Abstract
Replication of kDNA in the mitochondrion of the kinetoplastid protozoan is an essential process. One of the proteins that may be required for the kDNA replication is the ribonuclease H (RNase H; EC 3.1.26.4). We have identified four distinct ribonuclease H genes in Leishmania, one type I (LRNase HI) and three type II (LRNase HIIA, LRNase HIIB and LRNase HIIC). We detail here molecular characterization of LRNase HIIC. The coding sequence of LRNase HIIC is 1425 bp in length encoding a 474-amino acid protein with a calculated molecular mass of ~53 kDa. While LRNase HIIC shares several conserved domains with mitochondrial RNase H from other organisms, it has three extra patches of amino acid sequences unique to this enzyme. Functional identity of this protein as an RNase H was verified by genetic complementation in RNase H-deficient Escherichia coli. The precursor protein may be enzymatically inactive as it failed to complement the E. coli mutant. The mitochondrial localization signal in LRNase HIIC is within the first 40 amino acid residues at the N-terminus. In vitro import of the protein by the mitochondrial vesicles showed that the precursor protein is processed to a 49-kDa protein. Antisense ablation of LRNase HIIC gene expression is lethal to the parasite cells both in vitro and in vivo. This study not only reveals the significance of the LRNase HIIC in the kinetoplast biology but also identifies a potential molecular target for antileishmanial chemotherapy.
Keywords: Leishmania, Kinetoplast, Ribonuclease H, Antisense oligonucleotide
1. Introduction
Ribonuclease H (RNase H; EC 3.1.26.4) is a ubiquitous endoribonuclease that hydrolyzes the RNA in a RNA/DNA heteroduplex [1–3]. RNase H activity has been suggested to be involved in DNA replication, recombination repair and transcription [4,5]. Genes encoding RNase H are ubiquitously present in all the kingdoms of life starting from viruses to the humans [1].
RNase H enzymes were initially characterized from Escherichia coli [3]. Depending upon their amino acid sequence similarity with the two E. coli RNase H enzymes and requirement of either Mg2+ or Mn2+, RNases H are classified into three families: types I, II and III [6]. Enzymes that require Mg2+ for their activity and share conserved amino acid residues/domains with that of E. coli RNase HI are classified as type I RNase H [6]. Those sharing conserved domains with E. coli RNase HII and require Mn2+ for their activity are classified as type II [6]. The third type of RNase H share conserved domains with E. coli RNase HII but requires Mg2+ for their activities [6]. Apart from the difference in the divalent metal ion requirement RNases H from different families have differences in their specific activities and specificities for cleavage sites on the target [4–6]. RNase H has been shown to be essential in mouse mitochondria [7] and in Drosophila [8]. This enzyme also has been shown to play a critical role in the antisense effects of oligodeoxynucleotides in the ablation of the sense mRNA function [1,9–11]. We report here the characterization of a putative mitochondrial RNase H from the parasitic protozoan Leishmania. As RNase H is involved in the vital metabolism of the cell, any differential biochemistry and molecular biology of these enzymes in a parasite as compared to that of its host could be exploited for rational drug design against the parasite infection.
The kinetoplastid protozoan parasite Leishmania are transmitted by the phlebotomine sandfly and cause different clinical forms of leishmaniasis [12]. Visceral leishmaniasis is often fatal if untreated, mucocutaneous leishmaniasis is a mutilating disease, diffuse cutaneous leishmaniasis is disabling and cutaneous leishmaniasis can result in an unaesthetic stigma if multiple lesions occur [12]. Disability-adjusted life years (DALY) lost due to leishmaniasis are close to 2.4 million; there is 1.0–1.5 million cases of cutaneous and 500,000 cases of visceral leishmaniasis each year and a population of 350 million is at risk [12]. Leishmania is transmitted from the infected sandfly vectors to the mammalian hosts during blood meal [13,14]. Leishmania exists as the extracellular flagellated promastigotes in the gut of the fly vector and as the intracellular non-motile amastigote in the macrophages of the human host [13,15]. Amastigotes multiply in the macrophages, come out of them by killing the macrophages and then infect the neighboring macrophages thereby establishing the infection.
Kinetoplastids are also important as one of the earliest diverging eukaryotic groups that contain a mitochondrion [16,17]. They only have one mitochondrion per cell. The mitochondrial DNA creates a unique structure, known as the kinetoplast [16,17]. The kinetoplast contains several thousands of double stranded circular DNA catenated into a single disc-shaped network (kDNA), which is placed near the basal body of the flagellum in the mitochondrial matrix [18,19].
The mechanism of kDNA replication has been explored for many years, as it is very important for the survival of the parasite [17,20,21]. Kinetoplast DNA replication has been shown in Trypanosoma brucei to be critical for the survival of the parasite cells [20,21]. In Crithidia, protozoan cells related to Leishmania, Ferguson et al. [22] have shown the involvement of two antipodal sites in k-DNA replication. Thus, various enzymes involved in the kDNA replication are strategically placed around the kDNA disc. The DNA polymerase β [20], topoisomerase II [21], DNA ligase [23] and SSE1 [24] are localized in two antipodal sites, while the universal minicircle sequence binding protein [25] is found in the kinetoflagellar zone (KFZ). Primase was found to be on both the sides of the kDNA disc [26]. During the time of replication, the DNA circles are released from the kDNA network in the KFZ, where they replicate and newly formed progeny appear to migrate towards the antipodal sites where they are attached to the network periphery by topoisomerase II [27].
Since RNase H should be important in the replication of kDNA, we explored whether this enzyme in the Leishmania mitochondrion is essential for the survival of the parasite. We report here molecular cloning and characterization of a mitochondrial RNase H from Leishmania (we call it LRNase HIIC). Our data reveal that this is a type II RNase H that is apparently synthesized as an inactive proenzyme. This proenzyme is possibly activated upon the entry into the mitochondrion. We have also shown that this enzyme is essential for the survival of the parasite and thus it could be a potential target for antileishmanial chemotherapy.
2. Materials and methods
2.1. Cell culture
The promastigotes and the amastigotes of L. major (Friedlin) and L. donovani (DD8) were used in this study. The promastigotes were grown at 25 °C in medium M199 with 10% heat-inactivated fetal bovine serum [10,11]. The amastigotes were maintained in mouse tail-base lesions or in infected J774G8 cultures [11]. Virulent promastigotes were differentiated from the amastigotes freshly isolated from infected macrophages [28]. We used the mouse macrophage cell line J774A1 for our study. These cells were grown in tissue culture flasks in RPMI 1640 with 20% heat-inactivated (56 °C, 30 min) fetal bovine serum at 37 °C, as described [11,29].
2.2. Analysis of DNA and RNA
Genomic libraries of L. donovani and L. major in λZAP Express (Stratagene, La Jolla, CA) were screened to obtain their LRNase HIIC genes. Leishmania genomic library was constructed from Sau3AI-digested genomic DNA fragments (2–6 kb) in λZAP Express vector (Stratagene) following standard protocols [30]. We searched the Sanger Center L. major genomic sequence database with T. brucei RNase HII protein sequence as bait to get a homologous probe. An ‘end sequence’ was found with significant (P/N: 2.5e−55) similarity. Primers P1/P2 (Table 1) were used to amplify the genomic DNA fragment which was used as probe for library screening. After repeated screening of the library, the LRNase HIIC gene was rescued following standard protocols [30]. Restriction endonuclease digestions followed by agarose gel electrophoresis and Southern blotting revealed the restriction map of the clones [30]. RNA was isolated from Leishmania promastigotes and axenic amastigotes using Trizol reagent (Invitrogen, Carlsbad, CA). Total RNA was fractionated on a formaldehyde-agarose (1%) gel, transferred to a nylon membrane and probed with LRNase HIIC ORF [30]. Sequencing of the full-length clones was done using the T7 and SP6 universal primers in an automated DNA sequencer.
Table 1.
Nucleotide sequences of the primers and gapmers used in this study
| Primer | Sequence (5′–3′) | Description |
|---|---|---|
| P1 | GACTGAGCGGAGCGGTGC | Forward primer to amplify the LRNase HIIC probe |
| P2 | GCACCCGAGCAGTTTCAG | Reverse primer to amplify the LRNase HIIC probe |
| P7 | GGATCCATGGTCCACGCATGGCGC | Forward primer to amplify the full length of the LRNase HIIC ORF with Nco I site at the 5′ end |
| P8 | GGATCCATGGTCACTTCCTCTGCAGCC | Reverse primer to amplify the full length of the LRNase HIIC ORF with Nco I site at the 5′ end |
| P9 | GGATCCATGGTGCCGCCGACATGGAA-CGCGCTG | Forward primer to amplify the LRNase HIIC ORF without MLS |
| P10 | CACCATGCTCCACGCATGGCGC | Forward primer with 5′-CACC-3′ sequence to clone LRNase HIIC in pET100/D |
| P11 | CACCCCGCCGACATGGAACGCGCTG | Forward primer with 5′-CACC-3′ sequence to clone LRNase HIICΔMLS in pET100/D |
| ASN | GTGGAGCATGATAGGCAG | Antisense ODN against LRNase HIIC |
| SSN | CTGCCTATCATGCTCCAC | Sense ODN to ASN |
| INV | GACGGATAGTACGAGGTG | Inverted ODN to ASN |
| SCR | TACAGAGGTAGGCGAGGT | Scrambled ODN to ASN |
| ASM | CTGATACTTATATAGCG | Antisense ODN against miniexon |
2.3. Complementation of conditional-lethal E. coli rnh− mutant
E. coli mutant MIC2067 [F−λ− IN(rrnD-rrnE)1 rnhA339::cat rnhB716::kam] was a generous gift from Dr. M. Itaya, Tokyo. These cells form colonies at 30 °C but do not grow at 42 °C due to the absence of functionally active RNase H and can be complemented to grow at 42 °C with the functional expression of a homo- or heterologous RNase H [31]. For genetic complementation, LRNase HIIC coding region were amplified with (primer P7/P8) or without (primer P9/P8) the putative mitochondrial localization signal (MLS) and cloned behind the arabinose-inducible promoter at the Nco 1 site of pBAD/His A vector (Invitrogen). Chemically competent E. coli MIC2067 cells were transformed [30] with the pBAD, pBAD-LRNase HIIC or pBAD-LRNase HIICΔMLS constructs and were plated on LB-agar containing 100 μg/ml ampicillin, 25 μg/ml kanamycin, 5 μg/ml chloramphenicol and 0.2% arabinose and incubated at 30 °C. After the growth at 30 °C, the transformants were streaked in the above mentioned antibiotic-agar plates, one set of plates were incubated at 30 °C and the duplicate set of plates were incubated at 42 °C for 16 h. The control plates had all the antibiotics but no arabinose.
2.4. Expression of recombinant LRNase HIIC and immunoblot analysis
The LRNase HIIC ORF was amplified and cloned in pET100/D-TOPO (Invitrogen). The recombinant protein was expressed after induction of the recombinant E. coli BL21 Star (DE3) cells with IPTG (0.2 mM, 3 h) and was purified from the lysate by immobilized-metal affinity chromatography using Ni-NTA resin (Qiagen, Valencia, CA) under denaturing conditions following the supplier’s protocols. Polyclonal antibodies against recombinant LRNase HIIC were custom developed (Antibody Solutions, Mountain View, CA) in rabbits. Affinity purified antibodies were used for immunoblot analysis following standard protocols [32]. The purified antibodies were immobilized onto a Protein A-Sepharose column (SeizeX Protein A Immunoprecipitation kit, Pierce Biotechnology, Rockford, IL) and the LRNase HIIC or LRNase HIICΔMLS protein was affinity purified following the protocol recommended by the supplier.
2.5. RNase H assay
We evaluated LRNase HIIC activity by zymogram analysis using the synthetic 32P-labeled RNA:DNA hybrid as the substrate [1]. [32P]Poly(rA):poly(dT) was synthesized using E. coli RNA polymerase on poly (dT), as described before [10,11]. Bovine pancreatic RNase A (Type 1A, Sigma Chemical Co.) and E. coli RNase H (Invitrogen) were used as negative and positive control, respectively, to verify the efficacy and specificity of the substrate.
2.6. Isolation of mitochondrial vesicles and in vitro import assay
Mitochondrial vesicles were obtained from the promastigotes after lysis of the cells by nitrogen cavitation, as described [33]. We used in vitro synthesized 35S-labeled precursor LRNase HIIC protein to study its in vitro uptake by the isolated mitochondrial vesicles. Full-length LRNase HIIC ORF or its ΔMLS derivative was cloned at the Nde I site in the pET100/D vector behind the T7 RNA polymerase promoter. Radiolabeled LRNase HIIC protein was synthesized using T7 RNA polymerase in the TNT Quick coupled transcription/translation system (Promega, Madison, WI) and [35S]-methionine. The 35S-methionine-labeled precursor protein was incubated with isolated mitochondrial vesicles for in vitro import study, as described [33]. Reaction was started by the addition of 10 μl of rabbit reticulocyte lysate containing 35S-methionine labeled LRNase HIIC precursor protein and incubated at 30 °C. After 30 min, samples were collected, and divided into two parts, one part was treated with proteinase K (30 μg/ml) and the other part left untreated. Proteinase K reaction was stopped by the addition of PMSF (2 mM). In one set of experiment, valinomycin (0.5 μM) was added to disrupt the membrane potential. Mitochondrial vesicles were pelleted by centrifugation, washed with 200 μl of SME buffer (0.25 M sucrose, 10 mM MOPS/KOH at pH 7.2 and 2 mM EDTA). Radiolabeled proteins in the pellet were analyzed by SDS-PAGE and autoradiography [33].
2.7. Antisense phosphorothioate gapmer-mediated knock down of LRNase HIIC expression in the promastigotes and the amastigotes
To knock down the LRNase HIIC mRNA, we employed an antisense deoxyribonucleotide (ODN) gapmer designed against this mRNA. We designed the 18-mer antisense ODN (ASN) ‘gapmer’ from the predicted translation start site (−9 to +9) in the mRNA nucleotide sequence. Five nucleotides from each terminus in the gapmer contained 2′-O-methyl ribose whereas the middle eight nucleotides were connected via phosphorothioate backbones but had 2′-deoxyribose. The control ODNs included in the study were SSN (sense ODN), INV (inverted sequence), and SCR (scrambled sequence) (Table 1). The oligonucleotides were custom synthesized from Trilink Biotech (San Diego, CA) and purified by reverse-phase HPLC. We also used anti-miniexon oligonucleotide ASM as a positive control [34]. The promastigotes (1 × 106 cells/ml) were incubated in complete growth medium in 24-well plates with different concentrations (0–30 μM) of specific ODN, which were added directly in the culture media, at 26 °C, for 72 h and counted the number of cells/ml using a hemocytometer as a measure of growth [10,11]. To study the effects of these ODNs against the amastigotes of L. major in J774A1 macrophage cells, we employed the scavenger receptor-mediated drug targeting techniques, described before [34]. J774A1 cells were infected with virulent stocks of L. major (derived from mouse tail-base lesions) by incubating the cells for 5 h in the growth medium with 1:10 macrophage to Leishmania promastigotes ratio. The infected J774A1 cells were washed in PBS and incubated with medium containing the liposome encapsulated ODN. The numbers of Leishmania amastigotes per 100 macrophages were counted after 5 days of incubation with the ODN-liposome preparations [34].
3. Results and discussion
3.1. Leishmania has four ribonuclease H genes
Our objective was to identify the mitochondrial RNase H from Leishmania and to explore whether this enzyme can be targeted for the development of rational chemotherapy against the diseases caused by Leishmania and other related pathogenic parasites. We have amplified, cloned and characterized four RNase H genes from L. major and L. donovani (Table 2). BLAST searches were performed at either the Sanger center (http://www.sanger.ac.uk/cgi-bin/blast/submitblast/l_major/omni) or National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/BLAST/) web sites to search for the RNase H genes in L. major data bank. Query RNase H protein sequences used were RNase HI, -HII of E. coli, Saccharomyces pombe, S. cerevisiae, Crithidia, T. brucei and human. Nucleotide and derived amino acid sequence analyses revealed that one of these RNase H is a type I enzyme (LRNase HI) and the other three enzymes are type II (LRNase HIIA, LRNase HIIB and LRNase HIIC, named according to increasing molecular sizes). T. brucei and T. cruzi have homologues for RNase HI, RNase HIIB and RNase HIIC (Table 2) that were found by searching their respective gene databases. RNase HIIA is apparently present only in Leishmania. The nucleotide sequences of the RNase H genes in Leishmania do not share any significant similarities among each other. Majority of organisms so far studied have two RNase H genes: one of type I and other of type II [1]. Recently, genome database analysis revealed multiple type II RNase H genes in organisms like Bacillus subtilis [6] and Caenorhabditis elegans [35]. Thus, the presence of multiple types II RNase H in Leishmania and related protozoa may not be unusual, but the biological roles played by many of these enzymes are yet to be determined.
Table 2.
Summary of the identified RNase H proteins in Leishmania and Trypanosoma
| AA | L. majora | T. brucei | T. cruzi |
|---|---|---|---|
| RNase HI | 42 kDa (381 aa) LmjF06.0290 (AY835988) | 33 kDa (301 aa) Tb07.26A24.40 | 36 kDa (323 aa) Tc00.1047053506351.40 |
| RNase HIIA | 33 kDa (296 aa) LmjF13.0050 (AY835989) | Absent | Absent |
| RNase HIIB | 33 kDa (301 aa) LmjF36.0640 (AY835990) | 36 kDa (326 aa) Tb10.70.2140 | 36 kDa (324 aa) Tc00.1047053508661.50 |
| RNase HIIC | 53.3 kDa (474 aa) LmjF36.0330 (AF441859) | 54 kDa (477 aa) Tb10.70.2470 | 55 kDa (479 aa) Tc00.1047053503891.70 |
The lengths of the proteins (number of amino acid residues) are given in the parenthesis following the calculated molecular weight. Temporary systematic IDs for the L. major sequences are retrieved from its GeneDBs (release 5.1 for Friedlin, the reference strain MHOM/IL/80/Friedlin, zymodeme MON-103), T. brucei sequences are retrieved from its GeneDBs (Release 4 of T. brucei strain TREU927/4 GUTat10.1 genome; http://www.genedb.org/genedb/tryp/index.jsp) and for the T. cruzi CL Brener (TIGR-Seattle Biomedical Research Institute-Karolinska Institute T. cruzi Sequencing Consortium (TSK-TSC); http://www.genedb.org/genedb/tcruzi/index.jsp) are written below the molecular sizes.
DNA sequences showing the 5′-UTR, transplicing signal, coding sequence and the 3′-UTR for all of the ribonucleases H of L. major are deposited to GenBank. Corresponding accession numbers are shown for each of RNase H in the parenthesis.
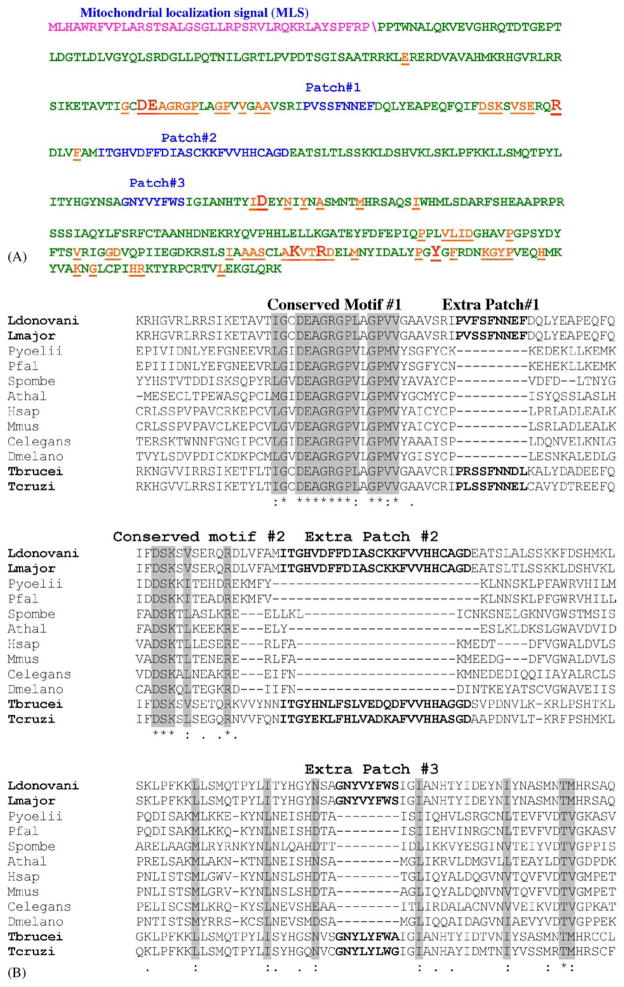
Computational analysis of the LRNase H amino acid sequences using web-based programs like Mitoprot and PSORTII suggested that only LRNase HIIC has the potential to be the mitochondrial enzyme with a putative mitochondrial localization signal (MLS), which is located within the N-terminal 40 amino acid residues (Fig. 1A). The predicted cleavage site for the signal peptide is given as FRP/PP (Fig. 1A). The results of the PSORT II prediction were 82.6% mitochondrial. Mitoprot predicted the probability of LRNase HIIC to be imported to mitochondria as 0.9862. We experimentally verified that the 40 amino acid residues in the N-terminal sequences are responsible for mitochondrial localization of LRNase HIIC (see below). Other than moderate similarity (49%) with the nucleotide sequences of T. brucei and T. cruzi RNase HIIC, there is no significant similarity of the LRNase HIIC nucleotide sequence with any other RNase HII nucleotide sequences available in the Entrez database, as was revealed by BLASTn analysis. Since the nucleotide sequence of LRNase HIIC mRNA has no significant similarity with that of the human ortholog, this parasite molecule can potentially be targeted for the development of antileishmanial antisense nucleic acid based chemotherapy, provided this is essential for the growth of the parasite (see below).
Fig. 1.
Analysis of LRNase HIIC amino acid sequence. (A) The amino acid sequence of LRNase HIIC highlighting the MLS (purple)\shows the predicted cleavage site, conserved amino acids (orange) and the extra patches of sequences (blue). The conserved amino acid residues that are essential for the enzymatic activity of the protein are shown in larger fonts. (B) ClustalW alignment of part of amino acid sequences of L. donovani LRNase HIIC (residues 119–298) with corresponding homologous RNase HII sequences from selected eukaryotes to show the relative location of the extra patches of amino acid sequences. The names of the organisms and the accession numbers of their RNase HII proteins are as follows: Ldonovani: L. donovani, AAL57850; Lmajor: L. major, AAL32059; Pyoellii: Plasmodium yoellii, EAA18794; Pfal: Plasmodium falciparum, NP 703896; T. brucei, AAD53318; Spombe: Schizosaccharomyces pombe, Q10236; Athal: Arabidopsis thalina, AAO64017; Hsap: Homo sapiens, AAQ64005; Mmus: Mus musculus, NP 081463; Celegans: Caenorhabditis elegans, CAC70103; and Dmelano: Drosophila melanogaster, Q9VPP5. The sequences for T. brucei and T. cruzi RNase HIIC were retrieved from the respective GeneDB (see the footnote of Table 2).
3.2. Along with the signature sequences, LRNase HIIC protein also has extra patches of amino acid sequences unique to this protein
Amino acid sequences of LRNase HIIC were aligned with homologous sequences from many eukaryotes including Trypanosoma, Plasmodium, yeast, nematode, plant and mammal (Fig. 1B). Highly conserved motifs, previously identified as hallmarks of RNase HII [36] are present in LRNase HIIC (Fig. 1A and B). One remarkable feature in the LRNase HIIC amino acid sequence is the presence of three unique patches of amino acid sequences (patch#1, residues 189–167; patch#2, residues 197–220, and patch#3, residues 264–271) which are significantly conserved in the RNase HIIC of the kinetoplastid flagellates but are absent in the RNase HII of plasmodia as well as in the non-protozoan cells (Fig. 1A and B). The antipodal sites of the kinetoplast are thought to be very critical for its proposed role in the replication of kDNA [22] and the mitochondrial RNase H of Leishmania thus may be located at these sites to aid in the kDNA replication process. It is thus possible that the unique patches in the kinetoplastid mitochondrial RNase H are participating in this proposed localization at the kinetoplast disk. Whether LRNase HIIC is localized at the antipodal nodes of the kinetoplast and whether this proposed strategic localization of LRNase HIIC in the kinetoplast is mediated through the extra patches of the identified sequences are yet to be determined.
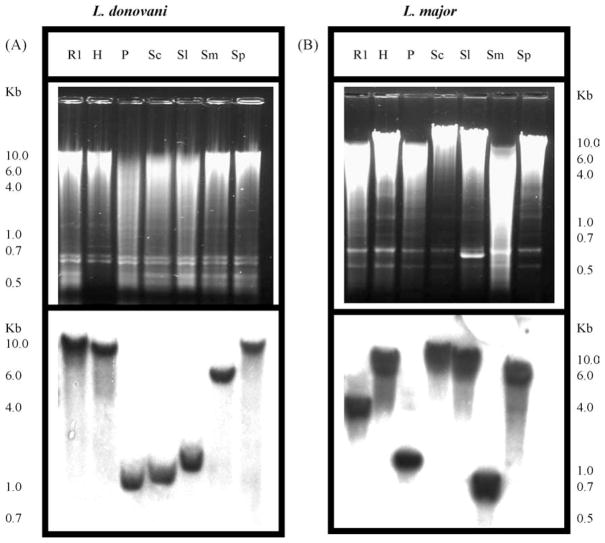
3.3. LRNase HIIC is a single copy gene
Southern analysis of Leishmania genomic DNA restriction digests with LRNase HIIC probe showed only single band (Fig. 2A and B), indicating single copy of the gene in the haploid genome of these diploid cells. In Leishmania and related other protozoa, the genes are transcribed as poly-cistronic messages. So determination of the copy number of a gene may help to understand how it is regulated. This information will also help to design experiments to knock out the gene by homologous recombination to study the contribution of the enzyme in parasite metabolism.
Fig. 2.
Gene copy number of LRNase HIIC. Southern blot analysis of the restriction digests of total DNA isolated from L. donovani (A) and L. major (B). Total DNA (5 μg for L. donovani and 10 μg for L. major) was digested with R1, Eco RI; H, Hind III; P, Pst I; Sc, Sac II; Sl, Sal I; Sm, Sma I and Sp, Spe I. The fragments were analyzed by agarose gel (0.8%) electrophoresis. The DNA fragments from the gel were transferred to Nytran membrane and hybridized with 32P-labeled RNase HII cDNA as probe. One kb plus DNA ladder (Invitrogen) was used as molecular size marker. Upper panels show the photographs of ethidium bromide-stained agarose gels and the lower panels show the corresponding autoradiograms.
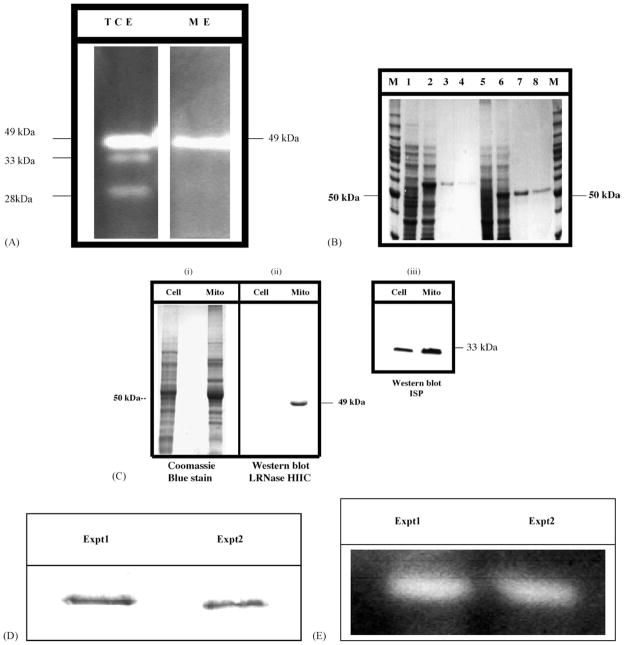
3.4. LRNase HIIC is enriched in the mitochondrial fraction as a 49-kDa protein
Analysis of RNase H activities in the total cellular extracts from the promastigotes using 32P-Poly(rA):Poly(dT) as substrate [10,11] in a gel assay revealed three bands (Fig. 3A). These activities are Mn2+-stimulated RNase H activities. Omitting Mn2+ or replacement of Mn2+ with Mg2+ abrogated or inhibited these activities (data not shown). The major Mn2+-stimulated RNase H activity (49-kDa) is enriched in the isolated mitochondrial fraction (Fig. 3A). The molecular size of the top-most band is 49-kDa. At this point we do not know the identity of the other two activity bands. They may represent the other two type II RNase H enzymes present in Leishmania. While E. coli RNase H cleaved the substrate into acid-soluble radioactive products, RNase A did not cleave the substrate under the conditions described. As stated above (Table 2), the calculated molecular size of LRNase HIIC protein should be 53-kDa. We did not see any 53-kDa RNase H activity band in the zymogram. In many other cells the precursor form of a mitochondrial protein does not accumulate in the cell and the predominant form of the protein is the proteolytically processed mature form [37]. Thus, the 49-kDa band may represent the mature ΔMLS form of LRNase HIIC.
Fig. 3.
RNase H activities in Leishmania. (A) Zymogram for the RNase H activities in the cell extracts from L. donovani. Cells and mitochondrial enriched fraction were lysed and boiled in SDS-containing denaturation solution for 3 min before loading. Proteins equivalent to 2 × 107 cells were loaded per lane (TCE, ME). The separating gel was made with the RNase H substrate 32P-poly(rA)/poly(dT). These activities were solely dependent upon Mn2+ and addition of Mg2+ instead did not yield any such band. The sizes of the bands shown are approximate. The major Mn2+-dependent RNase H activity is enriched in the mitochondrial fraction. TCE: total cellular extract; ME: mitochondrial extract. (B) Expression and purification of N-terminal His6-tagged L. major LRNase HIIC precursor protein and the mature LRNase HIICΔMLS in E. coli. Recombinant cells were induced with 0.2 mM IPTG. The proteins were separated in a 10% SDS-PAGE and stained with Coomasie Blue R250. Optimal induction was observed in 3 h at 37 °C. We followed up to 21 h. Lane M: size marker. Lanes 1–4 are for LRNase HIIC and lanes 5–8 are for LRNase HIICΔMLS. Lanes 1 and 5, lysates from uninduced cells; lanes 2 and 6, lysates from IPTG induced cells; lanes 3, 4, 7 and 8, fractions eluted from washed columns with buffer E (100 mM NaH2PO4, 10 mM Tris.Cl pH 4.5, and 8 M urea). (C) Western blot analysis of LRNase HIIC in L. donovani promastigotes. Left panel (i), Coomassie Brilliant Blue stained protein gel showing L. donovani total cellular protein (Cell; 50 μg) and total mitochondrial protein (Mito; 50 μg). Middle panel (ii), Western blot analysis with anti-LRNase HIIC antibodies (1:1000 dilution). The LRNase HIIC antibody detected a 49-kDa protein in the mitochondrial extract. The faint band in the total cellular extract is not visible in the photograph of the chemiluminiscence autoradiogram. Right panel (iii), Western blot with anti-ISP monoclonal antibody, showing mitochondrial enrichment. (D) Pull-down of the 49-kDa radiolabeled LRNase HIIC protein by antibody. Immunoprecipitation was done with the reagents from the SeizeX Protein A immunoprecipitation kit (Pierce Biotechnology). The size of the bands is 49-kDa. Experiment 1 and Experiment 2 are duplicates for same experiment. (E) Zymogram for the activity gel assay of the proteins isolated from L. donovani mitochondrial extracts by immunocapture. Assay was done as described above. The sizes of the bands correspond to 49-kDa.
We expressed recombinant LRNase HIIC protein in E. coli BL21 Star (DE3) cells (Invitrogen) and purified the hexa-histidine-tagged recombinant fusion protein by Ni++-NTA agarose chromatography (Fig. 3B). Purified LRNase HIIC protein was used to develop the antisera to further evaluate the mitochondrial enrichment of this protein by Western blotting. We loaded 50 μg of total cellular proteins or mitochondrial proteins per lane. There was a very faint band at the ~49 kDa in the total cellular extract and the same band is prominent (~30 fold) in the mitochondrial extract (Fig. 3C). This data is not in contradiction to the zymogram analysis data as is presented in Fig. 3A. In the zymogram analysis experiment, we loaded proteins from 2 × 107 cells per lane in both the total cellular extract and the mitochondrial extract. Thus, the amount of LRNase HIIC in each lane more or less remained the same. On the other hand, in the Western blot analysis we took equal amount of protein in each lane from total cell and mitochondrial extract. Thus, the relative amount of LRNase HIIC protein in the total cellular extract will be much less than that in the mitochondrial extract as this enzyme is enriched in the mitochondrial fraction. Enrichment of the iron-sulfur protein (ISP) (Fig. 3C) in the mitochondrial fraction indicated the enrichment of the organelle in that fraction [33].
To verify further that the antibody raised against the recombinant LRNase HIIC indeed binds to the Leishmania mitochondrial RNase H protein, we immunoprecipitated the protein from the mitochondrial fraction and evaluated the RNase H activity by zymogram analysis. The affinity purified and immobilized antibody was bound onto a Protein A-Sepharose column (SeizeX Protein A Immunoprecipitation kit, Pierce Biotechnology, Rockford, IL). This column pulled down a 49-kDa protein (Fig. 3D) from mitochondrial protein extract isolated from the biosynthetically radiolabeled proteins of L. donovani promastigotes. The experiment was repeated (Experiment 1 and Experiment 2, Fig. 3D) and in both the cases the size of the immunoprecipitated protein was 49 kDa. The flow through did not have the 49-kDa protein. This protein also did not bind to a similar column anchoring an unrelated antibody raised against a human protein (SLUG).
To evaluate the RNase H activity, we similarly pulled down the proteins from unlabeled Leishmania mitochondrial protein extracts using the antibody-coated beads. The eluted proteins (50 μg) were analyzed for RNase H activity. RNase H activity was detected as a 49-kDa band (Fig. 3E). Again, the flow through from the LRNase HIIC antibody bound column, as well as the eluted fraction from the human SLUG antibody column did not have any detectable RNase H activity. All these data thus strongly suggest that LRNase HIIC gene indeed codes for the mitochondrial RNase H in Leishmania and the active protein is enriched in the mitochondria as a 49-kDa molecule.
3.5. LRNase HIIC can complement rnh− E. coli only when it is without MLS
To evaluate whether the LRNase HIIC precursor protein has functional RNase H activity we performed a genetic complementation experiment using an rnh− temperature-sensitive E. coli strain. Activity gel assay with this E. coli MIC2067 strain in our laboratory did not show any activity band under the conditions of the assay. We cloned the coding sequences of LRNase HIIC or its ΔMLS derivative at the Nco I site of the plasmid pBAD/HisA (Invitrogen), selected the clones with forward orientation of the insert with respect to the ara operon promoter and transformed E. coli MIC2067 cells with the recombinant plasmid. In the presence of arabinose, chloramphenicol, kanamycin and ampicillin in the growth medium, the cells with the plasmid containing the ORF of ΔMLS derivative of LRNase HIIC only gave us growth at 42 °C. The precursor form failed to complement the growth defect of the E. coli. These data further suggest that the 53-kDa-precursor form is enzymatically inactive and the 49-kDa ΔMLS form is the enzymatically active form.
3.6. LRNase HIIC enters the mitochondrial vesicles isolated from Leishmania and is processed into the LRNase HIICΔMLS form
Mitochondrial vesicles with membrane potential, isolated from L. major promastigotes, were able to take up and process LRNase HIIC precursor (53-kDa) into the mature protein (49 kDa; Fig. 4). The 35S-labeled precursor protein was synthesized and incubated with isolated mitochondrial vesicles for the import study. Lane 1 of Fig. 4 shows 10% of the input of the radiolabeled precursor protein. When incubated with mitochondrial vesicles, a small portion of the 53-kDa precursor protein is apparently processed into a 49-kDa protein (lane 2, Fig. 4). To evaluate whether the 49-kDa protein is the processed protein located inside the mitochondrial vesicles, we treated the vesicles after the incubation with the radiolabeled precursor protein (as in lane 1) with proteinase K. Proteinase K should degrade any precursor form of protein that is bound to the mitochondrial vesicles but are not imported inside [33]. As is shown in lane 2 of Fig. 4, the 53-kDa precursor protein was not protected from the proteinase K digestion where as the 49-kDa protein was protected, further indicating that the 49-kDa protein is the mature form of LRNase HIIC that is retained in the mitochondrion of Leishmania.
Fig. 4.
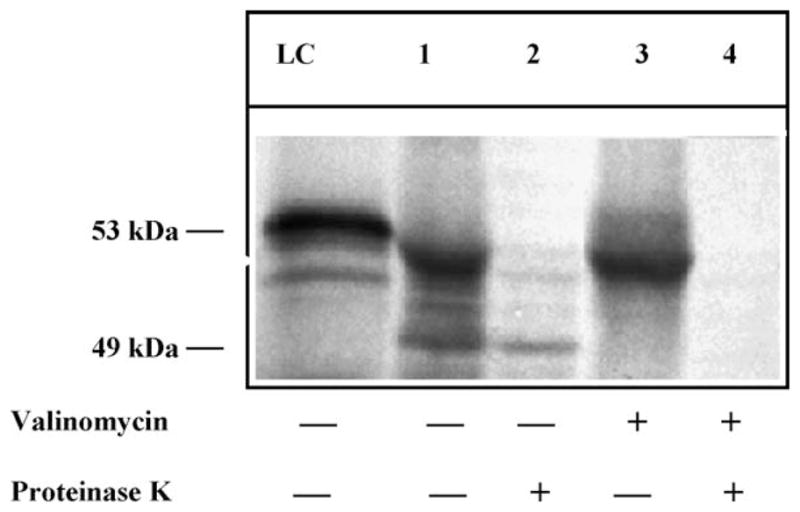
Import and processing of LRNase HIIC precursor protein into isolated mitochondrial vesicles from L. major. Similar results were obtained from mitochondrial vesicles isolated from L. donovani. Lane LC, the 35S-labeled-LRNase HIIC-precursor protein (10% of the input). Lane 1, protein associated with the mitochondrial vesicles after 30 min. Lane 2, corresponding 35S-labeled proteins inside the mitochondrial vesicles after proteinase K digestion. Lane 3, proteins associated with the 0.5 μM valinomycin-treated mitochondrial vesicles after 30 min. Lane 4, same as lane 3 but after proteinase K digestion. The 53-kDa precursor protein (LRNase HIIC) and the 49-kDa mature proteins (LRNase HIICΔMLS) are indicated. LRNase HIIC ΔMLS did not show any significant import to the mitochondrion (data not shown).
The import of LRNase HIIC into the mitochondrial vesicles was inhibited, when the vesicles were pretreated with proteinase K indicating that the import of LRNase HIIC is probably mediated through a receptor (data not shown). This import reaction was also inhibited when the import reaction contained 0.5 μM valinomycin (a potassium ionophore), which disrupts the membrane potential (lanes 3, 4, Fig. 4). These data indicated that the import of LRNase HIIC into Leishmania mitochondrion is dependent on membrane potential, a characteristic of typical receptor-mediated mitochondrial protein import process [33]. There was no import of the protein when the vesicles were incubated with the 35S-labeled LRNase HIICΔMLS, further suggesting that the signal needed for the mitochondrial targeting of LRNase HIIC resides within the N-terminal 40 amino acids of the protein (data not shown).
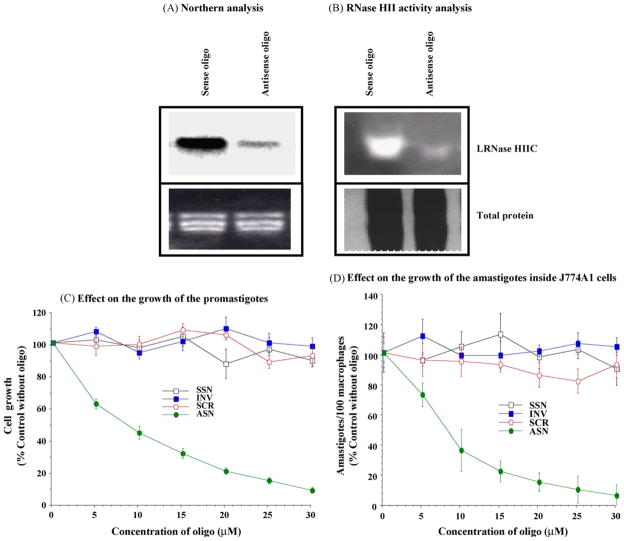
3.7. LRNase HIIC is essential for the growth of Leishmania promastigotes and amastigotes
We have evidence to suggest that LRNase HIIC is essential for Leishmania. We used mixed backbone oligonucleotide (16-mers) gapmers to knock down LRNase HIIC transcripts in L. donovani. The levels of LRNase HIIC mRNA (as was revealed by Northern analysis, Fig. 5A) and the level of LRNase HIIC enzyme (as was revealed by zymogram analysis, Fig. 5B) decreased during the treatment of the cells with the antisense oligo. The antisense oligo against LRNase HIIC affected the survival of L. donovani promastigotes in a dose dependent manner (Fig. 5C). These suggest that LRNase HIIC is essential for the survival of the parasite cells. As Leishmania exist as the amastigote form in the macrophages, we then tested the effects of the anti-sense oligos against the L. major amastigotes inside infected J774A1 cells [10,11,34]. To study the effect of the antisense oligo on the amastigotes, we mixed the antisense oligo or the control oligo with poly-L-lysine and encapsulated them inside the MBSA-coated liposomes [10,11,34]. When presented to the Leishmania-infected macrophages, these coated vesicles bound to the scavenger receptor (SR-A) present on the macrophage cell surface and they were endocytosed to the phagolysosomes containing amastigotes [34]. There was a steady decrease of amastigote count in the cultured macrophages (Fig. 5D) with the antisense ODN treatment as opposed to the treatments with the control ODNs, further suggesting the essentiality of LRNase HIIC for the survival of Leishmania.
Fig. 5.
Mitochondrial ribonuclease H (LRNase HIIC) is essential for the survival of Leishmania. (A) Northern analysis of RNA isolated from L. donovani promastigotes treated for 72 h with the sense or the antisense oligonucleotide (25 μM). Top panel is the autoradiogram and the lower panel corresponds to the ethidium bromide stained gel showing rRNA bands. (B) Activity gel assay with the proteins (100 μg) from the mitochondrial fraction of L. donovani promastigotes treated for 72 h with sense or antisense oligonucleotide (25 μM). Top panel is the zymogram and lower panel is the corresponding Coommasie blue-stained gel. (C) Inhibition of the growth of L. donovani promastigotes by antisense gapmer oligonucleotides against LRNase HIIC. Results are mean ± S.E. (n = 6). Similar results were obtained with axenic amastigotes and L. major promastigotes (not shown). (D) Inhibition of the growth of L. major amastigotes inside cultured macrophages (J774A1) by antisense gapmer oligonucleotides against LRNase HIIC. Results are mean ± S.E. (n = 6). SSN, the sense oligo; INV, oligo with the inverted sequence; SCR, the oligo with the sequence scrambled, and ASN, the antisense oligo. The amastigotes were counted 5-days post-treatment. The counts of amastigotes in the control macrophages (no oligo treatment) were 587 ± 12 cells/100 macrophages.
We thus conclude that LRNase HIIC is a mitochondrial enzyme. LRNase HIIC mRNA is essential for the survival of the parasite both in vitro and in vivo. As LRNase HIIC mRNA is distinct in its nucleotide sequence as compared to its human ortholog and thus may be targeted for the development of differential chemotherapy against antisense nucleic acid drugs.
Acknowledgments
We thank Dr. S. Hajduk for antibody against ISP. We also thank Drs. R. Couch and M. Itaya for E. coli MIC2067. This research was supported by National Institute of Health, USA Grant 5-R01AI42327-03 to GC.
Abbreviations
- MLS
Mitochondrial localization signal
- LRNase HIIC
Leishmania mitochondrial RNase HII precursor form
- LRNase HIICΔMLS
Leishmania mitochondrial RNase HII mature form lacking the MLS
- kDNA
kinetoplast DNA
- PMSF
phenylmethylsulfonyl fluoride
- IPTG
isopropylthio-β-D-galactoside
- ab
antibody
- KFZ
kinetoflagellar zone
References
- 1.Crouch RJ, Toulme JJ, Ribonuclease H. INSERM. Paris: 1998. [Google Scholar]
- 2.Wu H, Lima WF, Crooke ST. Properties of cloned and expressed human RNase H1. J Biol Chem. 1999;274:28270–8. doi: 10.1074/jbc.274.40.28270. [DOI] [PubMed] [Google Scholar]
- 3.Pileur F, Tolume JJ, Cazenave C. Eukaryotic ribonucleases HI and HII generate characteristic hydrolytic patterns on DNA–RNA hybrids: further evidence that mitochondrial RNase H is an RNase HII. Nucleic Acids Res. 2000;28:3674–83. doi: 10.1093/nar/28.18.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crouch RJ. Ribonuclease H: from discovery to 3D structure. New Biol. 1990;2:771–7. [PubMed] [Google Scholar]
- 5.Kao HI, Bambara RA. The protein components and mechanism of eukaryotic Okazaki fragment maturation. Crit Rev Biochem Mol Biol. 2003;38:433–52. doi: 10.1080/10409230390259382. [DOI] [PubMed] [Google Scholar]
- 6.Ohtani N, Haruki M, Morikawa M, Crouch RJ, Itaya M, Kanaya S. Identification of the genes encoding Mn2+-dependent RNase HII and Mg2+-dependent RNase HIII from Bacillus subtilis: classification of RNases H into three families. Biochemistry. 1999;38:605–18. doi: 10.1021/bi982207z. [DOI] [PubMed] [Google Scholar]
- 7.Cerritelli SM, Frolova EG, Feng C, Grinberg A, Love PE, Crouch RJ. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol Cell. 2003;11:807–15. doi: 10.1016/s1097-2765(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 8.Filippov V, Filippov M, Gill SS. Drosophila RNase H1 is essential for development but not for proliferation. Mol Genet Genomics. 2001;265:771–7. doi: 10.1007/s004380100483. [DOI] [PubMed] [Google Scholar]
- 9.Mirabelli CK, Crooke ST. Antisense research and applications. New York: CRC Press; 1993. pp. 7–35. [Google Scholar]
- 10.Mishra M, Bennett JR, Chaudhuri G. Increased efficacy of antileishmanial antisense phosphorothioate oligonucleotides in Leishmania amazonensis overexpressing ribonuclease H. Biochem Pharmacol. 2001;61:465–74. doi: 10.1016/s0006-2952(00)00568-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishra M, Porter-Kelley J, Singh PK, Bennett JR, Chaudhuri G. Enhanced activity of antisense phosphorothioate oligos against Leishmania amastigotes: augmented uptake of oligo, ribonuclease H activation, and efficient target intervention under altered growth conditions. Biochem Pharmacol. 2001;62:569–80. doi: 10.1016/s0006-2952(01)00695-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leishmaniasis Desjeux P. Nat Rev Microbiol. 2004;2:692–3. doi: 10.1038/nrmicro981. [DOI] [PubMed] [Google Scholar]
- 13.Alexander J, Satoskar AR, Russell DG. Leishmania species: models of intracellular parasitism. J Cell Sci. 1999;112:2993–3002. doi: 10.1242/jcs.112.18.2993. [DOI] [PubMed] [Google Scholar]
- 14.Hammarton TC, Mottram JC, Doerig C. The cell cycle of parasitic protozoa: potential for chemotherapeutic exploitation. Prog Cell Cycle Res. 2003;5:91–101. [PubMed] [Google Scholar]
- 15.Gull K. The biology of kinetoplastid parasites: insights and challenges from genomics and post-genomics. Int J Parasitol. 2001;31:443–52. doi: 10.1016/s0020-7519(01)00154-0. [DOI] [PubMed] [Google Scholar]
- 16.Sogin ML, Silberman JD. Evolution of the protists and protistan parasites from the perspective of molecular systematics. Int J Parasitol. 1998;28:11–20. doi: 10.1016/s0020-7519(97)00181-1. [DOI] [PubMed] [Google Scholar]
- 17.Shapiro TA, Englund PT. The structure and replication of kinetoplast DNA. Annu Rev Microbiol. 1995;49:117–43. doi: 10.1146/annurev.mi.49.100195.001001. [DOI] [PubMed] [Google Scholar]
- 18.Gull K. The cytoskeleton of trypanosomatid parasites. Annu Rev Microbiol. 1999;53:629–55. doi: 10.1146/annurev.micro.53.1.629. [DOI] [PubMed] [Google Scholar]
- 19.Morris JC, Drew ME, Klingbeil MM, et al. Replication of kinetoplast DNA: an update for the new millennium. Int J Parasitol. 2001;31:453–8. doi: 10.1016/s0020-7519(01)00156-4. [DOI] [PubMed] [Google Scholar]
- 20.Klingbeil MM, Motyka SA, Englund PT. Multiple mitochondrial DNA polymerases in Trypanosoma brucei. Mol Cell. 2002;10:175–86. doi: 10.1016/s1097-2765(02)00571-3. [DOI] [PubMed] [Google Scholar]
- 21.Melendy T, Sheline C, Ray DS. Localization of a type II DNA topoisomerase to two sites at the periphery of the kinetoplast DNA of Crithidia fasciculata. Cell. 1988;55:1083–8. doi: 10.1016/0092-8674(88)90252-8. [DOI] [PubMed] [Google Scholar]
- 22.Ferguson M, Torri AF, Ward DC, Englund PT. In situ hybridization to the Crithidia fasciculata kinetoplast reveals two antipodal sites involved in kinetoplast DNA replication. Cell. 1992;70:621–9. doi: 10.1016/0092-8674(92)90431-b. [DOI] [PubMed] [Google Scholar]
- 23.Sinha KM, Hines JC, Downey N, Ray DS. Mitochondrial DNA ligase in Crithidia fasciculate. Proc Natl Acad Sci USA. 2004;101:4361–8466. doi: 10.1073/pnas.0305705101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engel ML, Ray DS. The kinetoplast structure-specific endonuclease I is related to the 5′ exo/endonuclease domain of bacterial DNA polymerase I and colocalizes with the kinetoplast topoisomerase II and DNA polymerase beta during replication. Proc Natl Acad Sci USA. 1999;96:8455–60. doi: 10.1073/pnas.96.15.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abu-Elneel K, Robinson DR, Drew ME, Englund PT, Shlomai J. Intramitochondrial localization of universal minicircle sequence-binding protein, a trypanosomatid protein that binds kinetoplast minicircle replication origins. J Cell Biol. 2001;153:725–34. doi: 10.1083/jcb.153.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C, Englund PT. A mitochondrial DNA primase from the trypanosomatid Crithidia fasciculata. J Biol Chem. 1997;272:20787–92. doi: 10.1074/jbc.272.33.20787. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Englund PT. RNA interference of a trypanosome topoisomerase II causes progressive loss of mitochondrial DNA. EMBO J. 2001;20:4674–83. doi: 10.1093/emboj/20.17.4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heard PL, Lewis CS, Chaudhuri G. Leishmania mexicana amazonensis: differential display analysis and cloning of mRNAs from attenuated and infective forms. J Eukaryot Microbiol. 1996;43:409–15. doi: 10.1111/j.1550-7408.1996.tb05052.x. [DOI] [PubMed] [Google Scholar]
- 29.Seay MB, Heard PL, Chaudhuri G. Surface Zn-proteinase as a molecule for defense of Leishmania mexicana amazonensis promastigotes against cytolysis inside macrophage phagolysosomes. Infect Immun. 1996;64:5129–137. doi: 10.1128/iai.64.12.5129-5137.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 31.Itaya M, Omori A, Kanaya S, Crouch RJ, Tanaka T, Kondo K. Isolation of RNase H genes that are essential for growth of Bacillus subtilis 168. J Bacteriol. 1999;181:2118–23. doi: 10.1128/jb.181.7.2118-2123.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harlow ED, Lane D. Using antibodies: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- 33.Priest JW, Hajduk SL. In vitro import of the Rieske iron–sulfur protein by trypanosome mitochondria. J Biol Chem. 1996;271:20060–9. doi: 10.1074/jbc.271.33.20060. [DOI] [PubMed] [Google Scholar]
- 34.Chaudhuri G. Scavenger receptor-mediated delivery of antisense mini-exon phosphorothioate oligonucleotide to Leishmania-infected macrophages selective and efficient elimination of the parasite. Biochem Pharmacol. 1997;53:385–91. doi: 10.1016/s0006-2952(96)00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arudchandran A, Cerritelli SM, Bowen NJ, Chen X, Krause MW, Crouch RJ. Multiple ribonuclease H-encoding genes in the Caenorhabditis elegans genome contrasts with the two typical ribonuclease H-encoding genes in the human genome. Mol Biol Evol. 2002;19:1910–9. doi: 10.1093/oxfordjournals.molbev.a004015. [DOI] [PubMed] [Google Scholar]
- 36.Chapados BR, Chai Q, Hosfield DJ, Qui J, Shen B, Tainer JA. Structural biochemistry of a type 2 RNase H: RNA primer recognition and removal during DNA replication. J Mol Biol. 2001;307:541–56. doi: 10.1006/jmbi.2001.4494. [DOI] [PubMed] [Google Scholar]
- 37.Gakh O, Cavadini P, Isaya G. Mitochondrial processing peptidases. Biochim Biophys Acta. 2002;1592:63–77. doi: 10.1016/s0167-4889(02)00265-3. [DOI] [PubMed] [Google Scholar]