used. In some cases, low thermostability may correlate with the presence of unstructured loops or termini and consequently the construct-optimization strategies, as described above, are likely to yield a more crystallizable variant with a concomitantly increased stability. For example, a study of MAPKAP kinase 2 showed that truncated variants with increased thermal stability also showed higher crystallization propensity (Malawski et al., 2006 ▶). However, it is uncertain whether the relationship is causal or serendipitous. A recent analysis of large-scale data from a structural genomics project showed that when partly or fully unfolded proteins and hyperstable proteins (with melting temperatures T m of greater than 363 K) are excluded from comparisons, thermostability per se does not correlate with propensity for crystallization (Price et al., 2009 ▶). Consequently, it appears that in a general case of a well behaving protein attempts to increase thermostability by site-directed mutagenesis may not necessarily yield variants with enhanced crystallization properties, although when the prospective crystallization target is inherently unstable engineering more stable variants may be helpful. In fact, this strategy has been successfully used for membrane proteins, which are often unstable in detergent environments. The first structure of a recombinant G-protein-coupled receptor (GPCR), i.e. bovine rhodopsin in complex with 11-cis retinal, was obtained using a thermostable variant with an engineered disulfide bond (Standfuss et al., 2007 ▶). In the more recent case of the turkey adrenergic β2 receptor, 318 variants were screened and six mutations were identified that increased thermostability. A variant containing all six mutations had an apparent T m that was 21 K higher than that of the native protein in dodecylmaltoside (DDM), was more stable in short-chain detergents and was successfully crystallized (Warne et al., 2008 ▶, 2009 ▶). The effort required for such vast screening is substantial, but it appears that within protein families (such as GPCRs) the pattern of mutations enhancing thermostability is preserved, thus making it possible to transfer the mutations from one family member to another (Serrano-Vega & Tate, 2009 ▶).
Finally, it should be noted that protein thermostability is strongly dependent on solution parameters such as the ionic strength. In the case of ribonuclease SA, the melting temperature (T m) increased from ∼313 to ∼333 K on transfer from pure 50 mM diglycine buffer to 0.9 M ammonium sulfate (Trevino et al., 2007 ▶). Similarly, binding small molecules, either physiological or nonphysiological ligands, typically promotes stability (Matulis et al., 2005 ▶). High-throughput screening methods have been developed to aid in screening for conditions and ligands that enhance stability (Vedadi et al., 2006 ▶; Mezzasalma et al., 2007 ▶) and in a general case this strategy appears to hold better promise than attempts to engineer higher stability through mutagenesis.
9. Surface-entropy reduction (SER)
For well ordered intrinsically stable proteins which show none of the problems addressed above, the propensity of the molecules to associate together and form crystals mediated by weak but specific interactions is ultimately defined by the physical chemistry and topology of the molecular surface. As already pointed out, large flexible amino acids on the surface, such as Lys, Glu and Gln, constitute an impediment to intermolecular interactions and consequently to protein crystallization (Cieślik & Derewenda, 2009 ▶; Price et al., 2009 ▶). In fact, it has been suggested that these residues and the ‘entropy shield’ that they form play a role in protein evolution which, given the high average concentration of proteins in cells, disfavors protein–protein interactions unless they are biologically functional (Doye, 2004 ▶). Thus, an intuitively obvious way to generate crystallizable variants is to replace selected large and surface-exposed residues with smaller residues such as alanine. This crystal-engineering strategy based on the surface-entropy reduction (SER) concept was extensively tested using as a model system the globular domain of the human Rho-specific guanine nucleotide-dissociation inhibitor (RhoGDI), which is recalcitrant to crystallization in its wild-type form owing to a high content of Lys and Glu residues, which constitute more than 20% of the sequence (Longenecker, Garrard et al., 2001 ▶; Mateja et al., 2002 ▶; Derewenda, 2004 ▶; Cooper et al., 2007 ▶). These experiments established that in order to be most effective the SER strategy requires simultaneous mutations of clusters of two to three solvent-exposed high-entropy amino acids, typically Lys, Glu or Gln, located in close sequence proximity. These amino acids are replaced with alanine, although threonine and tyrosine, which is known to make a positive contribution at protein–protein interfaces such as antibody–antigen complexes (Fellouse et al., 2006 ▶), can also be used (Cooper et al., 2007 ▶). Engineered low-surface-entropy variants of RhoGDI produced new and unique crystal forms, many with superior diffraction quality when compared with the wild-type protein. Importantly, in the vast majority of these crystals the mutated surface patches mediated crystal contacts, suggesting that SER engineering directly drives crystallization in a rational fashion by creating suitable crystal-contact-forming interfaces. The general utility of the method was further established by the crystallization of several novel protein targets found to be recalcitrant to crystallization in their wild-type form (Longenecker, Lewis et al., 2001 ▶; Derewenda et al., 2004 ▶; Devedjiev et al., 2004 ▶; Janda et al., 2004 ▶).
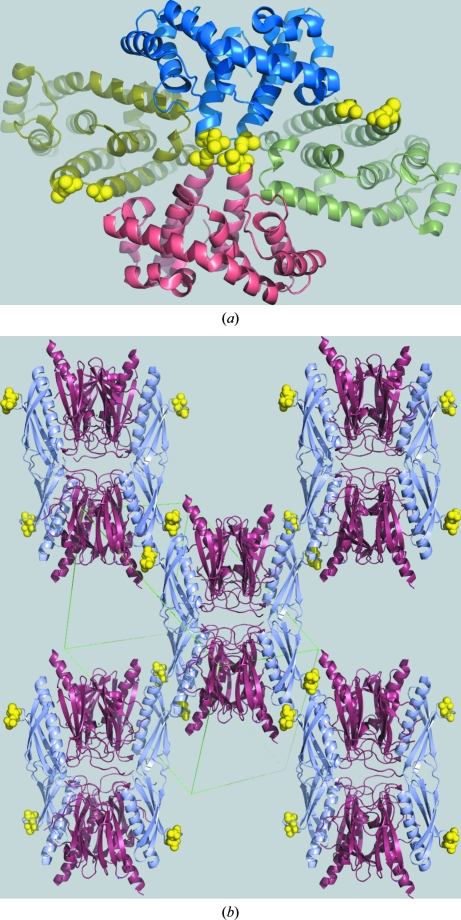
SER is quickly becoming a method of choice for engineering crystallizable variants of both individual proteins and protein–protein complexes. To date (December 2009), there have already been more than 100 depositions made to the Protein Data Bank (Berman et al., 2007 ▶) based on diffraction studies of crystals generated by SER and corresponding to 47 novel structures, seven novel protein–protein complexes, several studies of proteins in complexes with drug leads aimed at rational drug development and two membrane proteins. The current list of crystal structures obtained using SER crystals includes a number of cases of exceptionally high biological interest. For example, the EscJ protein from enteropathogenic E. coli, the oligomerization of which initiates assembly of the type III bacterial secretion system, was crystallized with three entropy-reducing mutations (E62A, K63A and E64A) forming a key contact in the crystal structure (Yip et al., 2005 ▶). Likewise, the HIV CcmK4 capsid protein only crystallized after an entropy-reducing mutation (E104Y) was introduced into the protein (Pornillos et al., 2009 ▶). The protein–protein complexes solved to date underscore the utility of the method, which extends beyond individual proteins because high-entropy patches occur outside complex interfaces. For example, the complex of c-Src and its inactivator Csk was crystallized using a variant of Csk carrying K361A and K362A mutations (Levinson et al., 2008 ▶). Similarly, it was possible to crystallize the complex of two pseudopilins EpsI and EpsJ from the type 2 secretion system of Vibrio vulnificus when a variant of EpsI carrying two mutations (E128T and K129T) was used (Yanez et al., 2008 ▶; Fig. 4 ▶).
Figure 4.
Two examples of proteins crystallized by the surface-entropy reduction (SER) method. (a) The RGSL domain of the PDZRhoGEF nucleotide-exchange factor (PDB code 1htj; Longenecker, Lewis et al., 2001 ▶); the yellow spheres show the alanines introduced by mutagenesis, which mediate an isologous crystal contact across a crystallographic twofold axis. (b) The crystal structure of EpsI complexed with EpsJ (PDB code 2ret; Yanez et al., 2008 ▶); the EpsI protein (pale blue) contains two surface mutations, shown by yellow spheres, which mediate heterologous crystal contacts.
An interesting variation of the SER method was used in the investigation of the RACK1 protein, which was crystallized as an in-line fusion with an MBP variant carrying D82A, K83A and K239A mutations (Ullah et al., 2008 ▶). This is the first example of the application of the surface-entropy reduction strategy to a carrier protein and not the crystallization target itself.
The SER strategy is attractive not only because of its efficacy but also because of its simplicity: once an expression construct for a target protein is available several rounds of mutagenesis can easily create variants with systematically enhanced crystallizability. To assist in the design of crystallizable variants, a server has been developed that uses the amino-acid sequence of the target to identify suitable mutation sites (Goldschmidt et al., 2007 ▶).
10. Improvement of crystal quality
In most cases, protein engineering is used as a tool of last resort to obtain variants for proteins for which no crystals can be grown using the wild-type form. However, it may sometimes be necessary to obtain a new, different crystal form even when the wild-type protein does crystallize. Such a need may arise, for example, in drug-design investigations, where high-resolution structures are particularly critical for evaluation of the interactions between lead compounds and the target protein and may not always be possible using wild-type crystals. A novel crystal form may also be necessary if the wild-type crystals contain the target protein in an orientation in which the active site is obscured by crystal contacts, making it impossible to soak in drug lead compounds and screen small-molecule libraries by high-throughput crystallography (Blundell & Patel, 2004 ▶).
One possible strategy for obtaining a new crystal form is to modify the existing crystal contacts by replacing some of the participating amino acids. While this approach occasionally leads to improvement of the X-ray data resolution (Liu et al., 2007 ▶; Mizutani et al., 2008 ▶), modification of crystal contacts is typically counterproductive as it abolishes the propensity of the target to crystallize in one form but does not necessarily induce another (Charron et al., 2002 ▶). A more successful strategy is to generate a novel crystal form by engineering new crystal contacts through SER. For example, a novel crystal form of the insulin-like growth factor 1 receptor kinase domain, a putative drug target, was obtained using a double mutant (E1067A and E1069A); the new form diffracted to 1.5 Å resolution, whereas the wild-type crystals only diffracted to 2.7 Å resolution (Munshi et al., 2003 ▶). In the case of the catalytic domain of activated factor XI, a key enzyme in the blood coagulation cascade and another potential drug target, a single K437A mutation allowed the preparation of a crystal form that diffracted to 2.0 Å resolution (Jin et al., 2005 ▶). Entropy-reducing mutations were also key in the preparation of a crystal form of HIV-1 reverse transcriptase for structure-based drug design that diffracted to 1.8 Å resolution, in contrast to the typical 2.5–3.0 Å range observed for the wild-type protein crystals (Bauman et al., 2008 ▶).
11. Conclusions
Protein engineering has become a routine tool that is used to generate crystallizable macromolecules and their complexes. While some approaches may only apply to very specific targets, a number of strategies offer general applicability. Among these, gene-construct optimization or surface-entropy reduction are quickly gaining popularity as methods of choice. However, it should be stressed that none of the methods described here offer a guarantee that the target protein can be coerced to crystallize. To maximize the chances of success, one must frequently attack the problem on multiple fronts based on an understanding of the chemical and physical properties of a specific protein. This is particularly true of technically difficult targets such as membrane proteins. A classic example illustrating this principle is the study of the HIV gp120 envelope glycoprotein (Kwong et al., 1998 ▶, 1999 ▶). The construct that was ultimately used in successful crystallization screens had deletions of 52 and 19 residues from the N- and C-termini and two flexible loops replaced by Gly-Ala-Gly linkages; additionally, the protein was 90% deglycosylated compared with the wild type. Moreover, this engineered gp120 was only crystallized in the form of a ternary complex with the CD4 receptor and a Fab fragment from a neutralizing antibody. In the recent case of the ATP-gated P2X4 ion channel, a crystallizable variant was obtained after a series of N- and C-terminal deletions were screened to identify the smallest functional unit and the introduction of three mutations (C51F/N78K/N187R) to eliminate both aggregation arising from oxidation and N-glycosylation (Kawate et al., 2009 ▶).
The rapidly expanding database of macromolecular structures greatly enhances our understanding of the physical chemistry of proteins, ultimately enhancing our ability to predict the behavior of a protein in solution from its sequence. It is therefore increasingly possible to rely on such theoretical predictions in lieu of tedious experimental screens. A number of online tools have been developed for this purpose. The propensity of a protein target to crystallize can be evaluated using the XtalPred server (http://ffas.burnham.org/XtalPred-cgi/xtal.pl), which offers insights into potential sources of problems arising from sequence features (Slabinski et al., 2007 ▶). Automated design of optimally truncated constructs for structural analysis has been made possible by the ProteinCCD meta-server (http://xtal.nki.nl/ccd), which uses the cDNA sequence of the target (Mooij et al., 2009 ▶). This server collects information about secondary structure, disorder, putative coiled coils, transmembrane segments, domains and domain linkers and suggests constructs so that the user can interactively choose suitable options and obtain sequences of oligonucleotides needed for appropriate PCR amplification (Mooij et al., 2009 ▶). For proteins recalcitrant to crystallization in their wild-type form, surface mutations enhancing crystallizability can be designed using the surface-entropy reduction server (http://nihserver.mbi.ucla.edu/SER/; Goldschmidt et al., 2007 ▶).
As the focus of macromolecular crystallography shifts from the principles of protein architecture to increasingly complex biological questions, the approach to crystallization is also undergoing dramatic evolution. As we gain better understanding of the microscopic nature of protein crystallization, we will be able to develop rational protein-engineering strategies that systematically and significantly improve the success rate of crystallization.
Acknowledgments
The research on protein crystallization in my laboratory is supported by the National Institute for General Medical Sciences. I am greatly indebted to Drs Urszula Derewenda, Anthony Kossiakoff, Alexander McPherson, Geoff Waldo and David Cooper for discussions and comments on the early version of the manuscript.
References
- Abramson, J., Smirnova, I., Kasho, V., Verner, G., Kaback, H. R. & Iwata, S. (2003). Science, 301, 610–615. [DOI] [PubMed]
- Al-Ayyoubi, M., Gettins, P. G. & Volz, K. (2004). J. Biol. Chem. 279, 55540–55544. [DOI] [PubMed]
- Aleshin, A. E., Hoffman, C., Firsov, L. M. & Honzatko, R. B. (1994). J. Mol. Biol. 238, 575–591. [DOI] [PubMed]
- Bandeiras, T. M. et al. (2008). Acta Cryst. D64, 339–353. [DOI] [PubMed]
- Bauman, J. D., Das, K., Ho, W. C., Baweja, M., Himmel, D. M., Clark, A. D. Jr, Oren, D. A., Boyer, P. L., Hughes, S. H., Shatkin, A. J. & Arnold, E. (2008). Nucleic Acids Res. 36, 5083–5092. [DOI] [PMC free article] [PubMed]
- Berman, H., Henrick, K., Nakamura, H. & Markley, J. L. (2007). Nucleic Acids Res. 35, D301–D303. [DOI] [PMC free article] [PubMed]
- Birtley, J. R. & Curry, S. (2005). Acta Cryst. D61, 646–650. [DOI] [PubMed]
- Blundell, T. L. & Patel, S. (2004). Curr. Opin. Pharmacol. 4, 490–496. [DOI] [PubMed]
- Brewer, S. J., Haymore, B. L., Hopp, T. P. & Sassenfeld, H. M. (1991). Bioprocess Technol. 12, 239–266. [PubMed]
- Brooun, A., Foster, S. A., Chrencik, J. E., Chien, E. Y., Kolatkar, A. R., Streiff, M., Ramage, P., Widmer, H., Weckbecker, G. & Kuhn, P. (2007). Protein Expr. Purif. 53, 51–62. [DOI] [PMC free article] [PubMed]
- Cabantous, S., Pedelacq, J. D., Mark, B. L., Naranjo, C., Terwilliger, T. C. & Waldo, G. S. (2005). J. Struct. Funct. Genomics, 6, 113–119. [DOI] [PubMed]
- Cabantous, S. & Waldo, G. S. (2006). Nature Methods, 3, 845–854. [DOI] [PubMed]
- Campbell, J. W., Duée, E., Hodgson, G., Mercer, W. D., Stammers, D. K., Wendell, P. L., Muirhead, H. & Watson, H. C. (1972). Cold Spring Harb. Symp. Quant. Biol. 36, 165–170. [DOI] [PubMed]
- Canaves, J. M., Page, R., Wilson, I. A. & Stevens, R. C. (2004). J. Mol. Biol. 344, 977–991. [DOI] [PubMed]
- Carter, C. W. Jr & Carter, C. W. (1979). J. Biol. Chem. 254, 12219–12223. [PubMed]
- Carugo, O. & Argos, P. (1997). Protein Sci. 6, 2261–2263. [DOI] [PMC free article] [PubMed]
- Center, R. J., Kobe, B., Wilson, K. A., Teh, T., Howlett, G. J., Kemp, B. E. & Poumbourios, P. (1998). Protein Sci. 7, 1612–1619. [DOI] [PMC free article] [PubMed]
- Charron, C., Kern, D. & Giegé, R. (2002). Acta Cryst. D58, 1729–1733. [DOI] [PubMed]
- Cherezov, V., Rosenbaum, D. M., Hanson, M. A., Rasmussen, S. G. F., Thian, F. S., Kobilka, T. S., Choi, H. J., Kuhn, P., Weis, W. I., Kobilka, B. K. & Stevens, R. C. (2007). Science, 318, 1258–1265. [DOI] [PMC free article] [PubMed]
- Cieślik, M. & Derewenda, Z. S. (2009). Acta Cryst. D65, 500–509. [DOI] [PMC free article] [PubMed]
- Cohen, S. L., Ferré-D’Amaré, A. R., Burley, S. K. & Chait, B. T. (1995). Protein Sci. 4, 1088–1099. [DOI] [PMC free article] [PubMed]
- Cooper, D. R., Boczek, T., Grelewska, K., Pinkowska, M., Sikorska, M., Zawadzki, M. & Derewenda, Z. (2007). Acta Cryst. D63, 636–645. [DOI] [PubMed]
- Corsini, L., Hothorn, M., Scheffzek, K., Sattler, M. & Stier, G. (2008). Protein Sci. 17, 2070–2079. [DOI] [PMC free article] [PubMed]
- Dale, G. E., Broger, C., Langen, H., D’Arcy, A. & Stuber, D. (1994). Protein Eng. 7, 933–939. [DOI] [PubMed]
- Derewenda, U., Mateja, A., Devedjiev, Y., Routzahn, K. M., Evdokimov, A. G., Derewenda, Z. S. & Waugh, D. S. (2004). Structure, 12, 301–306. [DOI] [PubMed]
- Derewenda, Z. S. (2004). Structure, 12, 529–535. [DOI] [PubMed]
- Derewenda, Z. S. & Vekilov, P. G. (2006). Acta Cryst. D62, 116–124. [DOI] [PubMed]
- Devedjiev, Y., Surendranath, Y., Derewenda, U., Gabrys, A., Cooper, D. R., Zhang, R. G., Lezondra, L., Joachimiak, A. & Derewenda, Z. S. (2004). J. Mol. Biol. 343, 395–406. [DOI] [PMC free article] [PubMed]
- Dong, A. et al. (2007). Nature Methods, 4, 1019–1021. [DOI] [PMC free article] [PubMed]
- Doye, J. P. K. (2004). Phys. Biol. 1, P9–P13. [DOI] [PubMed]
- Dutzler, R., Campbell, E. B. & MacKinnon, R. (2003). Science, 300, 108–112. [DOI] [PubMed]
- Dyda, F., Hickman, A. B., Jenkins, T. M., Engelman, A., Craigie, R. & Davies, D. R. (1994). Science, 266, 1981–1986. [DOI] [PubMed]
- Eichinger, A., Nasreen, A., Kim, H. J. & Skerra, A. (2007). J. Biol. Chem. 282, 31068–31075. [DOI] [PubMed]
- Engel, C. K., Chen, L. & Privé, G. G. (2002). Biochim. Biophys. Acta, 1564, 38–46. [DOI] [PubMed]
- Erdemir, D., Lee, A. Y. & Myerson, A. S. (2009). Acc. Chem. Res. 42, 621–629. [DOI] [PubMed]
- Farinas, E. T. (2006). Comb. Chem. High Throughput Screen. 9, 235–236. [DOI] [PubMed]
- Farinas, E. T., Bulter, T. & Arnold, F. H. (2001). Curr. Opin. Biotechnol. 12, 545–551. [DOI] [PubMed]
- Fellouse, F. A., Barthelemy, P. A., Kelley, R. F. & Sidhu, S. S. (2006). J. Mol. Biol. 357, 100–114. [DOI] [PubMed]
- Fellouse, F. A., Wiesmann, C. & Sidhu, S. S. (2004). Proc. Natl Acad. Sci. USA, 101, 12467–12472. [DOI] [PMC free article] [PubMed]
- Finkelstein, A. V. & Janin, J. (1989). Protein Eng. 3, 1–3. [DOI] [PubMed]
- Georgalis, Y., Umbach, P., Raptis, J. & Saenger, W. (1997). Acta Cryst. D53, 691–702. [DOI] [PubMed]
- Gilbreth, R. N., Esaki, K., Koide, A., Sidhu, S. S. & Koide, S. (2008). J. Mol. Biol. 381, 407–418. [DOI] [PMC free article] [PubMed]
- Gliko, O., Neumaier, N., Pan, W., Haase, I., Fischer, M., Bacher, A., Weinkauf, S. & Vekilov, P. G. (2005). J. Am. Chem. Soc. 127, 3433–3438. [DOI] [PubMed]
- Goeddel, D. V., Kleid, D. G., Bolivar, F., Heyneker, H. L., Yansura, D. G., Crea, R., Hirose, T., Kraszewski, A., Itakura, K. & Riggs, A. D. (1979). Proc. Natl Acad. Sci. USA, 76, 106–110. [DOI] [PMC free article] [PubMed]
- Goldschmidt, L., Cooper, D. R., Derewenda, Z. S. & Eisenberg, D. (2007). Protein Sci. 16, 1569–1576. [DOI] [PMC free article] [PubMed]
- Gordon, K., Redelinghuys, P., Schwager, S. L., Ehlers, M. R., Papageorgiou, A. C., Natesh, R., Acharya, K. R. & Sturrock, E. D. (2003). Biochem. J. 371, 437–442. [DOI] [PMC free article] [PubMed]
- Hamuro, Y., Coales, S. J., Southern, M. R., Nemeth-Cawley, J. F., Stranz, D. D. & Griffin, P. R. (2003). J. Biomol. Tech. 14, 171–182. [PMC free article] [PubMed]
- Hawkins, R. E., Russell, S. J. & Winter, G. (1992). J. Mol. Biol. 226, 889–896. [DOI] [PubMed]
- He, B., Wang, K., Liu, Y., Xue, B., Uversky, V. N. & Dunker, A. K. (2009). Cell Res. 19, 929–949. [DOI] [PubMed]
- Horan, T. P., Simonet, L., Jacobsen, R., Mann, M., Haniu, M., Wen, J., Arakawa, T., Kuwamoto, M. & Martin, F. (1998). Protein Expr. Purif. 14, 45–53. [DOI] [PubMed]
- Hunte, C. & Michel, H. (2002). Curr. Opin. Struct. Biol. 12, 503–508. [DOI] [PubMed]
- Itakura, K., Hirose, T., Crea, R., Riggs, A. D., Heyneker, H. L., Bolivar, F. & Boyer, H. W. (1977). Science, 198, 1056–1063. [DOI] [PubMed]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst. 24, 409–411.
- Janda, I., Devedjiev, Y., Derewenda, U., Dauter, Z., Bielnicki, J., Cooper, D. R., Graf, P. C., Joachimiak, A., Jakob, U. & Derewenda, Z. S. (2004). Structure, 12, 1901–1907. [DOI] [PMC free article] [PubMed]
- Janin, J. (1997). Nature Struct. Biol. 4, 973–974. [DOI] [PubMed]
- Janin, J. & Rodier, F. (1995). Proteins, 23, 580–587. [DOI] [PubMed]
- Jenkins, T. M., Hickman, A. B., Dyda, F., Ghirlando, R., Davies, D. R. & Craigie, R. (1995). Proc. Natl Acad. Sci. USA, 92, 6057–6061. [DOI] [PMC free article] [PubMed]
- Jiang, Y., Lee, A., Chen, J., Ruta, V., Cadene, M., Chait, B. T. & MacKinnon, R. (2003). Nature (London), 423, 33–41. [DOI] [PubMed]
- Jin, L., Pandey, P., Babine, R. E., Weaver, D. T., Abdel-Meguid, S. S. & Strickler, J. E. (2005). Acta Cryst. D61, 1418–1425. [DOI] [PubMed]
- Karpusas, M., Lucci, J., Ferrant, J., Benjamin, C., Taylor, F. R., Strauch, K., Garber, E. & Hsu, Y. M. (2001). Structure, 9, 321–329. [DOI] [PubMed]
- Kawate, T., Michel, J. C., Birdsong, W. T. & Gouaux, E. (2009). Nature (London), 460, 592–598. [DOI] [PMC free article] [PubMed]
- Ke, A. & Wolberger, C. (2003). Protein Sci. 12, 306–312. [DOI] [PMC free article] [PubMed]
- Kendrew, J. C., Parrish, R. G., Marrack, J. R. & Orlans, E. S. (1954). Nature (London), 174, 946–949. [DOI] [PubMed]
- Koide, A., Gilbreth, R. N., Esaki, K., Tereshko, V. & Koide, S. (2007). Proc. Natl Acad. Sci. USA, 104, 6632–6637. [DOI] [PMC free article] [PubMed]
- Koide, A., Tereshko, V., Uysal, S., Margalef, K., Kossiakoff, A. A. & Koide, S. (2007). J. Mol. Biol. 373, 941–953. [DOI] [PMC free article] [PubMed]
- Koide, S. (2009). Curr. Opin. Struct. Biol. 19, 449–457. [DOI] [PMC free article] [PubMed]
- Korotkov, K. V., Pardon, E., Steyaert, J. & Hol, W. G. (2009). Structure, 17, 255–265. [DOI] [PMC free article] [PubMed]
- Kovari, L. C., Momany, C. & Rossmann, M. G. (1995). Structure, 3, 1291–1293. [DOI] [PubMed]
- Kuge, M., Fujii, Y., Shimizu, T., Hirose, F., Matsukage, A. & Hakoshima, T. (1997). Protein Sci. 6, 1783–1786. [DOI] [PMC free article] [PubMed]
- Kwong, P. D., Wyatt, R., Desjardins, E., Robinson, J., Culp, J. S., Hellmig, B. D., Sweet, R. W., Sodroski, J. & Hendrickson, W. A. (1999). J. Biol. Chem. 274, 4115–4123. [DOI] [PubMed]
- Kwong, P. D., Wyatt, R., Robinson, J., Sweet, R. W., Sodroski, J. & Hendrickson, W. A. (1998). Nature (London), 393, 648–659. [DOI] [PMC free article] [PubMed]
- Lam, A. Y., Pardon, E., Korotkov, K. V., Hol, W. G. & Steyaert, J. (2009). J. Struct. Biol. 166, 8–15. [DOI] [PMC free article] [PubMed]
- Lawson, D. M., Artymiuk, P. J., Yewdall, S. J., Smith, J. M., Livingstone, J. C., Treffry, A., Luzzago, A., Levi, S., Arosio, P., Cesareni, G., Thomas, C. D., Shaw, W. V. & Harrison, P. M. (1991). Nature (London), 349, 541–544. [DOI] [PubMed]
- Lee, C. V., Liang, W. C., Dennis, M. S., Eigenbrot, C., Sidhu, S. S. & Fuh, G. (2004). J. Mol. Biol. 340, 1073–1093. [DOI] [PubMed]
- Lee, J. E., Fusco, M. L., Abelson, D. M., Hessell, A. J., Burton, D. R. & Saphire, E. O. (2009). Acta Cryst. D65, 1162–1180. [DOI] [PMC free article] [PubMed]
- Lee, J. E., Fusco, M. L., Hessell, A. J., Oswald, W. B., Burton, D. R. & Saphire, E. O. (2008). Nature (London), 454, 177–182. [DOI] [PMC free article] [PubMed]
- Lee, S. Y., Lee, A., Chen, J. & MacKinnon, R. (2005). Proc. Natl Acad. Sci. USA, 102, 15441–15446. [DOI] [PMC free article] [PubMed]
- Lee, X., Ahmed, F. R., Hirama, T., Huber, C. P., Rose, D. R., To, R., Hasnain, S., Tam, A. & Mort, J. S. (1990). J. Biol. Chem. 265, 5950–5951. [PubMed]
- Levinson, N. M., Seeliger, M. A., Cole, P. A. & Kuriyan, J. (2008). Cell, 134, 124–134. [DOI] [PMC free article] [PubMed]
- Lipovsek, D. & Pluckthun, A. (2004). J. Immunol. Methods, 290, 51–67. [DOI] [PubMed]
- Liu, B., Luna, V. M., Chen, Y., Stout, C. D. & Fee, J. A. (2007). Acta Cryst. F63, 1029–1034. [DOI] [PMC free article] [PubMed]
- Longenecker, K. L., Garrard, S. M., Sheffield, P. J. & Derewenda, Z. S. (2001). Acta Cryst. D57, 679–688. [DOI] [PubMed]
- Longenecker, K. L., Lewis, M. E., Chikumi, H., Gutkind, J. S. & Derewenda, Z. S. (2001). Structure, 9, 559–569. [DOI] [PubMed]
- Malawski, G. A., Hillig, R. C., Monteclaro, F., Eberspaecher, U., Schmitz, A. A., Crusius, K., Huber, M., Egner, U., Donner, P. & Muller-Tiemann, B. (2006). Protein Sci. 15, 2718–2728. [DOI] [PMC free article] [PubMed]
- Malhotra, A. (2009). Methods Enzymol. 463, 239–258. [DOI] [PubMed]
- Mark, B. L., Mahuran, D. J., Cherney, M. M., Zhao, D., Knapp, S. & James, M. N. (2003). J. Mol. Biol. 327, 1093–1109. [DOI] [PMC free article] [PubMed]
- Mateja, A., Devedjiev, Y., Krowarsch, D., Longenecker, K., Dauter, Z., Otlewski, J. & Derewenda, Z. S. (2002). Acta Cryst. D58, 1983–1991. [DOI] [PubMed]
- Matulis, D., Kranz, J. K., Salemme, F. R. & Todd, M. J. (2005). Biochemistry, 44, 5258–5266. [DOI] [PubMed]
- McElroy, H. H., Sisson, G. W., Schottlin, W. E., Aust, R. M. & Villafranca, J. E. (1992). J. Cryst. Growth, 122, 265–272.
- McPherson, A. (1982). The Preparation and Analysis of Protein Crystals. New York: John Wiley & Sons.
- Mezzasalma, T. M., Kranz, J. K., Chan, W., Struble, G. T., Schalk-Hihi, C., Deckman, I. C., Springer, B. A. & Todd, M. J. (2007). J. Biomol. Screen. 12, 418–428. [DOI] [PubMed]
- Mizutani, H., Saraboji, K., Malathy Sony, S. M., Ponnuswamy, M. N., Kumarevel, T., Krishna Swamy, B. S., Simanshu, D. K., Murthy, M. R. N. & Kunishima, N. (2008). Acta Cryst. D64, 1020–1033. [DOI] [PubMed]
- Mohanty, A. K., Fisher, A. J., Yu, Z., Pradeep, M. A., Janjanam, J. & Kaushik, J. K. (2009). Protein Expr. Purif. 64, 213–218. [DOI] [PubMed]
- Monne, M., Han, L., Schwend, T., Burendahl, S. & Jovine, L. (2008). Nature (London), 456, 653–657. [DOI] [PubMed]
- Mooij, W. T. M., Mitsiki, E. & Perrakis, A. (2009). Nucleic Acids Res. 37, W402–W405. [DOI] [PMC free article] [PubMed]
- Mullis, K., Faloona, F., Scharf, S., Saiki, R., Horn, G. & Erlich, H. (1986). Cold Spring Harb. Symp. Quant. Biol. 51, 263–273. [DOI] [PubMed]
- Munshi, S., Hall, D. L., Kornienko, M., Darke, P. L. & Kuo, L. C. (2003). Acta Cryst. D59, 1725–1730. [DOI] [PubMed]
- Muto, T., Tsuchiya, D., Morikawa, K. & Jingami, H. (2009). Acta Cryst. F65, 236–241. [DOI] [PMC free article] [PubMed]
- Nasreen, A., Vogt, M., Kim, H. J., Eichinger, A. & Skerra, A. (2006). Protein Sci. 15, 190–199. [DOI] [PMC free article] [PubMed]
- Nauli, S., Farr, S., Lee, Y.-J., Kim, H.-Y., Faham, S. & Bowie, J. U. (2007). Protein Sci. 16, 2542–2551. [DOI] [PMC free article] [PubMed]
- Neau, D. B., Gilbert, N. C., Bartlett, S. G., Dassey, A. & Newcomer, M. E. (2007). Acta Cryst. F63, 972–975. [DOI] [PMC free article] [PubMed]
- Nettleship, J. E., Ren, J., Rahman, N., Berrow, N. S., Hatherley, D., Barclay, A. N. & Owens, R. J. (2008). Protein Expr. Purif. 62, 83–89. [DOI] [PubMed]
- Niemann, H. H., Schmoldt, H. U., Wentzel, A., Kolmar, H. & Heinz, D. W. (2006). J. Mol. Biol. 356, 1–8. [DOI] [PubMed]
- Niessing, D., Huttelmaier, S., Zenklusen, D., Singer, R. H. & Burley, S. K. (2004). Cell, 119, 491–502. [DOI] [PubMed]
- Obradovic, Z., Peng, K., Vucetic, S., Radivojac, P., Brown, C. J. & Dunker, A. K. (2003). Proteins, 53, 566–572. [DOI] [PubMed]
- Ostermeier, C., Iwata, S., Ludwig, B. & Michel, H. (1995). Nature Struct. Biol. 2, 842–846. [DOI] [PubMed]
- Pai, E. F., Krengel, U., Petsko, G. A., Goody, R. S., Kabsch, W. & Wittinghofer, A. (1990). EMBO J. 9, 2351–2359. [DOI] [PMC free article] [PubMed]
- Pantazatos, D., Kim, J. S., Klock, H. E., Stevens, R. C., Wilson, I. A., Lesley, S. A. & Woods, V. L. Jr (2004). Proc. Natl Acad. Sci. USA, 101, 751–756. [DOI] [PMC free article] [PubMed]
- Patel, S. B., Cameron, P. M., Frantz-Wattley, B., O’Neill, E., Becker, J. W. & Scapin, G. (2004). Biochim. Biophys. Acta, 1696, 67–73. [DOI] [PubMed]
- Peabody, D. S. & Al-Bitar, L. (2001). Nucleic Acids Res. 29, E113. [DOI] [PMC free article] [PubMed]
- Pédelacq, J. D., Piltch, E., Liong, E. C., Berendzen, J., Kim, C.-Y., Rho, B.-S., Park, M. S., Terwilliger, T. C. & Waldo, G. S. (2002). Nature Biotechnol. 20, 927–932. [DOI] [PubMed]
- Pellicane, G., Smith, G. & Sarkisov, L. (2008). Phys. Rev. Lett. 101, 248102. [DOI] [PubMed]
- Petsev, D. N., Thomas, B. R., Yau, S. T., Tsekova, D., Nanev, C., Wilson, W. W. & Vekilov, P. G. (2001). J. Cryst. Growth, 232, 21–29.
- Pornillos, O., Ganser-Pornillos, B. K., Kelly, B. N., Hua, Y., Whitby, F. G., Stout, C. D., Sundquist, W. I., Hill, C. P. & Yeager, M. (2009). Cell, 137, 1282–1292. [DOI] [PMC free article] [PubMed]
- Price, W. N. II et al. (2009). Nature Biotechnol. 27, 51–57.
- Privé, G. G., Verner, G. E., Weitzman, C., Zen, K. H., Eisenberg, D. & Kaback, H. R. (1994). Acta Cryst. D50, 375–379. [DOI] [PubMed]
- Prongay, A. J., Smith, T. J., Rossmann, M. G., Ehrlich, L. S., Carter, C. A. & McClure, J. (1990). Proc. Natl Acad. Sci. USA, 87, 9980–9984. [DOI] [PMC free article] [PubMed]
- Robien, M. A., Nguyen, K. T., Kumar, A., Hirsh, I., Turley, S., Pei, D. & Hol, W. G. (2004). Protein Sci. 13, 1155–1163. [DOI] [PMC free article] [PubMed]
- Rosenbaum, D. M., Cherezov, V., Hanson, M. A., Rasmussen, S. G. F., Thian, F. S., Kobilka, T. S., Choi, H. J., Yao, X. J., Weis, W. I., Stevens, R. C. & Kobilka, B. K. (2007). Science, 318, 1266–1273. [DOI] [PubMed]
- Saiki, R. K., Gelfand, D. H., Stoffel, S., Scharf, S. J., Higuchi, R., Horn, G. T., Mullis, K. B. & Erlich, H. A. (1988). Science, 239, 487–491. [DOI] [PubMed]
- Saiki, R. K., Scharf, S., Faloona, F., Mullis, K. B., Horn, G. T., Erlich, H. A. & Arnheim, N. (1985). Science, 230, 1350–1354. [DOI] [PubMed]
- Sassenfeld, H. M. (1990). Trends Biotechnol. 8, 88–93. [DOI] [PubMed]
- Schwartz, T. U., Walczak, R. & Blobel, G. (2004). Protein Sci. 13, 2814–2818. [DOI] [PMC free article] [PubMed]
- Sennhauser, G., Amstutz, P., Briand, C., Storchenegger, O. & Grütter, M. G. (2007). PLoS Biol. 5, 106–113. [DOI] [PMC free article] [PubMed]
- Sennhauser, G. & Grütter, M. G. (2008). Structure, 16, 1443–1453. [DOI] [PubMed]
- Serrano-Vega, M. J. & Tate, C. G. (2009). Mol. Membr. Biol. 26, 385–396. [DOI] [PubMed]
- Sharma, S., Zheng, H., Huang, Y. P. J., Ertekin, A., Hamuro, Y., Rossi, P., Tejero, R., Acton, T. B., Xiao, R., Jiang, M., Zhao, L., Ma, L.-C., Swapna, G. V. T., Aramini, J. M. & Montelione, G. T. (2009). Proteins, 76, 882–894. [DOI] [PMC free article] [PubMed]
- Slabinski, L., Jaroszewski, L., Rychlewski, L., Wilson, I. A., Lesley, S. A. & Godzik, A. (2007). Bioinformatics, 23, 3403–3405. [DOI] [PubMed]
- Smyth, D. R., Mrozkiewicz, M. K., McGrath, W. J., Listwan, P. & Kobe, B. (2003). Protein Sci. 12, 1313–1322. [DOI] [PMC free article] [PubMed]
- Standfuss, J., Xie, G., Edwards, P. C., Burghammer, M., Oprian, D. D. & Schertler, G. F. (2007). J. Mol. Biol. 372, 1179–1188. [DOI] [PMC free article] [PubMed]
- Thornton, J. M. & Sibanda, B. L. (1983). J. Mol. Biol. 167, 443–460. [DOI] [PubMed]
- Tidor, B. & Karplus, M. (1994). J. Mol. Biol. 238, 405–414. [DOI] [PubMed]
- Trevino, S. R., Scholtz, J. M. & Pace, C. N. (2007). J. Mol. Biol. 366, 449–460. [DOI] [PMC free article] [PubMed]
- Trevino, S. R., Scholtz, J. M. & Pace, C. N. (2008). J. Pharm. Sci. 97, 4155–4166. [DOI] [PubMed]
- Uhlen, M., Forsberg, G., Moks, T., Hartmanis, M. & Nilsson, B. (1992). Curr. Opin. Biotechnol. 3, 363–369. [DOI] [PubMed]
- Ullah, H., Scappini, E. L., Moon, A. F., Williams, L. V., Armstrong, D. L. & Pedersen, L. C. (2008). Protein Sci. 17, 1771–1780. [DOI] [PMC free article] [PubMed]
- Uysal, S., Vasquez, V., Tereshko, V., Esaki, K., Fellouse, F. A., Sidhu, S. S., Koide, S., Perozo, E. & Kossiakoff, A. (2009). Proc. Natl Acad. Sci. USA, 106, 6644–6649. [DOI] [PMC free article] [PubMed]
- Vedadi, M. et al. (2006). Proc. Natl Acad. Sci. USA, 103, 15835–15840.
- Veesler, D., Dreier, B., Blangy, S., Lichière, J., Tremblay, D., Moineau, S., Spinelli, S., Tegoni, M., Plückthun, A., Campanacci, V. & Cambillau, C. (2009). J. Biol. Chem. 284, 30718–30726. [DOI] [PMC free article] [PubMed]
- Vekilov, P. G. (2003). Methods Enzymol. 368, 84–105. [DOI] [PubMed]
- Vekilov, P. G. (2004). Cryst. Growth Des. 4, 671–685.
- Vekilov, P. G., Feeling-Taylor, A., Yau, S.-T. & Petsev, D. (2002). Acta Cryst. D58, 1611–1616. [DOI] [PubMed]
- Vucetic, S., Brown, C. J., Dunker, A. K. & Obradovic, Z. (2003). Proteins, 52, 573–584. [DOI] [PubMed]
- Waldo, G. S. (2003). Curr. Opin. Chem. Biol. 7, 33–38.
- Waldo, G. S., Standish, B. M., Berendzen, J. & Terwilliger, T. C. (1999). Nature Biotechnol. 17, 691–695. [DOI] [PubMed]
- Warne, T., Serrano-Vega, M. J., Baker, J. G., Moukhametzianov, R., Edwards, P. C., Henderson, R., Leslie, A. G., Tate, C. G. & Schertler, G. F. (2008). Nature (London), 454, 486–491. [DOI] [PMC free article] [PubMed]
- Warne, T., Serrano-Vega, M. J., Tate, C. G. & Schertler, G. F. (2009). Protein Expr. Purif. 65, 204–213. [DOI] [PubMed]
- Wernimont, A. & Edwards, A. (2009). PLoS One, 4, e5094. [DOI] [PMC free article] [PubMed]
- Wiltzius, J. J., Sievers, S. A., Sawaya, M. R. & Eisenberg, D. (2009). Protein Sci. 18, 1521–1530. [DOI] [PMC free article] [PubMed]
- Yanez, M. E., Korotkov, K. V., Abendroth, J. & Hol, W. G. (2008). J. Mol. Biol. 375, 471–486. [DOI] [PMC free article] [PubMed]
- Yau, S.-T., Petsev, D. N., Thomas, B. R. & Vekilov, P. G. (2000). J. Mol. Biol. 303, 667–678. [DOI] [PubMed]
- Ye, J.-D., Tereshko, V., Frederiksen, J. K., Koide, A., Fellouse, F. A., Sidhu, S. S., Koide, S., Kossiakoff, A. A. & Piccirilli, J. A. (2008). Proc. Natl Acad. Sci. USA, 105, 82–87. [DOI] [PMC free article] [PubMed]
- Yip, C. K., Kimbrough, T. G., Felise, H. B., Vuckovic, M., Thomas, N. A., Pfuetzner, R. A., Frey, E. A., Finlay, B. B., Miller, S. I. & Strynadka, N. C. (2005). Nature (London), 435, 702–707. [DOI] [PubMed]
- Zhang, F., Basinski, M. B., Beals, J. M., Briggs, S. L., Churgay, L. M., Clawson, D. K., DiMarchi, R. D., Furman, T. C., Hale, J. E., Hsiung, H. M., Schoner, B. E., Smith, D. P., Zhang, X. Y., Wery, J.-P. & Schevitz, R. W. (1997). Nature (London), 387, 206–209. [DOI] [PubMed]