Abstract
For this study, we screened 1,152 signature-tagged mutagenesis mutants of Brucella melitensis 16M in a mouse model of infection and found 36 of them to be attenuated in vivo. Molecular characterization of transposon insertion sites showed that for four mutants, the affected genes were only present in Rhizobiaceae. Another mutant contained a disruption in a gene homologous to mosA, which is involved in rhizopine biosynthesis in some strains of Rhizobium, suggesting that this sugar may be involved in Brucella pathogenicity. A mutant was disrupted in a gene homologous to fliF, a gene potentially coding for the MS ring, a basal component of the flagellar system. Surprisingly, a mutant was affected in the rpoA gene, coding for the essential α-subunit of the RNA polymerase. This disruption leaves a partially functional protein, impaired for the activation of virB transcription, as demonstrated by the absence of induction of the virB promoter in the Tn5::rpoA background. The results presented here highlight the fact that the ability of Brucella to induce pathogenesis shares similarities with the molecular mechanisms used by both Rhizobium and Agrobacterium to colonize their hosts.
Brucella spp. are gram-negative, facultative, intracellular bacteria that cause abortion and sterility in domestic mammals and a chronic undulant fever in humans (6, 52). On the basis of ribosomal 16S sequence comparison, Brucella spp. are members of the alpha subdivision of the class Proteobacteria. Within the alpha subgroup, brucellae are specifically related to rickettsiae, agrobacteria, and rhizobiae, organisms that also have the ability or requirement to live in close association with eukaryotic cells (37). The complete genomes of Brucella melitensis and Brucella suis have been sequenced recently (11, 43); genomic analysis showed only pinpoint differences between the two species and suggested that Brucella may have evolved from a soil-plant-associated bacterium related to organisms like Rhizobium and Agrobacterium (43).
Brucella is able to survive in professional and nonprofessional phagocytes by subverting the intracellular trafficking of eukaryotic cells (3, 44, 45). Studies in epithelial cells have shown that the ability of Brucella to escape from the classical cellular trafficking pathway, which normally leads to the lysosome, needs at least the VirB system (homologous to a type IV secretion machinery) and the BvrR/BvrS two-component system (8, 10, 53). It has also been shown that Brucella recruits actin and activates small GTPases during its internalization in HeLa cells (20).
While genome analysis revealed some genes that could be related to virulence (e.g., adhesins, hemolysins, and invasins), it showed that Brucella lacks classical virulence-related sequences and genes, such as pathogenicity islands, type III secretion systems, toxins, pilus biogenesis genes, etc. (36). Therefore, to draw a complete map of the molecular basis of Brucella pathogenesis, unbiased approaches are still needed. Moreover, these approaches will help in the functional assignment of Brucella open reading frames (ORFs).
Recently, different methods have been used to detect genes which are induced or necessary in cellular models (16, 17, 27, 28), leading to the identification of several virulence genes. Although these models are relevant for Brucella virulence, they do not reflect the complexity of environments encountered by the bacterium in the host. In order to approach as much as possible the natural conditions, in vivo studies are required. One of the most powerful technologies for in vivo screening is signature-tagged mutagenesis (STM) (23), which allows screening for loss of virulence in an animal model of infection. The use of this technology led to the identification of new virulence genes in several pathogenic bacteria and fungi (for review, see reference 51). The method has been applied to Brucella in an acute infection model (29) as well as in a model of chronic persistence (25).
In this study, we present the results obtained by screening 1,152 new STM mutants during the acute phase of infection in a mouse model. We identified 36 attenuated mutants that were characterized regarding the ability to grow in different cellular models, sensitivity to oxidative and acidic stress, and lipopolysaccharide (LPS) phenotype. Molecular analysis of the mutants revealed that four of them are affected in genes that are only conserved in Rhizobiaceae. The presence of a fliF mutant allowed us to implicate a role for flagella in virulence. In this study, we focused on an unexpected mutant, which is disrupted in the essential gene rpoA. We showed that this mutant is impaired in the proper regulation of the virB operon and suggest that the C-terminal part of RpoA interacts with the virB transcriptional regulator.
MATERIALS AND METHODS
Bacterial strains and plasmids.
B. melitensis 16M Nalr (56) was used as the parental strain for all experiments. B. melitensis strains were grown on solid or liquid 2YT medium with appropriate antibiotics. The Escherichia coli strains used in this study were S17 λpir (recA thi pro hsd (r− m+) RP4::2-Tc::Mu::Km Tn7 lysogenized with λpir phage) (35), CC118 λpir [Δ(are-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE recA1 lysogenized with λpir phage] (9), and TOP10 [F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX14 recA1 deoR araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG] (Invitrogen). E. coli strains were grown on Luria-Bertani (LB) medium with appropriate antibiotics. Antibiotics were used at the following concentrations for E. coli and B. melitensis: ampicillin (Amp), 50 μg/ml; kanamycin (Kan), 50 μg/ml; nalidixic acid (Nal), 25 μg/ml. A modified minimal medium was used (nitrogen and carbon sources were supplied with glutamic acid, lactic acid, and glycerol) (18, 29). The plasmids used in this study were pUTmini-Tn5Km2 (23), pCR TOPO 2.1 (Invitrogen), and pBBR1-KGFP-virB (7).
Matings.
Matings were performed by mixing equal volumes (20 μl) of liquid cultures of E. coli S17 donor cells (optical density at 600 nm of 0.6) and the B. melitensis 16M Nalr recipient strain (overnight culture) on a 0.22-μm-pore-size filter. The filter was left for 1 h on an LB plate without antibiotics and then transferred onto a 2YT plate containing Kan and Nal. After 3 days of incubation at 37°C, the exconjugates were replicated on 2YT Nal Kan and on 2YT Nal Amp. The Amp-resistant clones (about 4% of the clones) were discarded, and the Amp-sensitive clones were transferred to 96-well plates.
Amplification and labeling of DNA tags. For amplification and labeling of tags, we modified a previously described protocol (24). For both the input and the output pools, bacteria from plates containing approximately 104 colonies were resuspended in phosphate-buffered saline (PBS) and then centrifuged, and genomic DNA from the pelleted bacteria was recovered by the cetyltrimethylammonium bromide method (4). Tags were initially amplified by PCR from genomic DNA with the primers P2 (5′-TACCTACAACCTCAAGCT-3′) and P4 (5′-TACCCATTCTAACCAAGC-3′). The amplicons were purified in Microbio spin columns (Bio-Rad), and a fraction was used as target for a second PCR including [32P]dCTP to radiolabel the tags. The cycling conditions for both PCRs were as follows: 5 min at 94°C followed by 20 cycles of 94°C for 30 s, 50°C for 45 s, and 72°C for 10 s. The reactions were performed with Taq polymerase from Biotools in a PTC-150 Minicycler (MJ Research).
Molecular techniques.
DNA manipulation was performed according to standard techniques (4). Restriction enzymes were purchased from Roche and primers were from Amersham Pharmacia. For dot blots, the selected pUTmini-Tn5Km2 was amplified from a plasmid midi-preparation using primers P3 (5′-CATGGTACCCATTCTAAC-3′) and P5 (5′-CTAGGTACCTACAACCTC-3′), with the following cycling conditions: 5 min at 94°C followed by 30 cycles of 94°C for 30 s, 50°C for 45 s, and 72°C for 10 s. One hundred microliters of a 1:10 dilution of the PCR products was transferred to a positively charged nylon membrane (Hybond N+; Amersham) with a dot blot apparatus (Bio-Rad). The blots and hybridization reactions were processed as described by Holden and Hensel (24).
Transposon insertion sites were amplified by arbitrary PCR (42). For the first round, the primers were ARB1 [5′-GGCCACGCGTCGACTAGTAC(N)9GATAT-3′], ARB3 [5′-GGCCACGCGTCGACTAGTAC(N)9CAGTC-3′], ARB4 [5′-GGCCACGCGTCGACTAGTAC(N)9TACGT-3′], and ARB5 [5′-GGCCACGCGTCGACTAGTAC(N)9GTTAC-3′], used with transposon-specific primers. P4 was used to amplify sequences from the I end and P7 (5′-GCACTTGTGTATAAGAGTCAG-3′) was used to amplify sequences from the O end. Ten microliters of the PCR product was used in the second round with the primers ARB2 (5′-GGCCACGCGTCGACTAGTAC-3′) and P9 (5′-CGGCCGCACTTGTGTATAAG-3′) for the product obtained with P4. The primers ARB2 (5′-GGCCACGCGTCGACTAGTAC-3′) and P8 (5′-CGCAGGGCTTTATTGATTC-3′) were used in the second round for the fragment obtained with P7. Arbitrary PCR was also performed with genomic DNA from the wild type as a negative control. The PCR products were cloned into pCR TOPO 2.1 (Invitrogen). The inserts were sequenced by the dye terminator method (Big Dye terminator kit; Perkin Elmer) with an ABI 377 sequencer. Sequences were analyzed by performing searches with the Blastx program (2) against the nonredundant peptide database (http://www.ncbi.nlm.nih.gov/) and against the B. melitensis database of the URBM bioinformatic group (http://serine.urbm.fundp.ac.be/∼seqbruce/GENOMES/).
Screening of the STM Library. Mutants were grown at 37°C in 200 μl of 2YT medium in 96-well microtiter plates with appropriate antibiotics for 48 h. The bacteria were then pooled, centrifuged at 4,000 rpm for 10 min in a Jouan centrifuge, and resuspended in 2 ml of 0.9% NaCl. The bacterial suspension was then diluted to a final concentration of 5 × 105 CFU in 100 μl of 0.9% NaCl. The number of bacteria was confirmed by plating dilutions on 2YT plates. The bacterial suspension (100 μl) was injected intraperitoneally into 5-week-old female BALB/c mice (n = 2). The remaining part of the suspension was plated onto medium for DNA isolation. Five days after the infection, animals were sacrificed and the spleens were removed aseptically. For recovery of bacteria, the spleens were homogenized in PBS-0.1% Triton X-100 (Roche) and dilutions were plated on 2YT medium. Plates containing approximately 104 clones were used for DNA extraction.
CI.
In competition experiments, mutant (Nalr Kanr) and wild-type (Nalr) bacteria were grown for 48 h in 2YT medium, and then equal amounts of bacteria (about 2.5 × 105 each in 100 μl of 0.9% NaCl) were mixed and injected intraperitoneally into mice (n = 2). Dilutions of the infecting doses were plated on 2YT and 2YT Kan media to estimate the ratio of mutant-to-wild-type bacteria in the inoculum. Mice were sacrificed after 5 days, and the spleens were removed and homogenized. To determine the proportion of mutant-to-wild-type bacteria, dilutions of spleen homogenates were plated on 2YT and 2YT Kan media. The competitive index (CI) was calculated as the proportion of mutant-to-wild-type bacteria recovered from the animals divided by the proportion of mutant-to-wild-type bacteria in the inoculum.
Phenotypical characterization of mutants. The crystal violet method (58) was used to stain B. melitensis 16M for the rough phenotype. Smooth colonies do not take up the dye, whereas rough colonies become colored. H2O2 sensitivity tests were performed as described previously (15).
For the pH sensitivity test, bacteria were grown on solid media buffered with acetate and citrate phosphate buffers (pHs 5, 5.4, and 7).
Infection of macrophages and HeLa cells.
Subconfluent monolayers (2 × 104) of bovine macrophages (54) or human HeLa cells were inoculated with bacteria diluted to 6 × 106 CFU ml−1 in cell culture medium. Plates were centrifuged for 10 min at 1,000 rpm at room temperature in a Jouan centrifuge and placed in a 5% CO2 atmosphere at 37°C. After 1 h, wells were washed three times and incubated for 48 h with cell culture medium supplemented with 50 μg of gentamicin per ml. At the end of the infection time, the monolayers were washed three times with cell culture medium and treated for 20 min with 200 μl of 0.1% Triton X-100 (Roche) in PBS. Serial dilutions of the lysates were plated onto 2YT plates for determination of CFU. Each infection was performed in triplicate.
Immunofluorescence assays.
Labeling of intracellular bacteria was performed as described previously (44). Briefly, a goat polyclonal anti-B. melitensis antibody was used to detect the bacterium and a Texas red-conjugated anti-goat immunoglobulin G was used as secondary antibody (Jackson ImmunoResearch Laboratories, Immunotech, Marseille, France). Glass coverslips (13-mm diameter) in 24-well plates were seeded with HeLa cells and macrophages suspended in culture medium (105 cells/well). The cells were inoculated with bacteria at a multiplicity of infection of 200 to 300 and washings were performed as described above. The infected cells were fixed for 20 min in 3% paraformaldehyde, pH 7.4, at room temperature at different times postinfection for immunofluorescence staining. Splenocytes from infected mice were collected at 5 days postinfection from spleens homogenized with a Potter-Elvehjem homogenizer in an NH4Cl solution. Cells were washed two times in PBS, pH 7.4, and resuspended in 2 ml of PBS. Five hundred microliters was deposed on glass coverslips saturated with poly-d-lysine and was left for 10 min at room temperature before fixation.
Fluorescence-activated cell sorting (FACS) analysis.
Flow cytometric analysis of bacterial virB expression was performed with a FACScalibur machine. About 10,000 individual events were excited with a 488-nm argon ion laser, and emission light was detected through a 530-nm bandpass filter. Flowjo software (Treestar) was used for the quantitation of fluorescence.
RESULTS
Selection of attenuated mutants.
For this study, we examined 12 uncharacterized pools from the signature-tagged mini-Tn5 library (29). Each pool was injected into two mice, and bacteria were recovered 5 days after infection. Of the 1,152 mutants screened, 37 gave consistently weak hybridization signals. The virulence defect of the 37 mutants was established by direct comparison against the wild type in mixed inoculum experiments. A fully virulent STM mutant was used as a negative control. This allowed us to confirm that 36 mutants had a significant colonization defect compared to B. melitensis 16M (Table 1).
TABLE 1.
Characterization of the mutantsa
| Functional family | Strain | Gene | Putative function | ORF no. | Mouse CI | Log CFU in HeLa cells/log CFU in macrophages |
|---|---|---|---|---|---|---|
| Transport | 19C3 | macA | Macrolide efflux | BMEI0359 | 1.0 × 10−3 | 2.4 ± 0.1/2.4 ± 0.1 |
| 16H1 | tig | Trigger factor | BMEI1069 | 4.0 × 10−3 | 2.2 ± 0.1/2.5 ± 0.4 | |
| 9A12 | ugpA | Glycerol transport | BMEII0591 | 1.1 × 10−2 | 1.8 ± 0.2/2.9 ± 1.1 | |
| 17F2 | ugpA | Glycerol transport | BMEII0624 | 3.5 × 10−3 | 2.5 ± 0.1/2.5 ± 0.1 | |
| Metabolism | 15B2 | dppA | Dipeptide uptake | BMEI0433 | 3.2 × 10−4 | 2.1 ± 0.4/3.3 ± 0.5 |
| 13D6 | ilvC | Val, Leu, and Ile synthesis | BMEI0624 | 3.1 × 10−2 | ND/NDb | |
| 8C7 | cobB | Cobalamin synthesis | BMEI0705 | 4.6 × 10−2 | 1.8 ± 0.2/3.3 ± 0.8 | |
| 8A8 | mosA | Inosamine methylase | BMEI1301 | 3.3 × 10−2 | 1.7 ± 0.1/2.4 ± 0.6 | |
| 16A11 | galcD | d-Galactarate dehydratase | BMEII0485 | 2.2 × 10−3 | 2.4 ± 0.1/3.0 ± 0.8 | |
| 15H1 | glcK | Glycerol kinase | BMEII0823 | 3.3 × 10−2 | 2.3 ± 0.3/3.1 ± 0.2 | |
| Regulation | 13B1 | asnC family | Transcriptional regulator | BMEI0357 | 5.4 × 10−4 | 0.9 ± 0.2/1.8 ± 0.2 |
| 16B12 | feuQ | Sensor | BMEI1336 | 8.0 × 10−4 | 2.4 ± 0.3/2.4 ± 0.1 | |
| 16D3 | gntR family | Transcriptional regulator | BMEII1066 | 1.0 × 10−1 | 2.4 ± 0.3/2.8 ± 0.1 | |
| DNA-RNA metabolism | 9A3 | rpoA | RNA polymerase α-subunit | BMEI0781 | 1.2 × 10−4 | 1.3 ± 0.1/0.5 ± 0.2 |
| 19A4 | xseA | Exodeoxyribonuclease | BMEII0527 | 3.7 × 10−3 | 1.2 ± 0.2/1.8 ± 0.3 | |
| LPS | 17F3 | pmm | Core biosynthesis | BMEI1396 | 5.7 × 10−3 | 2.3 ± 0.3/2.2 ± 0.2 |
| Flagella | 9C6 | fliF | MS ring | BMEII0151 | 6.6 × 10−2 | 2.0 ± 0.3/2.6 ± 0.7 |
| Oxidoreduction | 16B2 | pheB | Catechol dioxygenase | BMEII0136 | 3.2 × 10−4 | ND/ND |
| 19A2 | fdhA | Formate dehydrogenase | BMEII0378 | 1.3 × 10−2 | ND/ND | |
| 14G4 | norE | Nitric oxide reduction | BMEII1001 | 2.2 × 10−3 | 1.9 ± 0.1/2.5 ± 0.1 | |
| Unknown | 10B11 | Int. reg. | Intergenic region | BMEI0330-I0331 | 4.8 × 10−5 | 1.7 ± 0.1/2.0 ± 0.1 |
| 9A4 | Hypothetical protein | Coenzyme A-transferase III | BMEI0898 | 1.7 × 10−2 | 2.5 ± 0.6/2.0 ± 0.5 | |
| 8B9 | Hypothetical protein | Thioesterase | BMEI1167 | 2.5 × 10−2 | 2.5 ± 0.5/2.0 ± 0.2 | |
| 15F3 | Hypothetical protein | Brucella/Agrobacterium orphan gene | BMEI1229 | 4.5 × 10−1 | ND/ND | |
| 14D3 | Hypothetical protein | Brucella/Agrobacterium orphan gene | BMEI1339 | 3.6 × 10−4 | 1.5 ± 0.1/3.3 ± 0.2 | |
| 17E7 | Hypothetical protein | α-Proteobacteria orphan gene | BMEI1361 | 8.5 × 10−4 | ND/ND | |
| 13G7 | Hypothetical protein | α-Proteobacteria orphan gene | BMEI1433 | 3.2 × 10−4 | 1.9 ± 0.1/2.2 ± 0.1 | |
| 14B12 | Hypothetical protein | EAL domain | BMEI1448 | 4.5 × 10−3 | 2.8 ± 0.1/2.5 ± 0.5 | |
| 14B5 | Hypothetical protein | Tetratricopeptide repeat domain | BMEI1531 | 3.0 × 10−2 | 2.9 ± 0.5/0.8 ± 0.2 | |
| 17B2 | Hypothetical protein | ERFK/YBIS/YCFS/YNHG family | BMEI1809 | 1.0 × 10−3 | 1.7 ± 0.2/2.8 ± 0.2 | |
| 8B8 | Hypothetical protein | Rhizobiaceae orphan gene | BMEI1844 | 4.7 × 10−3 | 1.6 ± 0.4/2.0 ± 0.2 | |
| 9H5 | Hypothetical protein | Brucella/Mesorhizobium orphan gene | BMEI1879 | 1.8 × 10−2 | 1.7 ± 0.2/2.0 ± 0.1 | |
| 13G9 | Hypothetical protein | Beta-lactamase | BMEII0318 | 2.5 × 10−4 | 1.4 ± 0.2/2.4 ± 0.2 | |
| 13C9 | Hypothetical protein | Dipeptidase domain | BMEII0626 | 7.8 × 10−4 | 2.8 ± 0.2/2.4 ± 0.5 | |
| 8D7 | Hypothetical protein | Transmembrane protein | BMEII0935 | 1.7 × 10−2 | 1.9 ± 0.2/1.7 ± 0.2 | |
| 17B1 | Hypothetical protein | Halo-acid hydrolase domain | BMEII1045 | 1.3 × 10−2 | 2.6 ± 0.1/3.3 ± 0.4 |
For each mutant, homology searches were undertaken, using transposon flanking sequences against the genome of B. melitensis. The ORF number for each identified gene is given, and the putative function is based on homologies found by searching the BLAST database for the protein sequence deduced from each ORF. For the ORFs encoding proteins of unknown function, the domains detected by pfam are given. CIs were calculated relative to the wild-type strain B. melitensis 16M. The CI is defined as the output ratio (mutant/wild type) divided by the input ratio (mutant/wild type). Mouse CIs are the averages of CIs determined for two mice. Attenuation in a cellular model at 48 h postinfection is expressed as the difference in the log CFU between the wild type and the mutant. The standard deviations indicated are based on the mutants; the standard deviation for the wild type was ±0.03 for the two cellular models tested. Cellular infections were done individually for each mutant, and the experiments were performed two times in triplicate, with a ratio of 300 bacteria/cell.
ND, not done.
Molecular analysis of the mutants.
We demonstrated that each mutant contains a single copy of the transposon by Southern blotting with genomic DNA digested by EcoRV (data not shown). The insertion site of mini-Tn5 in the genomic DNA of attenuated mutants was amplified either by arbitrary PCR (42) or by inverse PCR. PCR products were cloned and sequenced. Homology searches were undertaken using the flanking sequences against the genome sequence of B. melitensis (11). The function of each disrupted gene was predicted by using genome annotation and by reusing the Blast software for each coding sequence against the nonredundant (NR) protein sequence database. Sequences with nonobvious similarities were analyzed with pfam (5) to detect conserved domains. The results of bioinformatic searches are shown in Table 1. Analysis of transposon insertion sites distribution on the two chromosomes (chr) showed that in 22 mutants, the transposon was located on chr I, and in 14 mutants, it was located on chr II. A total of 15 mutants had insertions in genes of unknown function, and among them, mutants 8B8, 9H5, 14D3, and 15F3 were affected in genes for which strong homologs are only found in bacteria belonging to the Rhizobiaceae and the mutated genes in mutants 17E7 and 13G7 were only shared by α-Proteobacteria (Table 1).
The 20 remaining mutants had insertions in genes for which the function assignment (based on homology) was obvious. These 20 genes can be classified into seven different functional classes (Table 1).
Of the four mutants affected in transport function, one had a transposon insertion in a macrolide efflux system, one had an insertion in a trigger factor that helps protein folding and secretion, and two had insertions in homologous genes coding for a UgpA-related protein.
Among the mutants classified by function as related to amino acid and sugar metabolism, two were related to amino acid metabolism, one was related to vitamin B-12 synthesis, and two were related to sugar metabolism. One of particular interest was mutated in a gene similar to mosA. In Rhizobium, mosA encodes a methylase involved in the synthesis of rhizopine, a compound synthesized in the plant nodule (39).
We found four mutants affected in regulation systems, three of them in classical transcriptional regulators and one in feuQ, encoding the sensor of a two-component system, FeuP/FeuQ. A feuP mutant has been described and was shown to be fully virulent in mice and macrophages (14). Our finding that a feuQ mutant of B. melitensis is attenuated in mice as well as in cellular models might suggest that feuP is not the sole response regulator interacting with feuQ or that this two-component system is involved in a different role in B. melitensis and B. suis.
The DNA-RNA metabolism class contains two mutants, one affected in xseA, a gene coding for an exodeoxyribonuclease, and one (9A3) in which the transposon is inserted at the 3′ end of the rpoA gene. rpoA codes for the α-subunit of the RNA polymerase, which is essential for transcription. Since the transposon is located close to the end of the gene, it is likely that the bacterium expresses a C-terminally truncated protein, allowing growth in laboratory medium.
The exopolysaccharide class contains one mutant, which has an insertion in the gene for phosphomannomutase, a protein involved in LPS biosynthesis (1).
We also found a mutant (9C6) in the fliF gene coding for the MS ring of the flagellar apparatus. It has been known for a long time that Brucella contains flagellar genes (22), and the genome sequence has revealed that it contains all of the genes necessary to build a flagellum but that it is deprived of any chemotactic system (30). It has been suggested that these genes are cryptic, because flagellated Brucella has never been observed. This is the first indication that the flagellar genes are not cryptic and may be involved in Brucella pathogenesis.
Three strains were mutated in sequences involved in oxidoreduction, including norE, which is involved in nitric oxide reduction.
One of the attenuated strains has a mutation in an intergenic region. In this mutant, the transposon is integrated at nucleotide 340509 of chr I and is therefore located 217 bp downstream of a gene that in Rhodobacter sphaeroides codes for an enzyme involved in succinylation of osmoregulated periplasmic glucans (7a) and 271 bp upstream of a gene coding for a protein of unknown function. The insertion of the transposon in such a place could have an effect on transcriptional termination of the upstream gene, or more likely, it might disrupt the promoter region of the downstream gene.
In vitro characterization of the attenuated mutants.
To further analyze the phenotypes of the mutants, we performed in vitro assays. Expression of the complete LPS molecule is known to be required for full virulence of Brucella (1, 19, 34, 46). Therefore, all mutants were tested for expression of a full LPS molecule by crystal violet colony staining. The sole rough mutant detected was strain 17F3, in which the gene coding for phosphomannomutase was disrupted. During its intracellular travel, Brucella is thought to resist oxidative stress as well as acidic pH (47), and to mimic these conditions, H2O2 and pH sensitivity of all mutants was studied. The mutants 8B9, 9A4, 13G7, and 16A11 showed a significant increase in sensitivity to H2O2, as tested by a disk sensitivity assay (diameters of 4.61 cm [P < 0.01], 4.15 cm [P < 0.05], 4.2 cm [P < 0.05], and 4.16 cm [P < 0.05], respectively, compared to 3.5 cm for the wild type). Interestingly, mutant 9A3 (Tn5::rpoA) was shown to be more resistant to oxidative stress. Mutants 8B9, 9A4, 14D3, and 16H1 were unable to grow at pH 5.4, unlike the wild type. Mutant 9A3 was the only strain that was able to grow at pH 5, whereas the wild type could not.
Intracellular growth of the mutants.
The pathogenicity of Brucella is critically dependent on its ability to infect and multiply in professional and nonprofessional phagocytes. We therefore established whether the attenuated mutants were also affected in the ability to invade into and survive within bovine macrophages and epithelial cells. The results showed that most mutants were attenuated in both bovine macrophages and HeLa cells (Table 1). However, mutant 9A3 (Tn5::rpoA) is attenuated only in HeLa cells.
RpoA is involved in virB regulation in vitro.
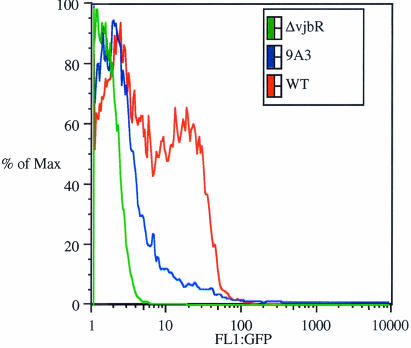
The transposon in the mutant 9A3 (Tn5::rpoA) is integrated at the 3′ end of the gene (at nucleotide 1007 of the 1,014-bp ORF). The mutant is predicted to express a protein in which the last residues of its C-terminal part are replaced by residues encoded by the I end of the transposon. It is known that the C-terminal part of the α-subunit is involved in the regulation of transcription through contact with cis or trans elements while the N-terminal domain is involved in the assembly of the polymerase (for review, see reference 48). Since it has been shown that RpoA is involved in the regulation of virB operon transcription in Agrobacterium tumefaciens (32, 33), we compared virB regulation in the Brucella rpoA::Tn5 mutant and wild-type strains. The plasmid pBBR1-KGFP-virB (7), containing the putative virB promoter fused with promoterless gfp, was shown to be suitable for virB expression studies (7). This plasmid was electroporated into B. melitensis 16M, into the rpoA transpositional mutant, and as a negative control into a characterized mutant (vjbR) known to be unable to activate transcription of the virB operon (R.-M. Delrue, personal communication). Green fluorescent protein production was monitored and measured by fluorescence-activated cell sorting (FACS). With the wild-type strain, fluorescence was detected during exponential growth in rich medium and after a 4-h shock in minimal medium, whereas no fluorescence was detected for the rpoA mutant nor for the negative control (Fig. 1).
FIG. 1.
Flow cytometry analysis of gfp expression from the virB promoter in rich medium. The plasmid pBBR1-KGFP-virB (7), containing the putative virB promoter fused to promoterless gfp, was electroporated into B. melitensis 16M, the rpoA transpositional mutant, and as a negative control a characterized mutant (vjbR) known to be unable to activate transcription of the virB operon. GFP production was monitored and measured by FACS after 24 h of growth. The graph shows the induction of the virB promoter after 24 h of growth in the wild-type background (optical density [OD], 1.38 at 600 nm) (red), in the rpoA mutant (OD, 1.6) (blue), and in the vjbR mutant (OD, 1.48) (green). FlowJo software (Treestar) was used for the quantitation of fluorescence. The percentage of the maximum is the number of cells in each bin divided by the number of cells in the bin that contains the largest number of cells.
RpoA is involved in virB regulation ex vivo and in vivo.
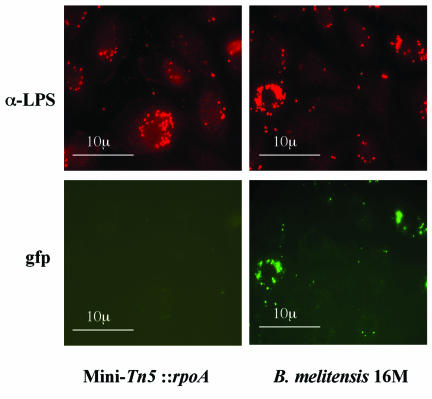
For B. suis, it was shown that virB is induced during growth in J774 macrophages (7). HeLa cells and bovine macrophages were infected with B. melitensis 16M and the rpoA mutant, both harboring pBBR1-KGFP-virB. Green fluorescent protein (GFP) production was monitored by fluorescence microscopy after 8, 24, and 48 h of infection. In both cellular models, we made the following observation: for the wild type, GFP-positive intracellular bacteria were detected each time. However, in the same cell, not all the bacteria were GFP positive, as shown in Fig. 2. For the rpoA mutant, no GFP-positive intracellular bacteria were detected at any time, suggesting that the mutant is unable to induce the virB promoter in either cellular model.
FIG. 2.
Expression of GFP from the virB promoter in macrophages at 24 h postinfection. Bovine macrophages were infected with B. melitensis 16M and the rpoA mutant, both harboring pBBR1-KGFP-virB. GFP production was monitored by fluorescence microscopy after 24 h of infection. GFP-positive intracellular bacteria were only detected in the cells infected by the wild type. With the rpoA mutant, no GFP-positive intracellular bacteria were detected at any time, suggesting that the mutant is unable to induce the virB promoter. Macrophages were infected at a ratio of 300 bacteria/cell. Bacteria were detected by specific antibodies (α-LPS).
Considering the complex phenotype of the rpoA mutant, we wanted to know if it was also unable to express virB in splenocytes. Mice were infected with B. melitensis 16M and the rpoA mutant transformed with pBBR1-KGFP-virB. Splenocytes were collected 5 days after intraperitoneal infection and GFP production was analyzed by fluorescence microscopy. Our results show that the virB operon is induced in the wild-type strain in vivo, whereas no GFP is detected in the rpoA mutant, suggesting that its inability to induce the virB operon leads to its rapid clearance from mice (data not shown).
DISCUSSION
The purpose of this study was to identify genes encoding virulence factors strictly required for the in vivo survival of B. melitensis 16M. Since Brucella is an intracellular pathogen, ex vivo assays have been developed to study the molecular mechanisms of its virulence (12, 13). The ex vivo studies allowed the discovery of several virulence factors (10, 16, 17, 27, 28, 41, 53). However, cellular models of infection only reflect part of the host-pathogen interactions. In vivo approaches are therefore required to extend our knowledge of bacterial pathogenesis. Two in vivo STM screens have already been performed with Brucella, allowing the discovery of several new virulence factors (25, 29). In this study, we performed an STM screen in a mouse model of infection in order to identify genes required during the acute phase of infection. This study allowed us to identify 36 new virulence genes of B. melitensis. The genes affected in the different mutants were classified into seven functional groups. Analysis of the different groups allowed us to draw the classical picture of what has recently been called the Brucella virulome (27), showing that amino acid and sugar metabolism is a major key for in vivo Brucella survival.
We found a mutant in norE, a gene coding for a component of the anaerobic respiratory chain that transforms nitrate to dinitrogen. This finding suggests two different possibilities: (i) the bacterium replicates in an environment deprived of oxygen and switches to anaerobic growth by using nitrate as an electron acceptor, as has already been suggested by the narG mutant described by Kohler et al. (27), with the absence of functional NorE leading to a toxic accumulation of NO in the bacterium, or (ii) the bacterium detoxifies nitric oxide produced by the cells as proposed by Wang et al. (57).
Genomic sequences from B. melitensis and B. suis revealed strong similarities with bacteria from the Rhizobium/Agrobacterium group (43). It has also been shown that the intracellular lifestyle of Brucella spp. requires genes that are also involved in the virulence of A. tumefaciens and in Rhizobium symbiosis (21, 26, 31, 53). The data presented here highlight the similarities between the two groups, as explained below.
The pathological and symbiotic processes developed by Agrobacterium and Rhizobium share many features, one of them being the ability to synthesize or induce synthesis of specific compounds known as opines for Agrobacterium and rhizopine for Rhizobium. Rhizopine (l-3-O-methyl-scyllo-inosamine [3-O-MSI]) is specifically synthesized by Rhizobium in the plant nodule (38, 39). The mos operon, composed of four genes (orf1, mosA, mosB, and mosC), encodes the enzymes of the 3-O-MSI synthesis pathway (38). It has been shown that MosA, MosB, and MosC are required for the synthesis of 3-O-MSI, whereas the function of orf1 still needs to be investigated (40). Rhizopine is a specific growth substrate for strains of Rhizobium carrying the moc operon and is thought to give a specific advantage to moc+ strains in the rhizosphere (49). In this study, we show that Brucella contains in its genome a gene that is highly homologous to mosA and that a mutation of this gene causes attenuation in vivo and in two cellular models, suggesting that Brucella might also produce an inositol derivative important for the colonization of its host. This result is reinforced by the finding that a gene homologous to mocC is specifically induced in macrophages (16). Moreover, analysis of the B. melitensis genome allowed us to find homologues of mocC, mocA, and mosC, and the implication of these genes in rhizopine metabolism and Brucella pathogenesis is under investigation in our lab. Preliminary results suggest that rhizopine might be a signal molecule affecting the growth of Brucella. However, we still do not know if Brucella does effectively synthesize rhizopine.
Bacteria from the Rhizobium/Agrobacterium group are usually motile and their flagella are involved in host colonization, whereas Brucella spp. are classically described as nonmotile. The presence of flagellar genes within the B. abortus genome has been described (22, 30, 50), but to date, no relation between these genes and virulence and/or motility has been reported. In this study, we have isolated an STM mutant in which the fliF gene is disrupted. This gene encodes a protein similar to the MS ring monomer, a basal component of the flagellum. This mutant is outcompeted by a wild-type strain in mice and is unable to replicate in macrophages and HeLa cells, suggesting an important role of the Brucella flagellar genes in virulence. Given the structural similarity between flagellar export machinery and the type III secretion system, Brucella flagellar genes might be involved in secretion rather than in motility. For example, for Yersinia (sensu stricto) it has been shown that flagellar components may act as a secretory apparatus involved in the export of phospholipase C (59).
The type four secretion system encoded by the Ti plasmid from the virB operon is an essential component of Agrobacterium virulence. In Agrobacterium, the transcription of the virB operon requires the interaction of the RpoA C-terminal domain with the transcriptional activator VirG (32, 33). In our study, we isolated a mutant of rpoA. Analysis of the transposon insertion site showed that the last 7 nucleotides of the gene are replaced by an unrelated sequence from the I end of the transposon. Our results show that this mutant is unable to induce transcription of the virB operon under all conditions tested. This led us to suggest that the change in the C-terminal domain of the protein impaired its interaction with different transcriptional regulators, among them the virB regulator. The VirB apparatus is known to be important for intracellular replication in both macrophages and HeLa cells; however, our mutant is attenuated in HeLa cells but not in macrophages and showed a rather puzzling phenotype, as it is resistant to acidic and oxidative stress. This complex phenotype may be due to a deregulation of transcription at some locus, as it is known, for example, that in E. coli the C-terminal part of the α-subunit interacts with OxyR, a transcriptional regulator for hydrogen peroxide-inducible genes (55). The persistence in macrophages is in contradiction with the absence of virB expression. However, in cellular models, we used inactivated macrophages, and infection study cultures stand for ≤48 h, a lapse of time during which its higher stress resistance might allow the mutant to persist in a place that is different from the natural Brucella replication vacuole. In mice, the infection lasts for a longer period of time and therefore macrophages may be activated. This is also illustrated by the observation that even with the wild type, it is rare to find a splenocyte full of Brucella, whereas in ex vivo models, HeLa cells and macrophages contain large numbers of bacteria after 48 h of infection. This may reflect less efficient replication in macrophages in vivo and emphasize the importance of using in vivo models of infection.
Finally, we also found six unknown virulence genes which, to date, are only shared among members of the α-proteobacteria. Studies of these genes might allow a better understanding of the phylogeny of the α-proteobacteria and of the story of their interaction with eukaryotic cells.
Acknowledgments
We are thankful to David Holden for the generous gift of the pool of tagged transposons and for scientific advice. We thank Christoph Tang and David Stroud for help in selecting the tags. We thank Christoph Tang for critical reading of the manuscript. We thank David O'Callaghan for pBBR1-KGFP-virB plasmid. We thank Jacques Godfroid, Isabelle Danese, and P. Michel for help in FACS analysis. We also thank Yves Poumay and Michel Herin for free access to their microscope.
This work was supported by the Commission of the European Communities, contract no. QLK2-CT-1999-00014.
Editor: D. L. Burns
REFERENCES
- 1.Allen, C. A., L. G. Adams, and T. A. Ficht. 1998. Transposon-derived Brucella abortus rough mutants are attenuated and exhibit reduced intracellular survival. Infect. Immun. 66:1008-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arenas, G. N., A. S. Staskevich, A. Aballay, and L. S. Mayorga. 2000. Intracellular trafficking of Brucella abortus in J774 macrophages. Infect. Immun. 68:4255-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. E. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1991. Current protocols in molecular biology. Green Publishing Associates, New York, N.Y.
- 5.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boschiroli, M. L., V. Foulongne, and D. O'Callaghan. 2001. Brucellosis: a worldwide zoonosis. Curr. Opin. Microbiol. 4:58-64. [DOI] [PubMed] [Google Scholar]
- 7.Boschiroli, M. L., S. Ouahrani-Bettache, V. Foulongne, S. Michaux-Charachon, G. Bourg, A. Allardet-Servent, C. Cazevieille, J. P. Liautard, M. Ramuz, and D. O'Callaghan. 2002. The Brucella suis virB operon is induced intracellularly in macrophages. Proc. Natl. Acad. Sci. USA 99:1544-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Cogez, V., E. Gak, A. Puskas, S. Kaplan, and J.-P. Bohin. 2002. The opgGIH and opgC genes of Rhodobacter sphaeroides form an operon that controls backbone synthesis and succinylation of osmoregulated periplasmic glucans. Eur. J. Biochem. 269:2473-2484. [DOI] [PubMed] [Google Scholar]
- 8.Comerci, D. J., M. J. Martinez-Lorenzo, R. Sieira, J. P. Gorvel, and R. A. Ugalde. 2001. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell. Microbiol. 3:159-168. [DOI] [PubMed] [Google Scholar]
- 9.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delrue, R. M., M. Martinez-Lorenzo, P. Lestrate, I. Danese, V. Bielarz, P. Mertens, X. De Bolle, A. Tibor, J. P. Gorvel, and J. J. Letesson. 2001. Identification of Brucella sp. genes involved in intracellular trafficking. Cell. Microbiol. 3:487-497. [DOI] [PubMed] [Google Scholar]
- 11.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H. Elzer, S. Hagius, D. O'Callaghan, J. J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Detilleux, P. G., B. L. Deyoe, and N. F. Cheville. 1990. Entry and intracellular localization of Brucella spp. in Vero cells: fluorescence and electron microscopy. Vet. Pathol. 27:317-328. [DOI] [PubMed] [Google Scholar]
- 13.Detilleux, P. G., B. L. Deyoe, and N. F. Cheville. 1990. Penetration and intracellular growth of Brucella abortus in nonphagocytic cells in vitro. Infect. Immun. 58:2320-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorrell, N., S. Spencer, V. Foulonge, P. Guigue-Talet, D. O'Callaghan, and B. W. Wren. 1998. Identification, cloning and initial characterisation of FeuPQ in Brucella suis: a new sub-family of two-component regulatory systems. FEMS Microbiol. Lett. 162:143-150. [DOI] [PubMed] [Google Scholar]
- 15.Elzer, P. H., R. W. Phillips, M. E. Kovach, K. M. Peterson, and R. M. Roop, 2nd. 1994. Characterization and genetic complementation of a Brucella abortus high-temperature-requirement A (htrA) deletion mutant. Infect. Immun. 62:4135-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eskra, L., A. Canavessi, M. Carey, and G. Splitter. 2001. Brucella abortus genes identified following constitutive growth and macrophage infection. Infect. Immun. 69:7736-7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foulongne, V., G. Bourg, C. Cazevieille, S. Michaux-Charachon, and D. O'Callaghan. 2000. Identification of Brucella suis genes affecting intracellular survival in an in vitro human macrophage infection model by signature-tagged transposon mutagenesis. Infect. Immun. 68:1297-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerhardt, P., L. A. Tucker, and J. B. Wilson. 1950. The nutrition of Brucellae; utilization of single amino acids for growth. J. Bacteriol. 59:777-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godfroid, F., B. Taminiau, I. Danese, P. Denoel, A. Tibor, V. Weynants, A. Cloeckaert, J. Godfroid, and J. J. Letesson. 1998. Identification of the perosamine synthetase gene of Brucella melitensis 16 M and involvement of lipopolysaccharide O side chain in Brucella survival in mice and in macrophages. Infect. Immun. 66:5485-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guzman-Verri, C., E. Chaves-Olarte, C. von Eichel-Streiber, I. Lopez-Goni, M. Thelestam, S. Arvidson, J. P. Gorvel, and E. Moreno. 2001. GTPases of the Rho subfamily are required for Brucella abortus internalization in nonprofessional phagocytes: direct activation of Cdc42. J. Biol. Chem. 276:44435-44443. [DOI] [PubMed] [Google Scholar]
- 21.Guzman-Verri, C., L. Manterola, A. Sola-Landa, A. Parra, A. Cloeckaert, J. Garin, J. P. Gorvel, I. Moriyon, E. Moreno, and I. Lopez-Goni. 2002. The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. Proc. Natl. Acad. Sci. USA 99:12375-12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halling, S. M. 1998. On the presence and organization of open reading frames of the nonmotile pathogen Brucella abortus similar to class II, III, and IV flagellar genes and to LcrD virulence superfamily. Microb. Comp. Genomics 3:21-29. [DOI] [PubMed] [Google Scholar]
- 23.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 24.Holden, D. W., and M. Hensel. 1998. Signature tagged mutagenesis. Methods Microbiol. 27:359-370. [Google Scholar]
- 25.Hong, P. C., R. M. Tsolis, and T. A. Ficht. 2000. Identification of genes required for chronic persistence of Brucella abortus in mice. Infect. Immun. 68:4102-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inon de Iannino, N., G. Briones, M. Tolmasky, and R. A. Ugalde. 1998. Molecular cloning and characterization of cgs, the Brucella abortus cyclic beta(1-2) glucan synthetase gene: genetic complementation of Rhizobium meliloti ndvB and Agrobacterium tumefaciens chvB mutants. J. Bacteriol. 180:4392-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohler, S., V. Foulongne, S. Ouahrani-Bettache, G. Bourg, J. Teyssier, M. Ramuz, and J. P. Liautard. 2002. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc. Natl. Acad. Sci. USA 99:15711-15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohler, S., S. Ouahrani-Bettache, M. Layssac, J. Teyssier, and J. P. Liautard. 1999. Constitutive and inducible expression of green fluorescent protein in Brucella suis. Infect. Immun. 67:6695-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lestrate, P., R. M. Delrue, I. Danese, C. Didembourg, B. Taminiau, P. Mertens, X. De Bolle, A. Tibor, C. M. Tang, and J. J. Letesson. 2000. Identification and characterization of in vivo attenuated mutants of Brucella melitensis. Mol. Microbiol. 38:543-551. [DOI] [PubMed] [Google Scholar]
- 30.Letesson, J. J., P. Lestrate, R. M. Delrue, I. Danese, F. Bellefontaine, D. Fretin, B. Taminiau, A. Tibor, A. Dricot, C. Deschamps, V. Haine, S. Leonard, T. Laurent, P. Mertens, J. Vandenhaute, and X. De Bolle. 2002. Fun stories about Brucella: the “furtive nasty bug.” Vet. Microbiol. 90:317-328. [DOI] [PubMed] [Google Scholar]
- 31.LeVier, K., R. W. Phillips, V. K. Grippe, R. M. Roop, 2nd, and G. C. Walker. 2000. Similar requirements of a plant symbiont and a mammalian pathogen for prolonged intracellular survival. Science 287:2492-2493. [DOI] [PubMed] [Google Scholar]
- 32.Lohrke, S. M., S. Nechaev, H. Yang, K. Severinov, and S. J. Jin. 1999. Transcriptional activation of Agrobacterium tumefaciens virulence gene promoters in Escherichia coli requires the A. tumefaciens RpoA gene, encoding the alpha subunit of RNA polymerase. J. Bacteriol. 181:4533-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lohrke, S. M., H. Yang, and S. Jin. 2001. Reconstitution of acetosyringone-mediated Agrobacterium tumefaciens virulence gene expression in the heterologous host Escherichia coli. J. Bacteriol. 183:3704-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McQuiston, J. R., R. Vemulapalli, T. J. Inzana, G. G. Schurig, N. Sriranganathan, D. Fritzinger, T. L. Hadfield, R. A. Warren, L. E. Lindler, N. Snellings, D. Hoover, S. M. Halling, and S. M. Boyle. 1999. Genetic characterization of a Tn5-disrupted glycosyltransferase gene homolog in Brucella abortus and its effect on lipopolysaccharide composition and virulence. Infect. Immun. 67:3830-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno, E., and I. Moriyon. 2002. Brucella melitensis: a nasty bug with hidden credentials for virulence. Proc. Natl. Acad. Sci. USA 99:1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreno, E., E. Stackebrandt, M. Dorsch, J. Wolters, M. Busch, and H. Mayer. 1990. Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the alpha-2 subdivision of the class Proteobacteria. J. Bacteriol. 172:3569-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy, P. J., N. Heycke, Z. Banfalvi, M. E. Tate, F. J. de Bruijn, A. Kondorosi, J. Tempe, and J. Schell. 1987. Genes for the catabolism and synthesis of an opine-like compound in Rhizobium meliloti are closely linked on the Sym plasmid. Proc. Natl. Acad. Sci. USA 84:493-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy, P. J., N. Heycke, S. P. Trenz, P. Ratet, F. J. de Bruijn, and J. Schell. 1988. Synthesis of an opine-like compound, a rhizopine, in alfalfa nodules is symbiotically regulated. Proc. Natl. Acad. Sci. USA 85:9133-9137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy, P. J., S. P. Trenz, W. Grzemski, F. J. De Bruijn, and J. Schell. 1993. The Rhizobium meliloti rhizopine mos locus is a mosaic structure facilitating its symbiotic regulation. J. Bacteriol. 175:5193-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Callaghan, D., C. Cazevieille, A. Allardet-Servent, M. L. Boschiroli, G. Bourg, V. Foulongne, P. Frutos, Y. Kulakov, and M. Ramuz. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 33:1210-1220. [DOI] [PubMed] [Google Scholar]
- 42.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 43.Paulsen, I. T., R. Seshadri, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, T. D. Read, R. J. Dodson, L. Umayam, L. M. Brinkac, M. J. Beanan, S. C. Daugherty, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, W. C. Nelson, B. Ayodeji, M. Kraul, J. Shetty, J. Malek, S. E. Van Aken, S. Riedmuller, H. Tettelin, S. R. Gill, O. White, S. L. Salzberg, D. L. Hoover, L. E. Lindler, S. M. Halling, S. M. Boyle, and C. M. Fraser. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA 99:13148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pizarro-Cerda, J., S. Meresse, R. G. Parton, G. van der Goot, A. Sola-Landa, I. Lopez-Goni, E. Moreno, and J. P. Gorvel. 1998. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect. Immun. 66:5711-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pizarro-Cerda, J., E. Moreno, V. Sanguedolce, J. L. Mege, and J. P. Gorvel. 1998. Virulent Brucella abortus prevents lysosome fusion and is distributed within autophagosome-like compartments. Infect. Immun. 66:2387-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Porte, F., A. Naroeni, S. Ouahrani-Bettache, and J. P. Liautard. 2003. Role of the Brucella suis lipopolysaccharide O antigen in phagosomal genesis and in inhibition of phagosome-lysosome fusion in murine macrophages. Infect. Immun. 71:1481-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rafie-Kolpin, M., R. C. Essenberg, and J. H. Wyckoff, 3rd. 1996. Identification and comparison of macrophage-induced proteins and proteins induced under various stress conditions in Brucella abortus. Infect. Immun. 64:5274-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rhodius, V. A., and S. J. Busby. 1998. Positive activation of gene expression. Curr. Opin. Microbiol. 1:152-159. [DOI] [PubMed] [Google Scholar]
- 49.Rossbach, S., G. Rasul, M. Schneider, B. Eardly, and F. J. de Bruijn. 1995. Structural and functional conservation of the rhizopine catabolism (moc) locus is limited to selected Rhizobium meliloti strains and unrelated to their geographical origin. Mol. Plant Microbe Interact. 8:549-559. [DOI] [PubMed] [Google Scholar]
- 50.Sanchez, D. O., R. O. Zandomeni, S. Cravero, R. E. Verdun, E. Pierrou, P. Faccio, G. Diaz, S. Lanzavecchia, F. Aguero, A. C. Frasch, S. G. Andersson, O. L. Rossetti, O. Grau, and R. A. Ugalde. 2001. Gene discovery through genomic sequencing of Brucella abortus. Infect. Immun. 69:865-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shea, J. E., J. D. Santangelo, and R. G. Feldman. 2000. Signature-tagged mutagenesis in the identification of virulence genes in pathogens. Curr. Opin. Microbiol. 3:451-458. [DOI] [PubMed] [Google Scholar]
- 52.Smith, L. D., and T. A. Ficht. 1990. Pathogenesis of Brucella. Crit. Rev. Microbiol. 17:209-230. [DOI] [PubMed] [Google Scholar]
- 53.Sola-Landa, A., J. Pizarro-Cerda, M. J. Grillo, E. Moreno, I. Moriyon, J. M. Blasco, J. P. Gorvel, and I. Lopez-Goni. 1998. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol. Microbiol. 29:125-138. [DOI] [PubMed] [Google Scholar]
- 54.Stabel, J. R., and T. J. Stabel. 1995. Immortalization and characterization of bovine peritoneal macrophages transfected with SV40 plasmid DNA. Vet. Immunol. Immunopathol. 45:211-220. [DOI] [PubMed] [Google Scholar]
- 55.Tao, K., C. Zou, N. Fujita, and A. Ishihama. 1995. Mapping of the OxyR protein contact site in the C-terminal region of RNA polymerase alpha subunit. J. Bacteriol. 177:6740-6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verger, J. M., M. Grayon, E. Chaslus-Dancla, M. Meurisse, and J. P. Lafont. 1993. Conjugative transfer and in vitro/in vivo stability of the broad-host-range IncP R751 plasmid in Brucella spp. Plasmid 29:142-146. [DOI] [PubMed] [Google Scholar]
- 57.Wang, M., N. Qureshi, N. Soeurt, and G. Splitter. 2001. High levels of nitric oxide production decrease early but increase late survival of Brucella abortus in macrophages. Microb. Pathog. 31:221-230. [DOI] [PubMed] [Google Scholar]
- 58.White, P. G., and J. B. Wilson. 1951. Differentiation of smooth and nonsmooth colonies of brucellae. J. Bacteriol. 61:239-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Young, G. M., D. H. Schmiel, and V. L. Miller. 1999. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. USA 96:6456-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]